Abstract
The rhizobium–legume symbiosis has been widely studied as the model of mutualistic evolution and the essential component of sustainable agriculture. Extensive genetic and recent genomic studies have led to the hypothesis that many distinct strategies, regardless of rhizobial phylogeny, contributed to the varied rhizobium–legume symbiosis. We sequenced 26 genomes of Sinorhizobium and Bradyrhizobium nodulating soybean to test this hypothesis. The Bradyrhizobium core genome is disproportionally enriched in lipid and secondary metabolism, whereas several gene clusters known to be involved in osmoprotection and adaptation to alkaline pH are specific to the Sinorhizobium core genome. These features are consistent with biogeographic patterns of these bacteria. Surprisingly, no genes are specifically shared by these soybean microsymbionts compared with other legume microsymbionts. On the other hand, phyletic patterns of 561 known symbiosis genes of rhizobia reflected the species phylogeny of these soybean microsymbionts and other rhizobia. Similar analyses with 887 known functional genes or the whole pan genome of rhizobia revealed that only the phyletic distribution of functional genes was consistent with the species tree of rhizobia. Further evolutionary genetics revealed that recombination dominated the evolution of core genome. Taken together, our results suggested that faithfully vertical genes were rare compared with those with history of recombination including lateral gene transfer, although rhizobial adaptations to symbiotic interactions and other environmental conditions extensively recruited lineage-specific shell genes under direct or indirect control through the speciation process.
Keywords: Glycine, prokaryote
The rhizobium–legume symbiosis is considered one of the models of mutualistic evolution and the essential component of sustainable agriculture and has been the subject of more than 2,500 papers in the past 5 y. As many as 90 species belonging to 12 genera of α- and β-proteobacterium can establish nodules, the mutualistic nitrogen-fixing structure formed with leguminous plants (1). How could such diverse bacteria be recruited in this mutualistic symbiosis with legumes? Like many other bacteria, rhizobial genomes are characterized by their core and accessory genome (2). The earlier belief that the transfer of key symbiosis loci including nod (nodulation) genes is the determining factor leading to symbiosis has been recently challenged to some extent by whole-genome analyses of two symbiotic Bradyrhizobium strains that lack the canonical nodABC genes (3). Extensive genetic studies and recent comparative genomics of eight available rhizobial genomes suggested that a unique shared genetic strategy did not support symbiosis of rhizobia with legumes (4, 5). However, it should be noted that the limited number of genomes for certain genera and species made it impossible to get meaningful insight into the historical microevolutionary forces driving the genomic evolution of rhizobia and was, thus, less likely to reveal the dynamic adaptive evolution of rhizobia to legume hosts or diverse environmental conditions. Addressing these topics will help uncover the underlying mechanisms of the survival ability and competitiveness of rhizobia, which are still major obstacles in the widespread application of rhizobial inoculants to plants.
Glycine max (soybean) is one of the most important legume crops in the world. It is thought to have originated in East Asia, and differentiated gene pools have been reported in different ecoregions of China (6). At least five rhizobial species have been reported as its microsymbionts by independent studies: Bradyrhizobium japonicum, B. elkanii, B. liaoningense, B. yuanmingense, and Sinorhizobium fredii (7–11). These microsymbionts showed clear biogeographic patterns, with S. fredii as the dominant species in alkaline–saline soil and Bradyrhizobium being widely distributed and dominant in neutral to acidic soil (7–11). Recently, S. sojae was also reported as the microsymbiont of soybean in the region of alkaline soil (10, 12). How could bacteria of these two contrasting genera (Bradyrhizobium and Sinorhizobium) evolve into the microsymbionts of the same legume plant? Here, we sequenced 26 genomes of representative strains nodulating soybean from different ecoregions and made a systematic comparative genomic analysis of these soybean microsymbionts and other rhizobia. This study revealed not only the distinct genomic features of Bradyrhizobium/Sinorhizobium related to their symbiotic capacity and environmental adaptations but also the general evolutionary strategy used by these soybean microsymbionts and other rhizobia to adapt to the varied symbiotic interactions and other environmental conditions. The implications of these findings to our understanding of the speciation process are also discussed.
