Abstract
The large protein superfamily of NADPH oxidases (NOX enzymes) is found in members of all eukaryotic kingdoms: animals, plants, fungi, and protists. The physiological functions of these NOX enzymes range from defense to specialized oxidative biosynthesis and to signaling. In filamentous fungi, NOX enzymes are involved in signaling cell differentiation, in particular in the formation of fruiting bodies. On the basis of bioinformatics analysis, until now it was believed that the genomes of unicellular fungi like Saccharomyces cerevisiae and Schizosaccharomyces pombe do not harbor genes coding for NOX enzymes. Nevertheless, the genome of S. cerevisiae contains nine ORFs showing sequence similarity to the catalytic subunits of mammalian NOX enzymes, only some of which have been functionally assigned as ferric reductases involved in iron ion transport. Here we show that one of the nine ORFs (YGL160W, AIM14) encodes a genuine NADPH oxidase, which is located in the endoplasmic reticulum (ER) and produces superoxide in a NADPH-dependent fashion. We renamed this ORF YNO1 (yeast NADPH oxidase 1). Overexpression of YNO1 causes YCA1-dependent apoptosis, whereas deletion of the gene makes cells less sensitive to apoptotic stimuli. Several independent lines of evidence point to regulation of the actin cytoskeleton by reactive oxygen species (ROS) produced by Yno1p.
Keywords: cell cycle, integral membrane reductase, wiskostatin, latrunculin
Reactive oxygen species (ROS) have multiple roles in physiology and pathophysiology, in particular during aging and induction of programmed cell death. This includes also nonmitochondrial sources, besides the long-studied mitochondrially generated ROS. These findings can be viewed as important additions to the classical “free radical theory of aging” (1) and theories developed thereafter (2, 3).
In higher organisms, among others, at least two major sources of superoxide other than mitochondria are known. On the one hand, xanthine oxidase, an enzyme in the catabolism of purines, which catalyses the oxidation of hypoxanthine to xanthine and to uric acid, produces superoxide (4). On the other hand, NADPH oxidases (NOX) catalyze the production of superoxide from oxygen and NADPH (5).
The NADPH oxidase superfamily of membrane-located enzymes of higher cells has been known for a decade (for review, ref. 5). Whereas the human NOX2 was discovered early on, other NOX (Nox1/3/4/5) as well as dual oxidase (DUOX) (Duox1/2) enzymes (displaying two domains: a NADPH oxidase domain and a peroxidase domain) have been found relatively recently in human cells. The human NOX2 was discovered as a defense enzyme of neutrophils and macrophages, which produce a burst of superoxide (O2·−) as a first line of defense against invading microorganisms. Although X-ray or NMR structure determinations are not available, we know from indirect evidence and bioinformatics that the catalytic subunit of the macrophage enzyme contains six transmembrane helices, is located in the plasma membrane, and produces superoxide in a vectorial way to the lumen of the phagosome, which is topologically outside the cytoplasmic space of the cell. In fact, all known NOX enzymes are membrane bound and deliver superoxide in a vectorial way. Not all NOX enzymes are located in the plasma membrane, however. For instance, human NOX4 was reported to be localized in the endoplasmic reticulum (ER), the nucleus, or in the mitochondria in several conflicting reports (6, for review, ref. 7). The NADPH oxidases require three different redox cofactors, namely two tightly bound b-type hemes, FADH2 and NADPH, which are bound by the dehydrogenase domain (5, 6). The general structural and mechanistic features of the NOX catalytic subunit seem to be relatively well conserved between the different NOX enzymes; however, they are regulated in different ways, either through regulatory subunits or by calcium ions. Calcium-dependent regulation is assumed and in some cases verified by experiments to occur via EF-hand motifs, which are present in NOX5 and the DUOX enzymes (8).
The immediate product (presumed or directly measured) of NOX (and DUOX) enzymes is the superoxide radical anion, which is, however, metabolized to other ROS molecules (peroxynitrite, H2O2, hypochlorite, and hydroxyl radical) relevant for defense in the case of NOX2 (9). The other NOX enzymes serve more specialized metabolic functions. A signaling function of ROS (superoxide and hydrogen peroxide) originating from NOX enzymes is increasingly discussed in the literature (5).
Fungal NOX enzymes of the NOXA, -B, and -C subfamilies and their physiological roles were discovered relatively recently (10, for review, ref. 11). The filamentous fungi Podospora anserina, Neurospora crassa (12) and the social amoeba Dictostelium discoideum (13) could be shown to express NADPH oxidases that actively produce superoxide (14). The deletion of PaNOX1 has no influence on growth, but completely blocks fruiting body formation (14). Fungal NOX enzymes mostly produce ROS as second messengers for coordination of multicellular communication (14). However, in hemiascomycetous yeasts no gene or enzyme has been assigned as a superoxide-generating NADPH oxidase up to now (10, 15).
In this work we present evidence that the yeast Saccharomyces cerevisiae produces a NADPH oxidase encoded by the ORF, YGL160W (AIM14). We show that this enzyme exhibits all biochemical properties of a fungal NADPH oxidase and have therefore renamed it YNO1 (yeast NADPH oxidase 1). Overexpression leads to production of ROS causing Yca1p-dependent apoptosis. Deletion of the gene confers resistance against apoptotic stimuli and hypersensitivity to wiskostatin and latrunculin B, indicating a role of Yno1p in regulation of the actin cytoskeleton.
Results and Discussion
Yeast Genome Codes for a Putative NADPH Oxidase Homolog: Yno1p.