Results and Discussion
Contrasting Genomic Features of Bradyrhizobium vs. Sinorhizobium.
Recently we reported that soybean nodules were dominated by Sinorhizobium at sampling sites with alkaline–saline soil conditions, whereas Bradyrhizobium were widely distributed and dominated in neutral to acidic soils (7, 9–11) (Fig. S1). According to their internal transcribed spacer–restriction fragment length polymorphism (ITS-RFLP) clusters, 26 representatives from more than 1,100 nodule isolates of soybean were chosen for genome sequencing (Tables S1 and S2).
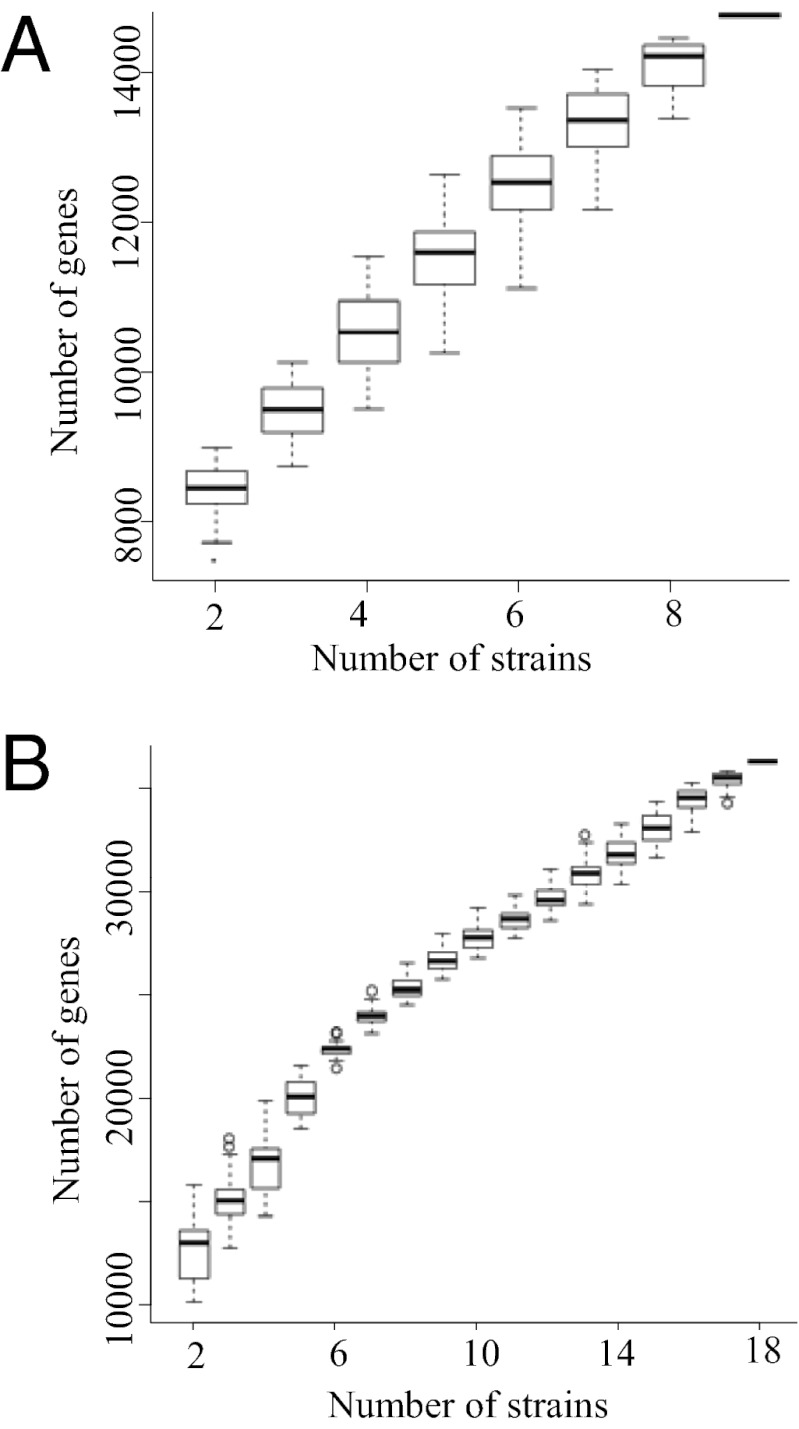
Noteworthy, the accumulation curves (Fig. 1 A and B) depicted that pan genomes of test strains from both Sinorhizobium and Bradyrhizobium are of the open type, for which the sequencing of more genomes adds more accessory genes. This result emphasizes the need for more extensive investigation of microsymbionts from soybean. The average genome size of 18 Bradyrhizobium strains was 9.8 ± 0.87 Mb (including the published genome of Bradyrhizobium sp. USDA110), which was significantly (P < 0.001) larger than that of 9 Sinorhizobium genomes (6.6 ± 0.30 Mb). To reveal the genomic features specific to each genus, we first computed the set of orthologous proteins shared by the test genomes from either Sinorhizobium (nine strains) or Bradyrhizobium (18 strains) by using the bidirectional best-hit (BBH) method. Sinorhizobium had the core genome of 3,133 orthologous genes, which made up 45–53% of the repertoire of protein coding genes in each strain. The core genome of Bradyrhizobium consisted of 2,570 genes, which code for 23–33% of the protein pool in each genome. Then, we compared the core genome of Sinorhizobium species and that of Bradyrhizobium by using Cluster of Orthologous Groups (COG) assignments to determine whether there were differences in the proportion of the core genome attributable to a particular cellular process (Table 1). Bradyrhizobium core genome was found to be disproportionally enriched in secondary metabolism