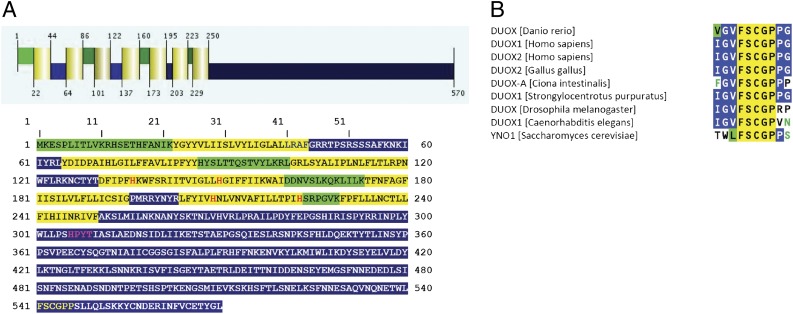
The common structural features of the NOX/integral membrane reductase (IMR) superfamily include in the N-terminal part six or seven transmembrane helices, two of which contain two strictly conserved histidine residues responsible for coordinating two spectroscopically nonidentical b-type heme groups. The C-terminal cytoplasmic globular domain contains binding sites for FADH2 and NADPH (Fig. 1A). Position-specific iterated (PSI)-BLAST analyses with the sequence of the catalytic subunit of the human NOX2 against the yeast genome showed that all features of NOX enzymes could be found in yeast ferric reductases, which are, like NADPH oxidases, proven flavocytochromes b558 (16). We conclude that within the large NOX protein superfamily functional diversity developed and a new function (ferric reductase or, more generally, IMR) arose (11). Among the yeast FRE subfamily the closest homolog to human NOX enzymes was found to be the yeast ORF YGL160W, bearing 32.1% sequence similarity (16.6% identity) to NOX5. The binding site for the nicotinamide moiety of NADPH in YNO1 is identical with the corresponding sequences of the DUOX enzymes (Fig. 1B).
Fig. 1.
Bioinformatic predictions for Yno1p (YGL160W). (A) Prediction of secondary structure for Yno1p obtained with the Phillius transmembrane prediction algorithm as described in SI Materials and Methods. The transmembrane helices are highlighted in yellow; the luminal regions, which reside presumably in the lumen of the ER, are highlighted in green; and the cytoplasmic regions are highlighted in blue. The four conserved histidine residues responsible for coordinating the two heme groups are marked with red letters. The NADPH binding site is marked with yellow letters and the FAD binding site is highlighted with purple letters. (B) Sequence alignment of the nicotinamide binding site of dual oxidases (DUOX) from various organisms. The sequence “FSCGP” is specific for DUOX enzymes and for Yno1p.
In a phylogenetic tree of the NOX/IMR superfamily, FRE1–FRE7 cluster together, whereas the closely related sequences of FRE8 and YNO1 are located somewhat farther away from the FRE genes and show a closer relationship to the animal NOX5 sequences (Fig. S1). A ferric reductase function has been shown for FRE1–FRE4 (17), and iron- (or copper)-dependent transcriptional control was found for FRE1–FRE7 (18, 19). FRE8 and YNO1 cluster together and are neither transcriptionally controlled by iron nor is the deletion dependent on increased iron for growth (18).
Yno1p Does Not Function as a Ferric Reductase.
To test for the possibility that Yno1p might have ferric reductase activity that aids iron uptake leading to superoxide production, we determined total iron as well as soluble iron in strains overexpressing YNO1 and compared the results with controls carrying the empty vector (Fig. S2). Clearly, overexpression of YNO1 did not lead to a significant accumulation of iron in the cells. Microsomes isolated from a strain overexpressing YNO1, which showed a high level of NADPH oxidase activity (see below) were only weakly active irrespective of the presence or absence of Yno1p when tested for iron reductase activity (20) (Fig. S3). Taken together, the iron reductase activity measurements, iron uptake data, phylogenetic tree construction, and the transcriptional regulation data cited above do not support a ferric reductase function for Yno1p.
Yno1p Produces Superoxide Anions.
To clarify whether the proteins encoded by YNO1 or any one of the eight yeast FRE genes has the capacity to produce superoxide (O2·−), as would be expected of a genuine NADPH oxidase, all nine genes were cloned into the expression vector pCM297 under the control of a tightly regulated but relatively weak doxycyline-inducible promoter (19, 21). All these constructs were transformed into the yeast strain BY4741, which was grown to midexponential phase. Thereafter, 100 mg/L doxycyline was added to induce the expression of the genes to be tested. After 6 and 16 h, respectively, the superoxide levels were measured by dihydroethidium (DHE) assay. Compared with the wild-type strain BY4741, which was transformed with the empty vector pCM297, YNO1 was the only one of the yeast genes tested that gave a small but significant positive signal indicated by a 50% increased oxidation of DHE (Fig. S4).
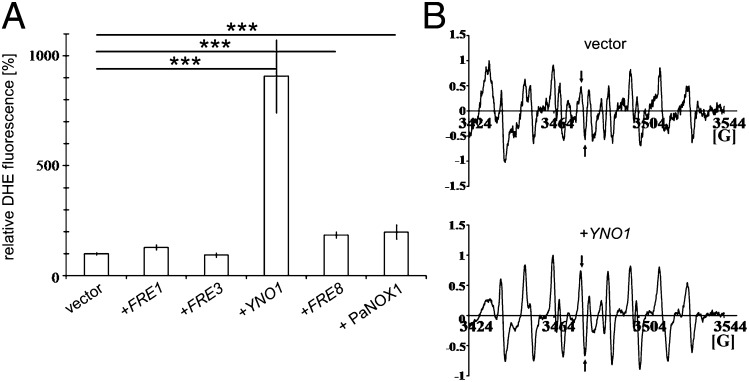
For further experiments, the yeast YNO1 as well as the PaNOX1 gene (positive control) and the FRE1, FRE3, and FRE8 genes were subcloned into the expression vector pYES2, harboring the strong GAL1 promoter to induce high expression of the selected genes. Expression of these constructs confirmed the results obtained by expression in pCM297. Overexpressing YNO1 led to an approximately five- to ninefold increase of the ROS level. The control strain with PaNOX1 showed a twofold increase in fluorescence signal intensity, as did the FRE8 ORF. FRE1 and FRE3 showed a ROS level comparable to the wild-type strain expressing the empty vector (Fig. 2A).
Fig. 2.
(A) Quantitative determination of fluorescence in live yeast strains overexpressing putative NADPH oxidases after galactose induction and in controls. ROS production was determined by measuring oxidized DHE in BY4741 cells transformed with: the pYES2 vector, pYES2-FRE1, pYES2-FRE3, pYES2-YNO1, pYES2-FRE8, and pYES2-PaNOX1. See Table S1 for determination of statistical significance. (B) Results of in vivo ESR measurements using the spin trap, DEPMPO (22). Lines marked with arrows are specific for the superoxide adduct. Upper spectrum corresponds to BY4741 transformed with the empty vector. Lower spectrum was obtained after galactose induction of pYES2-YNO1. Both spectra were recorded with three biological replicas each. Comparing the signal intensities of the superoxide-specific bands shows that they are significantly different with a two-tailed P value of 0.0057.
The DHE assay is very sensitive but not absolutely specific for superoxide (21). Therefore, we also performed ESR measurements for a more direct detection of superoxide with the spin trap 5-(Diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide (DEPMPO), which forms a stable radical adduct, the resonance spectrum of which is specific for superoxide (22). ESR spectra were recorded in vivo at room temperature. Wild-type cells overexpressing the empty vector only produce signals with DEPMPO at background level. However, cells of a strain overexpressing YNO1 produced a strong signal characteristic of the superoxide radical anion (Fig. 2B and Table S1).
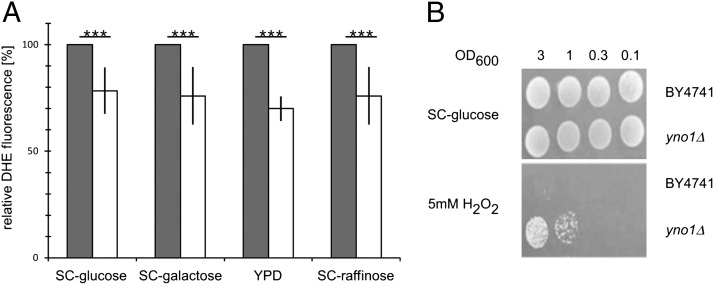
Using the same DHE fluorescence assay as described above, the yno1Δ strain also was tested under the same conditions after growth on YPD, SC −glucose, SC −raffinose, and SC −galactose. Reproducibly and significantly, the deletion strain produced a signal that was about 20% weaker than that of an untransformed WT strain (Fig. 3A). This indicates that Yno1p contributes about 20% to the total ROS production under exponential growth conditions and that other sources of ROS also exist. We hypothesized that the deletion strain could be resistant to externally applied oxidative stress. Ten-microliter aliquots of the deletion strain and the WT strain in different dilutions were spotted on SC plates containing hydrogen peroxide, diamide, or tert-butyl hydroperoxide. Indeed, the deletion strain showed a higher resistance than the wild-type strain against hydrogen peroxide (Fig. 3B). The deletion strain showed no change in sensitivity on plates containing diamide or tert-butyl hydroperoxide (Fig. S5).
Fig. 3.
(A) ROS production in WT (gray bars) and yno1Δ (open bars) cells. Four different growth media were used as indicated. In each of the four experiments, a statistically significant loss of ∼20% activity was consistently observed in yno1Δ compared with WT cells. (B) yno1Δ cells exhibit marked resistance to 5mM H2O2, which completely inhibits growth of WT cells. Serial dilutions of WT and yno1Δ cells were spotted on SC −glucose and SC −glucose +5 mM H2O2 media and scored after 2 d.
Yno1p-Mediated Production of Superoxide Depends on NADPH in Vivo.
We wanted to test whether the high superoxide signal in the YNO1 overexpressing strain was dependent on the NADPH level of the cells. In S. cerevisiae, ZWF1 codes for glucose-6-phosphate dehydrogenase, an enzyme catalyzing the first and rate-limiting step of the pentose phosphate pathway, a reaction that generates NADPH. Cells lacking ZWF1 show a reduced level of NADPH (23). A zwf1Δ strain overexpressing YNO1 showed no increase in ROS compared with wild type or the deletion strain carrying the empty vector, indicating that Yno1p activity very probably is NADPH dependent in vivo (Fig. S6).
Yno1p Localizes to the Perinuclear ER.