genes and lipid transport and metabolism genes (Fisher’s exact test; P value < 0.01). Similarly, genus-specific core genes of Bradyrhizobium were also enriched in these two categories (Fisher’s exact test; P value < 0.001) compared with those of Sinorhizobium. This is consistent with an earlier report that larger prokaryotic genomes preferentially accumulate genes directly or indirectly involved in metabolism (13). These genes could support a broader metabolic diversity, which, in turn, would improve the ecological success of Bradyrhizobium in more diverse soil conditions where resources are limited but varied and where there is little penalty for slow growth (13). Although the majority of Bradyrhizobium-specific core genes have not been experimentally characterized, fsrR encoding a protein that contains signal transduction histidine kinase and response regulator receiver domains has been shown to be involved in enhancing the viability of cells exposed to acid conditions in the acid-tolerant strain S. medicae WR101 (14). In contrast to the prevalence of Bradyrhizobium in soybean nodules in neutral and acid soils, Sinorhizobium strains dominated the nodulating microsymbionts of soybean in sites of alkaline–saline soils (9–11, 15) (Fig. S1). Interestingly, genus-specific core genes of Sinorhizobium (compared with those of Bradyrhizobium) contained genes known to be involved in alkaline–saline adaptations, for example, the pha2 gene cluster encoding a monovalent cation/proton antiporter involved in Na+ resistance and adaption to alkaline pH (16), the bet gene cluster in charge of the synthesis of osmoprotectant glycine betaine (17), and glycine betaine transporter genes. Therefore, these genus-specific genes likely contribute to the observed biogeographic patterns of Bradyrhizobium and Sinorhizobium nodulating soybean, although more systematic functional genomic analyses are needed to explore the working mechanisms related to these and other genus specific genes, especially those of unknown function.
Fig. 1.