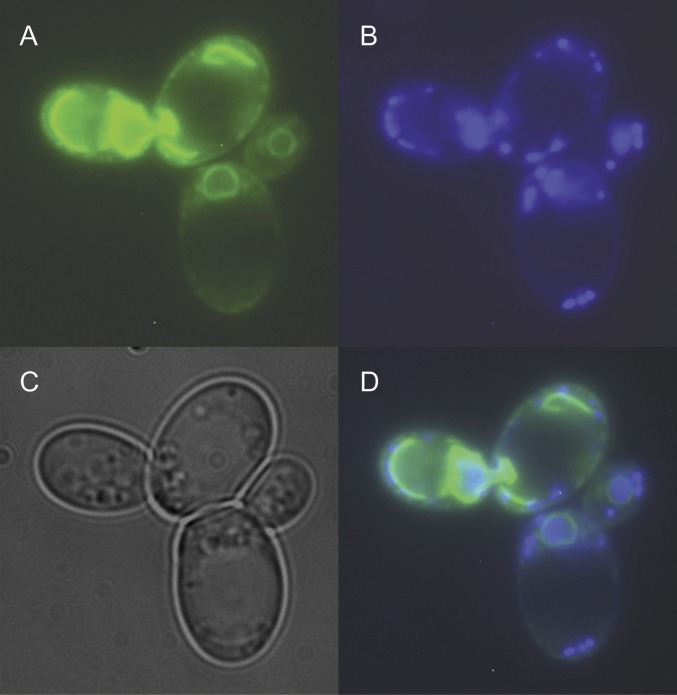
Previous studies of NADPH oxidases in human cells indicate that NOX enzymes are localized in various membranes such as the plasma membrane, ER, or the mitochondrial membranes (6, 7). To determine subcellular localization of Yno1p, we constructed a C-terminal fusion protein of Yno1p and eGFP expressed under the control of the constitutive MET25 promoter in the pUG35 vector. In cells expressing the fusion protein, a clear perinuclear GFP signal was observed (Fig. 4; compare with ref. 24). Simultaneously expressing HDEL-RFP (25) showed colocalization of Yno1-GFP with HDEL-RFP in the perinuclear ER (Fig. S7).
Fig. 4.
Fluorescence micrograph of two live cells with large daughters from midlog phase growth on rich media. (A) Localization of the Yno1–eGFP fusion protein. (B) Nuclear and mitochondrial DNA stained with DAPI. (C) The same cells visualized by phase contrast microscopy. (D) Overlay of A–C. The perinuclear location of the fusion protein is clearly visible.
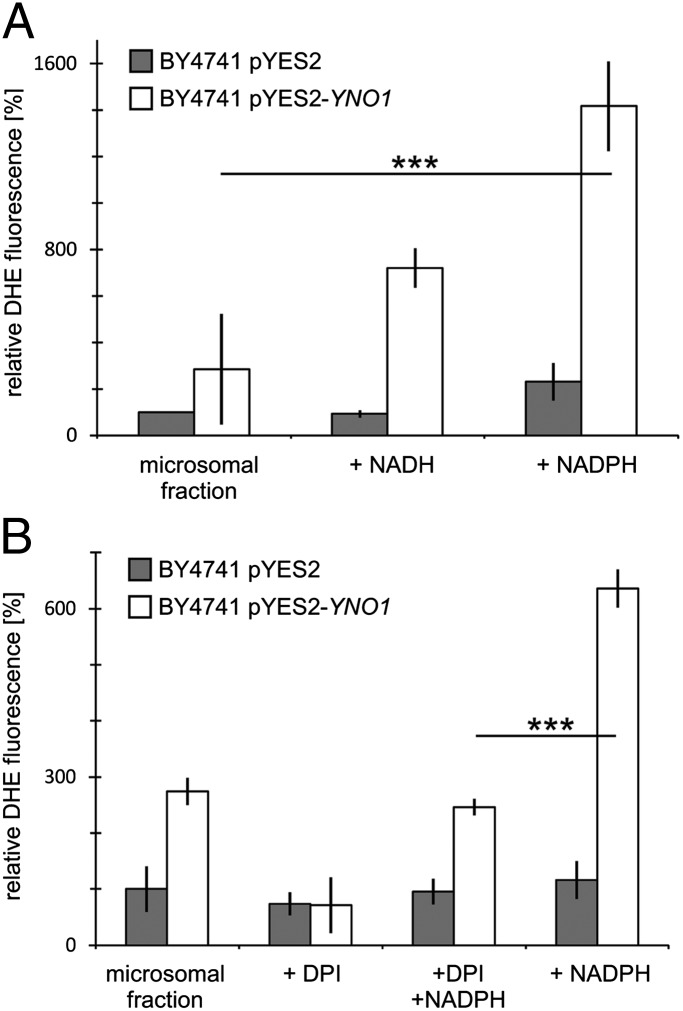
To biochemically localize the enzyme activity in cells of a strain overexpressing the full-length nonfusion Yno1p under galactose control, cells were broken with glass beads and fractions were obtained by differential centrifugation. DHE fluorescence assays were performed as described above to measure superoxide anions in microsomal fractions (100,000 × g pellet) from cells transformed with the empty vector and from cells overexpressing YNO1. Microsomes were characterized by electron microscopy (Fig. S8). As shown in Fig. 5, the microsomal fraction contains the Yno1p enzyme activity, which is NADPH dependent in vitro and is inhibited by diphenyleneiodonium chloride (DPI) (26), indicating a genuine NADPH oxidase and confirming the results seen with the Yno1–eGFP fusion protein. The microsomes displayed a residual activity with NADH as cofactor. Note that some flavocytochromes b, like the human NOX2 enzyme, also show a weak affinity for NADH, whereas NOX4 and NOX5 are specific for NADPH (27). The microsomes show a small residual NADPH-dependent activity that was not inhibited by DPI, which is likely due to the presence of microsomal oxidoreductases other than Yno1p. When we compared the specific activity of Yno1p (DHE fluorescence counts divided by protein amount) in the supernatant of the 4,500 × g centrifugation with the activity in the purified microsomes, we observed a significant enrichment (SI Results), indicating that the majority of the enzyme is located in microsomes. All other fractions obtained except the 18,000 × g supernatant containing microsomes showed reduced levels of DHE fluorescence (Fig. S9).
Fig. 5.
ROS measurements by DHE fluorescence in purified microsomes. (Gray bars) Microsomes from a strain containing only the vector (vector control). (Open bars) Strain expressing Yno1p under galactose control. (A) NADPH oxidase activity detected in the microsomal fraction in the presence of NADPH; a weaker activity is detected with NADH. (B) NADPH-dependent activity is inhibited by adding DPI. Residual DHE oxidation in the absence of added NADPH is also inhibited by DPI.
YNO1 Overexpression Causes Cell Death Depending on the Yeast Caspase YCA1.
To investigate the impact of Yno1p on yeast cell physiology, we first tested cell growth (Fig. S10). Compared with WT, a yno1 deletion strain exhibited normal growth on standard media (synthetic complete, SC) (Fig. S10A). Overexpression of Yno1p resulted in a substantial slowing of growth (Fig. S10B), which was caused by induction of apoptosis and necrosis, as shown below. However, growth was not altered by overexpression of YNO1 in a yca1Δ strain (28, 29), indicating that cell death by YNO1 overexpression depends on a functional Yca1p caspase (Fig. S10C).
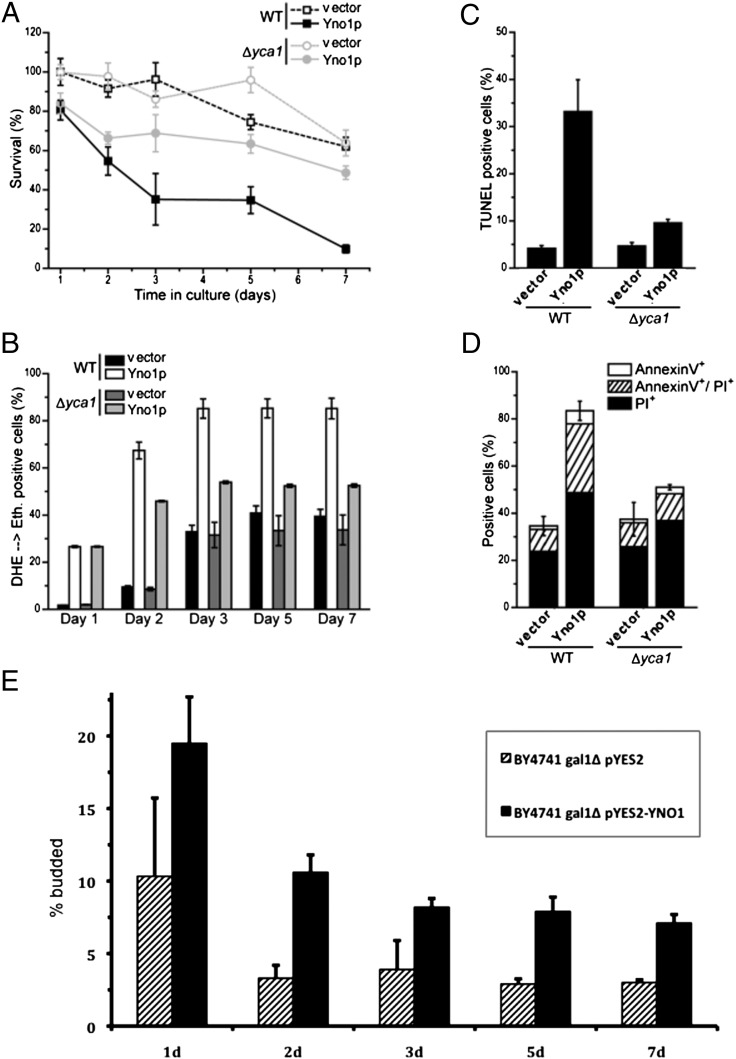
Next we tested apoptotic and necrotic markers in the same strains just discussed (Fig. 6). The results show that clonogenicity over 7-d culture in stationary phase decreased substantially when Yno1p was expressed, indicating increased chronological aging. However, this effect was substantially dampened by deleting YCA1 (Fig. 6A), confirming the results seen in the growth curves of Fig. S10. Measuring apoptotic and necrotic markers showed that the cells were rescued from both forms of death by the YCA1 deletion (Fig. 6 C and D), although ROS production was still high in the yca1Δ strain overexpressing YNO1 at day 2 (Fig. 6B). This latter effect was time dependent and at the end of the survival curve (day 7), ROS production in the yca1Δ YNO1 overexpressing strain appeared to be lower. Microscopic images of the apoptotic markers examined are shown in Fig. S11. Taken together, the results shown in Fig. 6 and Figs. S10 and S11 establish that Yca1p acts downstream of the ROS produced by Yno1p.
Fig. 6.
Survival, apoptotic phenotype, and bud index of stationary phase cells overexpressing YNO1. Cells were pregrown in SC raffinose; expression of YNO1 from the pYES2 plasmid was induced by 3% (wt/vol) galactose, and survival, apoptotic phenotypes, and bud index in stationary phase were determined. (A) Chronological aging (survival in stationary phase) over 7-d culture in spent medium and its inhibition by inactivation of YCA1. (B) ROS production measured over time in stationary phase for the same cells described in A. Up until day 2, ROS production in the yca1Δ, YNO1 overexpression strain was high, although apoptotic phenotypes are low (A, C, and D). Later in stationary phase, the yca1Δ strain displayed a lower ROS level. (C) TUNEL measurements in the same four strains on day 2 of stationary phase. TUNEL+ cells indicating DNA strand breaks are more numerous in the strain overexpressing YNO1, but reduced in number when YCA1 is deleted. (D) Measurements of staining with AnnexinV and propidium iodide (PI) in the same four strains in stationary phase (day 2). Overexpression of YNO1 led to high-level staining with both AnnexinV and PI, which indicates the existence of apoptotic as well as necrotic cells. Staining was not elevated if YCA1 was deleted. (E) Determination of bud index (% budded cells) in stationary phase. Overexpression of YNO1 was induced by shifting from medium containing 2% (wt/vol) raffinose to medium containing 2% (wt/vol) raffinose +3% (wt/vol) galactose, growing the cells to stationary phase and keeping them in spent medium for 7 d. Cells transformed with the empty vector were used as controls. After 3 d, ∼8% of cells in stationary cultures overexpressing YNO1 remained budded, in contrast to the ∼3% budded cells detected in controls.