Pan genome of Sinorhizobium or Bradyrhizobium. (A and B) Accumulation curves for the pan genome of Sinorhizobium (A) or Bradyrhizobium (B). Each box plot [interquartile range (IQR)] represents the 25–75% range, with the thick line placed at the sample median. Horizontal bars connected to the box by dash lines show the range of quartile 1 − 1.5 × IQR and quartile 3 + 1.5 × IQR. Open circles represent outliers of this range.
Table 1.
Comparison of COG assignments between Sinorhizobium and Bradyrhizobium
| Core genome |
Genus-specific genes† |
|||||
| Individual functional categories | Sino | Brady | P value* | Sino | Brady | P value |
| B: Chromatin structure and dynamics | 2 | 2 | 1 | 0 | 0 | — |
| J: Translation, ribosomal structure and biogenesis | 142 | 139 | 0.177 | 8 | 18 | 0.165 |
| K: Transcription | 115 | 99 | 0.834 | 38 | 41 | 0.731 |
| L: Replication, recombination and repair | 93 | 69 | 0.473 | 9 | 16 | 0.421 |
| D: Cell cycle control, cell division, chromosome partitioning | 24 | 17 | 0.64 | 5 | 4 | 0.74 |
| V: Defense mechanisms | 28 | 21 | 0.774 | 6 | 11 | 0.468 |
| O: Posttranslational modification, protein turnover, chaperones | 114 | 89 | 0.668 | 25 | 24 | 0.472 |
| M: Cell wall/membrane/envelope biogenesis | 126 | 91 | 0.299 | 45 | 40 | 0.184 |
| P: Inorganic ion transport and metabolism | 114 | 112 | 0.22 | 42 | 51 | 1 |
| U: Intracellular trafficking, secretion, and vesicular transport | 74 | 51 | 0.319 | 16 | 17 | 0.861 |
| N: Cell motility | 52 | 25 | 0.021 | 16 | 25 | 0.432 |
| T: Signal transduction mechanisms | 75 | 59 | 0.793 | 32 | 29 | 0.3 |
| F: Nucleotide transport and metabolism | 72 | 44 | 0.11 | 16 | 8 | 0.041 |
| G: Carbohydrate transport and metabolism | 133 | 104 | 0.642 | 46 | 41 | 0.189 |
| E: Amino acid transport and metabolism | 255 | 250 | 0.056 | 75 | 97 | 0.579 |
| H: Coenzyme transport and metabolism | 128 | 106 | 1 | 25 | 23 | 0.383 |
| I: Lipid transport and metabolism | 102 | 124 | 0.004 | 16 | 54 | <0.001 |
| C: Energy production and conversion | 161 | 164 | 0.067 | 39 | 55 | 0.46 |
| Q: Secondary metabolites biosynthesis, transport and catabolism | 54 | 78 | 0.002 | 10 | 45 | <0.001 |
| R: General function prediction only | 288 | 257 | 0.417 | 75 | 113 | 0.109 |
| S: Function unknown | 286 | 223 | 0.457 | 93 | 100 | 0.498 |
Brady, Bradyrhizobium; Sino, Sinorhizobium.
*Fisher's exact test.
†Genes common and specific in test Sinorhizobium/Bradyrhizobium genomes.
Phyletic Distribution of Symbiosis Genes Reflects the Species Phylogeny of Soybean Microsymbionts and Other Rhizobia.
Although test Sinorhizobium and Bradyrhizobium strains nodulate the same legume host, we did not find any gene that is both common and specific to these soybean microsymbionts compared with other legume microsymbionts (Table S1). This implied that there is no gene specifically shared by Sinorhizobium and Bradyrhizobium to establish symbiosis with soybean. This phenomenon is, to a certain extent, consistent with earlier finding that no common gene is specifically used by rhizobia to establish symbiosis with legumes (4). Moreover, many known symbiosis genes were found to be specific to certain strain/species (1). Given the diversity of the machinery involved in the varied rhizobia–legume symbiosis, it is likely that there is no single rhizobium recipe but, instead, several distinct strategies regardless of rhizobial phylogeny (1). However, the extent to which the rhizobial phylogeny is related to the phyletic distribution of symbiosis genes has not been investigated.