We next tested the budding index in stationary WT cells with or without overexpression of Yno1p. As shown in Fig. 6E, after 3 d, the WT exhibited a low number of budded cells, but in the YNO1 overexpressing strain, cells began new cell cycles despite the absence of nutrients. This is consistent with the lethal effect of YNO1 overexpression and suggests that the ROS produced by Yno1p creates a signal that starts a new cell cycle.
ROS Production by Yno1p Is Independent of the Mitochondrial Respiratory Chain and Does Not Contribute to ER Stress.
We wanted to determine whether the remarkable increase in ROS production in YNO1 overexpressing strains required a functional respiratory chain. The deletion of AFO1 (30), a mitochondrial ribosomal protein, leads to cells completely devoid of mitochondrial DNA (ρ0), which also display a very low level of ROS, in contrast to standard ρ0 strains isolated after treatment with ethidium bromide. This strain was therefore ideally suited for this measurement. The results indicate that a functional respiratory chain (which is absent in ρ0 strains) is not needed for ROS production by Yno1p (Fig. S12A).
Tunicamycin induces ER stress and a high level of ROS in WT yeast (31) that can lead to apoptosis. As shown in Fig. S12B, this effect is independent of Yno1p. Therefore, Yno1p is not a significant contributor to ER stress. This finding is also consistent with the observation that a yno1Δ strain shows neither resistance nor sensitivity to the reductant dithiothreitol (DTT), which leads to an accumulation of unfolded proteins in the ER and causes ER stress (Fig. S13). We conclude that ROS production by Yno1p is not related to mitochondrial respiration and mitochondrial ROS production. We also conclude that the direct influence of Yno1p on mitochondria and vice versa is weak or absent in this case, in contrast to higher cells where a cross-talk between NOX enzymes and mitochondria has been described (32). Furthermore, the production of ROS by this ER membrane-localized enzyme is independent of the ROS production due to ER stress and the accumulation of unfolded proteins in the ER.
Interaction of the Yno1p-Produced ROS with the Nucleation and Elongation Step in the Biosynthesis of F-Actin Cables.
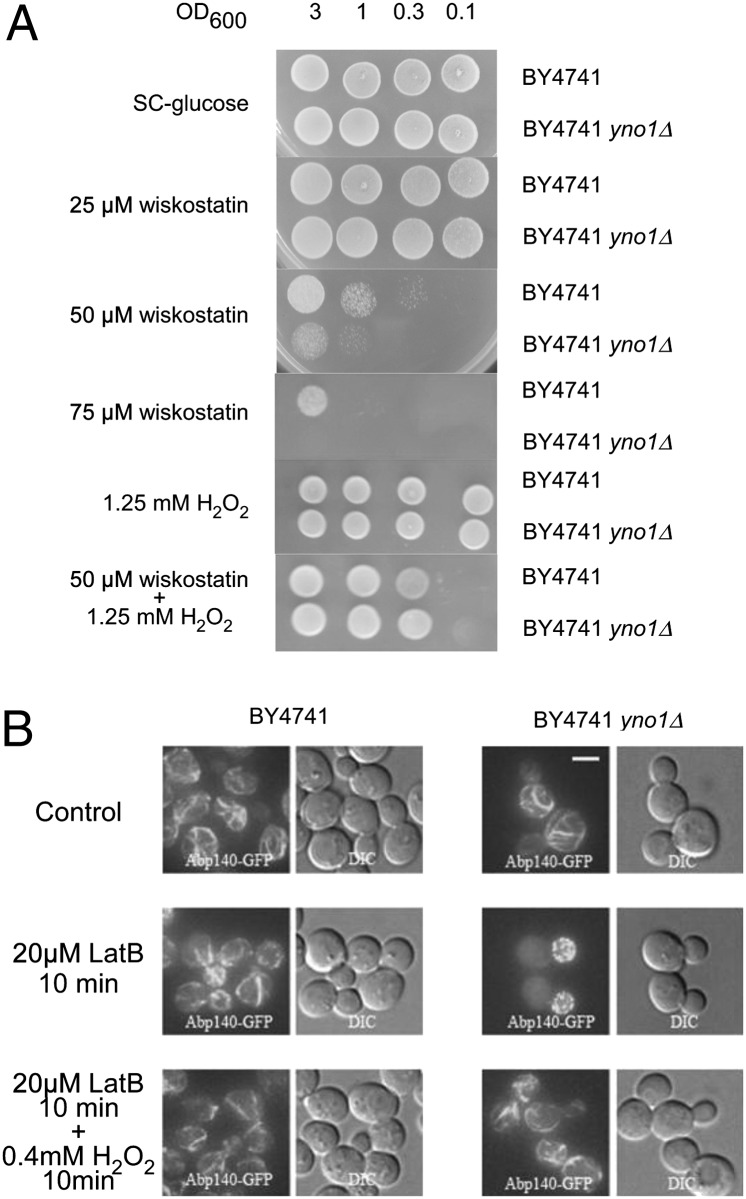
Previous whole genome screening (33) identified the yno1Δ strain as hypersensitive to wiskostatin and showed synthetic lethality of yno1Δ with a heterozygous deletion of act1, the only actin-encoding gene on the haploid genome of yeast (34). Wiskostatin is a drug that interacts with Wiskott-Aldrich syndrome protein (WASP), thereby preventing the nucleation of F-actin cables. We confirmed this high-throughput result and also determined that a nontoxic concentration of H2O2 partially reverses the toxic effect of wiskostatin (Fig. 7A). Note that H2O2 does not oxidize wiskostatin under the conditions used.