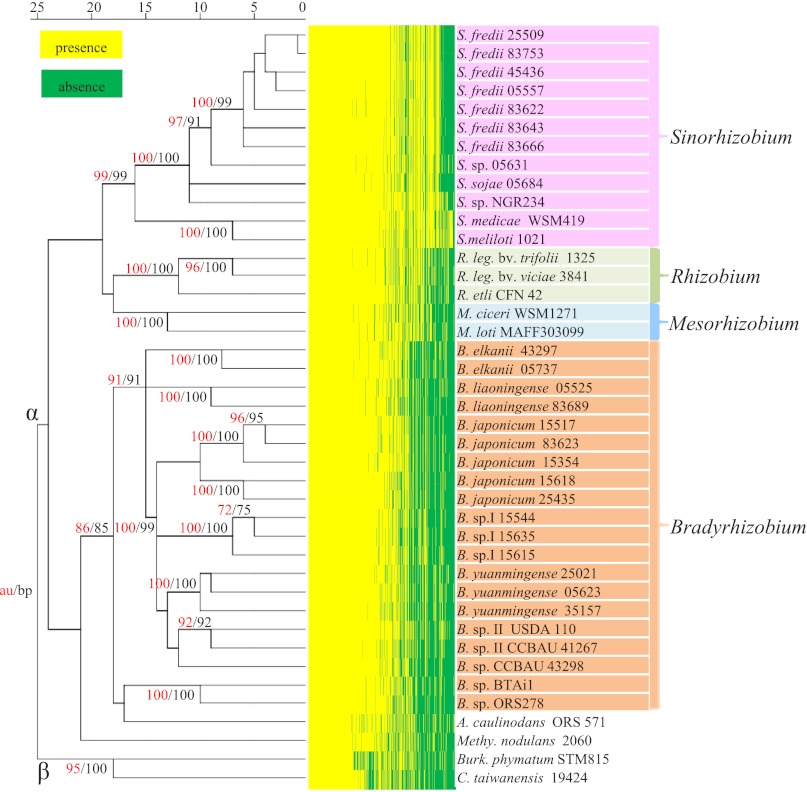
Based on extensive research on published genetics studies of the rhizobium–legume symbiosis, we collected 561 symbiosis genes (Table S3) from the literature and Nodulation Mutant Database (NodMutDB) (5). According to the presence and absence of these symbiosis genes across genomes of these soybean microsymbionts and other rhizobia, hierarchical clustering was used to build bifurcating trees for the identification of strains with similar gene content. As shown in Fig. 2, the hierarchical cluster derived from these data distinguished Sinorhizobium clearly from Bradyrhizobium among these soybean rhizobia. Notably, exoADFKLOQUX, rkpAIJZ, and lpsCDE coding the machinery required for the production of polysaccharides active in symbiosis (exopolysaccharide, capsular polysaccharide, and lipopolysaccharide) in the model strain Sinorhizobium meliloti 1021 were also found in Sinorhizobium strains nodulating soybean but absent in Bradyrhizobium. Moreover, β-rhizobia and main lineages in α-rhizobia (Bradyrhizobium, Rhizobium, Mesorhizobium, and Sinorhizobium) were also identified. (See Table S4 and Dataset S1 for more examples of lineage-specific genes.) When multiple genomes for the same species were available, the clustering results correspond well with the species assignments based on average nucleotide identity (ANI) using MUMmer (ANIm) (18) (Table S5). It is noteworthy that USDA110 should not be referenced as B. japonicum (ANIm < 91%) and that the widely studied broad-host strain Sinorhizobium sp. NGR234 may represent a species other than Sinorhizobium fredii (ANIm < 93%) (8, 19). To test whether the phylogeny derived from the heat map of symbiosis genes could reflect the phylogeny of the rhizobial species, we constructed the neighbor-joining tree based on concatenated sequences of housekeeping genes dnaK and rpoB, which shows congruent phylogeny with the species tree of rhizobia (Fig. S2). Global similarity between two trees was observed.