Fig. 7.
(A) Deletion of yno1 caused hypersensitivity to wiskostatin, which was suppressed by external exposure to H2O2. Dilutions of WT and yno1Δ strains were spotted onto SC −glucose, SC −glucose +wiskostatin, and SC −glucose +wiskostatin +H2O2. Addition of a low nontoxic amount of H2O2 to the media enabled the hypersensitive cells of the deletion strain to grow in the presence of an otherwise inhibitory concentration of wiskostatin. (B) F-Actin was visualized with Abp140-GFP; deletion of yno1 caused hypersensitivity to latrunculin B (inhibitor of F-actin cable elongation) at a concentration of 20 μM, which does not inhibit F-actin cable elongation in the WT. H2O2 reversed inhibition of actin cables by latrunculin B. Thus, ROS produced by Yno1p are required for F-actin regulation.
Additional inhibitors of the actin cytoskeleton have been identified (33), including latrunculin B, an inhibitor of F-actin cable elongation (35). After a 10-min incubation with 20 μM latrunculin B, the yno1Δ mutant completely lost F-actin cables, but not F-actin patches (in the daughter cell at this stage of the cell cycle). Under the same conditions, WT cells remain completely normal in appearance. The effect of latrunculin B on the mutant was efficiently reversed by adding a low concentration of H2O2, which did not inhibit cell growth (Fig. 7B). We conclude that Yno1p functionally interacts with F-actin by producing ROS and that the function of YNO1 in this interaction very probably is to supply H2O2 generated via its primary product, superoxide. This conclusion is strengthened by the fact that in the mammalian system, ROS created by NOX have been shown to regulate the actin cytoskeleton (36).
Concluding Remarks.
We show here that Yno1p of S. cerevisiae is a NADPH oxidase, which produces superoxide and indirectly H2O2. Two of the nine S. cerevisiae members of the NOX/IMR superfamily, Fre8p and Yno1p, the protein described in this communication, cluster together at a location different from the other seven Fre enzymes in phylogenetic trees constructed by two different methods (ref. 11, this paper, and Fig. S1). Yno1p and Fre8p are also different from Fre1-7p because they show no involvement in iron reduction and uptake. Another distinctive feature of Yno1p is that overproduction of this protein, but not overproduction of Fre1-Fre7p leads to slow growth as determined in a whole genome screen (37) and in this study (Fig. S10). Slow growth of these strains can be explained by apoptotic and necrotic cell death.
Additional criteria for identification of Yno1p as a genuine NADPH oxidase are its inhibition by DPI, dependence on NADPH, and sequence information that predicts seven transmembrane helices, four conserved histidine residues that can coordinate two heme b groups, and a globular C-terminal region containing binding sites for FAD and NADPH (Fig. 1). FRE8 overexpression leads to weak but significant superoxide production, whereas the other seven FRE genes after overexpression yield a level of ROS equal to the WT or uninduced yeast cells.
Strong (albeit indirect) evidence points to a physiological function of Yno1p in the regulation of the cell’s actin cytoskeleton. The yno1Δ mutant shows hypersensitivity to wiskostatin, which inhibits F-actin cable nucleation, and to latrunculin B, which inhibits F-actin cable elongation. In both cases, the phenotype can be rescued by addition of a relatively small nontoxic concentration of H2O2. This indicates that the function missing in yno1Δ cells is production of ROS that have a role in signaling. How is this possible considering that there are many other sources of ROS in the cell? Although the answer to this interesting question must await future research, it could be related to compartmentalization of different kinds of ROS in space and time during the cell cycle. This possibility is consistent with the observation that overexpression of YNO1 causes a significant increase of budded cells in stationary phase.
Materials and Methods
Strains BY4741 (MATα his3 Δ1 leu2Δ0 ura3Δ0 met15Δ0) and the respective knockout in the YNO1 gene (European Saccharomyces Cerevisiae Archive for Functional Analysis) were grown in SC medium (20) with 2% (wt/vol) glucose as carbon source and used for most of the experiments. Overexpression of the YNO1 and its homologs was accomplished by cloning it as a PmeI/PmeI fragment into the galactose-inducible pYES2 yeast expression plasmid (Invitrogen) and growing the strains on 3% galactose. For visualizing the actin cytoskeleton, the genomic integration of ABP140–eGFP in strain SEY (MATa his3 Δ200 leu2-3112 ura3-52 trp1-Δ901 lys2-801 suc2Δ9) was used. Latrunculin B (Sigma) was added to a final concentration of 20 μM to cultivation media. Fluorescence microscopy was performed using an Olympus IX-71 inverted microscope equipped with Hamamatsu Orca/ER digital camera and the Cell R detection system. ROS (superoxide) determination was performed in vivo with DHE in an Anthos Zenyth 3100 plate reader as described previously (20). Apoptotic markers and chronological lifespan were determined as described in ref. 27. A more detailed description of the materials and methods, including statistical analysis, is given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dan Kosman for his help with ferric reductase determinations and Philippe Silar for supplying the PaNOX1 cDNA. This work was supported by Austrian Science Fund (FWF) Grants S9302-B05 (to M.B.), T414-B09 (to S.B.), S9304-B05, LIPOTOX, DK-MCD W 1226-B18 and P23490-B12 (to F.M.); P24381-B20 (to F.M. and T.E.); the European Commission (Brussels) Role of Mitochondria in Conserved Mechanisms of Aging (MIMAGE) Project (Contract 512020, to M.B.) and a National Cancer Institute Support Grant (P30CA016056) to Roswell Park Cancer Institute; and the Medical Research Council (United Kingdom) for Career Development Fellowship 78573 (to C.W.G). J.H. and O.B. were supported by CSF204/09/1924, MEB 060902, 7AMB12ATOO2; and T.G. and I.F. were supported by LC545 (Czech Republic).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201629109/-/DCSupplemental.