Fig. 2.
Hierarchical clustering of rhizobia based on heat map of 561 symbiosis genes. Presence and absence of the homolog for each symbiosis gene are indicated in yellow and green, respectively. Approximately unbiased probability and bootstrap probability (au/bp) values of >70% were indicated at each node. Four genera for which multiple genomes were available are shown in different colors. α, the branch of α-proteobacteria; β, the branch of β-proteobacteria.
However, this finding did not challenge the well-known story of lateral transfer of nodulation genes (nod) among bacteria (20). Indeed, the only exceptions are found in photosynthetic and nonphotosynthetic bacteria belonging to Aeschynomene–Bradyrhizobium CI-group 3 (cross-inoculation group) (3, 21). However, occasionally lateral acquisition of nod genes has led to the adaptation of some CI-group 3 strains to more Aeschynomene species (3, 21). nod genes might have been first subject to multiple lateral transfers into members of different bacterial groups (Fig. S3), followed by vertical descent and/or being swapped between closely related strains/species within each group, such as the group of B. liaoningense, B. japonicum, Bradyrhizobium sp. I and sp. II, the group of S. fredii, S. sojae, Sinorhizobium sp. CCBAU05631 and Sinorhizobium sp. NGR234, and the group of Rhizobium leguminosarum bv. viciae and R. fabae (22, 23).
Consistent with the great divergence of nod genes between these rhizobial groups, it was recently demonstrated that transfer of nod-nif itself is just the starting point for bacterial evolution into rhizobia (24). The experimental evolution procedure described by these authors (24) highlighted the importance of gene inactivation in the short-term evolution (nodulation and infection) of nonrhizobia into legume microsymbionts (1), although it was difficult to recapture the long-term evolutionary events (symbiotic performance, host range, and competitiveness). In addition to those nod-nif genes, around 500 other symbiosis genes involved in both short-term and long-term evolution have been documented (Table S3 and references therein). We have demonstrated that more than two-thirds of 561 symbiosis genes were not shared by all of the 41 analyzed strains, and the phyletic pattern of these shell genes is consistent with the species phylogeny of rhizobia, indicating extensive recruitment of lineage-specific genes in symbiotic interactions of bacteria with legumes. It should also be mentioned that one-third of known symbiosis genes with universal distributions in rhizobial genomes include 84 genes involved in metabolism (glnBK, hemAH, ilvICD2, leuCB, phbAC, etc.), 30 genes belonging to the transcription category (nifA, nodD1D2D3, rpoBC, rpoH1H2, etc.), 29 genes of signal transduction (actR, chvI, dctD, fixJK1K2, ntrCX, phoB, relA, regR, ttsI, etc.), 16 belonging to the functional category of posttranslational modification, protein turnover chaperones (dnaJ, glnD, groEL1234, groES123, etc.) (Dataset S1). Interestingly, earlier estimates date the divergence between bradyrhizobia and the fast-growing rhizobia at 507–553 million years ago (MYA) and that among the fast-growing rhizobial genera (Sinorhizobium–Rhizobium–Mesorhizobium) at 203–324 MYA, which is before the emergence of legume, i.e., 60 MYA (25, 26). It seems that the innovation of rhizobial symbiotic interactions with legumes depends on the integrations of shell genes with core functions; however, it remains elusive how many shell genes preexist before the innovation of rhizobium–legume nodulation.
Recombination Including Lateral Gene Transfer in Rhizobial Long-Term Evolution.
The abovementioned phylogeny based on the conservation of symbiosis genes implies that gene content information in the shell genome of rhizobia could reflect the species phylogeny. Interestingly, when the same procedure was used to analyze all of those 887 experimentally demonstrated functional genes, including 326 nonsymbiosis genes (Table S3), the resulting tree (Fig. S4) was similar to both the tree for symbiosis genes and the species tree. This is noteworthy because the evolution of bacterial shell genome is believed to be dominated by lateral gene transfer (LGT), the nongenealogical transmission of genetic material from one organism to another (27, 28). Generally speaking, recombination including LGT has been considered to preclude the reconstruction of phylogenetic relationships in the microbial world, and, on the other hand, phylogenetic incongruence has been widely used to detect recombination, particularly with ancient events (28, 29). Then, how could phyletic patterns of these shell genes reflect the species phylogeny of rhizobia? To answer this question, it was essential to get a global view about the role of recombination in rhizobial evolution, so we further investigated its extent and relative importance in the rhizobial core genome.
Firstly, we obtained 295 core genes of 26 representative genomes from 22 rhizobial species by using the BBH method. These 295 core genes/proteins were used to construct the supertree (Fig. S5A) and maximum likelihood (ML) phylogenetic tree (Fig. S5B) of rhizobia. Both of these two widely used species-tree reconstruction methods produced similar tree topology that supported the known phylogeny of rhizobia. However, the phylogenetic positions of Methylobacterium nodulans and Azorhizobium caulinodans were inconsistent between the trees. This is not likely to be the cause of low signal-to-noise ratio, because alignments were all treated with Gblocks (30) and all of them passed the permutation tail probability (PTP) test integrated in PAUP* 4.0b10 (31). Instead, the lack of additional species that could be used to break up the long branch leading to either M. nodulans or A. caulinodans lineage may be responsible for their unstable phylogenetic placements, i.e., the long branch attraction (LBA) effect (32). Interestingly, the Shimodaira–Hasegawa (SH) test (33) between each gene tree (ML) and the reference tree based on 295 core genes (ML) uncovered only four genes with phylogenies congruent with the reference tree. However, this observation could also be attributable to the influence of LBA (32). To test the potential effect of LBA, we removed four taxa that have a long terminal branch (i.e., Cupriavidus taiwanensis, Burkholderia phymatum, A. caulinodans, and M. nodulans). The reliability of resulting ML tree of 22 rhizobial genomes was evidenced by its congruent topology (SH-test; P = 1.0) with the corresponding strict consensus tree of 295 single gene trees and was used as the reference species tree. Subsequently, SH test revealed 247 or 262 gene trees to be incongruent with this species tree when the confidence interval was 99% or 95%. It should be noted that the big confidence interval of 99% clearly increased the rate of false negatives, i.e., the number of undetected events of LGT in this study as revealed by manual examinations of single gene trees. In contrast, only few false negatives were observed at 95% confidence interval, and around 90% of the core genes were, thus, found to have undergone LGT or intergenic recombination in their history. In addition to the intergenic recombination, intragenic recombination was recently found in several widely studied housekeeping genes for various rhizobia (8, 23). To evaluate the extensiveness of intragenic recombination, a combination of single break point (SBP) screening method with small sample Akaike information criterion (AICc) (34) and the phi (Фw) statistic (35) was used. This combination of methods increased sensitivity and detected 142 genes to have been affected by intragenic recombination (P < 0.05). Taken together, significant indications of (intergenic and/or intragenic) recombination were obtained for 93.2% of the rhizobial core genes (275/295) tested. When similar analyses were carried out for Sinorhizobium and Bradyrhizobium (Table S6), 92.2% and 86.3% of core genes, respectively, in Sinorhizobium and Bradyrhizobium were found to have been affected by recombination. Interestingly, among these recombined genes, both the core genes and genus-specific core genes of Bradyrhizobium were found to be enriched in secondary metabolism genes and lipid transport and metabolism genes (Fisher’s exact test; P value < 0.01 or P value < 0.001) compared with those of Sinorhizobium (Table S6). Moreover, most of the genus-specific genes known to be involved in alkaline–saline adaptations of Sinorhizobium, including the entire betICBA gene cluster and 5/7 of pha2 genes, were affected by recombination. However, biased codon use was only found in 3.2%/3.2% of recombined genes in Sinorhizobium/Bradyrhizobium and has no preference to lineage-specific genes or any functional category. These findings suggested that the majority of these extensive recombinant events could be very ancient.