References
- 1.Harman D. Aging: A theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. The biologic clock: The mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Harman D. The aging process: Major risk factor for disease and death. Proc Natl Acad Sci USA. 1991;88:5360–5363. doi: 10.1073/pnas.88.12.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porras AG, Olson JS, Palmer G. The reaction of reduced xanthine oxidase with oxygen. Kinetics of peroxide and superoxide formation. J Biol Chem. 1981;256:9006–9103. [PubMed] [Google Scholar]
- 5.Nauseef WM. Biological roles for the NOX family NADPH oxidases. J Biol Chem. 2008;283:16961–16965. doi: 10.1074/jbc.R700045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause KH. Tissue distribution and putative physiological function of NOX family NADPH oxidases. Jpn J Infect Dis. 2004;57:S28–S29. [PubMed] [Google Scholar]
- 7.Block K, Gorin Y, Abboud HE. Subcellular localization of Nox4 and regulation in diabetes. Proc Natl Acad Sci USA. 2009;106:14385–14390. doi: 10.1073/pnas.0906805106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara T, Lambeth JD. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol Biol. 2007;7:178. doi: 10.1186/1471-2148-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valko M, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: Diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–1076. doi: 10.1016/j.fgb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Grissa I, Bidard F, Grognet P, Grossetete S, Silar P. The Nox/Ferric reductase/Ferric reductase-like families of Eumycetes. Fungal Biol. 2010;114:766–777. doi: 10.1016/j.funbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Cano-Domínguez N, Alvarez-Delfín K, Hansberg W, Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 2008;7:1352–1361. doi: 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lardy B, et al. NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum. Biochim Biophys Acta. 2005;1744:199–212. doi: 10.1016/j.bbamcr.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Malagnac F, Lalucque H, Lepère G, Silar P. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol. 2004;41:982–997. doi: 10.1016/j.fgb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol. 2007;7:109. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatwell KP, Dancis A, Cross AR, Klausner RD, Segal AW. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J Biol Chem. 1996;271:14240–14244. doi: 10.1074/jbc.271.24.14240. [DOI] [PubMed] [Google Scholar]
- 17.Georgatsou E, Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3065–3073. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgatsou E, Alexandraki D. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast. 1999;15:573–584. doi: 10.1002/(SICI)1097-0061(199905)15:7<573::AID-YEA404>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Martins LJ, Jensen LT, Simon JR, Keller GL, Winge DR. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J Biol Chem. 1998;273:23716–23721. doi: 10.1074/jbc.273.37.23716. [DOI] [PubMed] [Google Scholar]
- 20.Wyman S, Simpson RJ, McKie AT, Sharp PA. Dcytb (Cybrd1) functions as both a ferric and a cupric reductase in vitro. FEBS Lett. 2008;582:1901–1906. doi: 10.1016/j.febslet.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Klinger H, et al. Quantitation of (a)symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol. 2010;45:533–542. doi: 10.1016/j.exger.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Stolze K, Udilova N, Nohl H. Spin trapping of lipid radicals with DEPMPO-derived spin traps: Detection of superoxide, alkyl and alkoxyl radicals in aqueous and lipid phase. Free Radic Biol Med. 2000;29:1005–1014. doi: 10.1016/s0891-5849(00)00401-9. [DOI] [PubMed] [Google Scholar]
- 23.Ralser M, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen CJ, Young BP, Tavassoli S, Levine TP. Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol. 2007;179:467–483. doi: 10.1083/jcb.200708205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell BV, Tew DG, Jones OT, England PJ. Studies on the inhibitory mechanism of iodonium compounds with special reference to neutrophil NADPH oxidase. Biochem J. 1993;290:41–49. doi: 10.1042/bj2900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 28.Madeo F, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 29.Khan MA, Chock PB, Stadtman ER. Knockout of caspase-like gene, YCA1, abrogates apoptosis and elevates oxidized proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:17326–17331. doi: 10.1073/pnas.0508120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heeren G, et al. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging (Albany NY) 2009;1:622–636. doi: 10.18632/aging.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauptmann P, et al. Defects in N-glycosylation induce apoptosis in yeast. Mol Microbiol. 2006;59:765–778. doi: 10.1111/j.1365-2958.2005.04981.x. [DOI] [PubMed] [Google Scholar]
- 32.Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol Ther. 2005;4:1367–1373. doi: 10.4161/cbt.4.12.2233. [DOI] [PubMed] [Google Scholar]
- 33.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. Modeling complex genetic interactions in a simple eukaryotic genome: Actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 2007;21:148–159. doi: 10.1101/gad.1477507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: Novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 36.San Martín A, Griendling KK. Redox control of vascular smooth muscle migration. Antioxid Redox Signal. 2010;12:625–640. doi: 10.1089/ars.2009.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.