To calculate the ratio of rates at which recombination and mutation occur (ρ/θ) and relative contribution of recombination and mutation in the creation of the sample from a common ancestor (r/m), the longest 100 blocks in the whole-genome alignments of Sinorhizobium or Bradyrhizobium nodulating soybean were analyzed by using ClonalFrame (36). The value of ρ/θ was 0.55 ± 0.02 and 0.25 ± 0.02 for Sinorhizobium and Bradyrhizobium, respectively. Interestingly, the r/m values were 36.76 ± 1.49 and 4.77 ± 0.26 for Sinorhizobium and Bradyrhizobium, respectively. These values of ρ/θ in genus-core of Sinorhizobium/Bradyrhizobium were close to the lowest value of the boundary (0.25–2), above which clusters no longer diverge, but were constantly reabsorbed into the parent population by the cohesive effect of recombination (37). These imply that the core genome in either Sinorhizobium or Bradyrhizobium has undergone a considerable level of recombination but were short of panmixis in their evolutionary history. Although the rate of recombination was clearly lower than mutation, recombination could introduce a segment of foreign DNA into the genome rather than the point substitution caused by unfaithful replication or DNA damage. Consequently, recombination (compared with point mutations) contributed to the observed diversity of core genome of Bradyrhizobium/Sinorhizobium nodulating soybean by a factor of 4.77-/36.76-fold.
Therefore, the tiny vertically descended core (less than 15% of core genome) might have been marginalized by the powerful recombination, as hypothesized earlier (27). Then, how could phyletic distributions of functional genes including symbiosis genes in the shell genome reflect the species phylogeny of rhizobia? It is widely accepted that lateral sharing of genetic innovations would have triggered an explosion of genetic diversity (27). However, only those successfully integrated foreign genetic materials could become new members of the vertical core in the recipient genomes, and these successful integration events in the evolutionary history could have contributed to the observed species/genus specific functional genes. This process itself might be one of the key steps in the speciation of rhizobia. Interestingly, when phyletic distributions of all genes in the known rhizobial pan genome (66,150 genes) were analyzed (Fig. S6), we could not get the tree of similar topology as the species phylogeny: β-rhizobia, Mesorhizobium, Rhizobium, and Sinorhizobium formed a cluster of high bootstrap support (99%); Bradyrhizobium sp. BTAi1 and Bradyrhizobium sp. ORS278 were incorporated into the group of B. japonicum–B. liaoningense–B. yuanmingense, etc. This could be attributable to many transiently acquired genes in the pan genome that have not been removed by the selective pressure of speciation.
Conclusion
Twenty-six representative genomes of Sinorhizobium and Bradyrhizobium nodulating soybean are valuable resources for the investigation of rhizobial evolution in terms of their contrasting genomic features. During the adaptations to symbiotic interactions and other environmental conditions, lineage-specific genes have been extensively recruited by these microsymbionts of soybean and other legume hosts. The tree based on the gene content of functional genes rather than that based on the total set of pan genome is consistent with the rhizobial phylogeny, whereas recombination is the dominant microevolutionary force in rhizobial evolution and leads to few core genes with the congruent phylogenetic topology as the species tree. Thus, successful integrations of functional genes seem to play an essential role in the speciation process. Rhizobia, living as either saprophytic or symbiotic bacteria, are characterized with their large genomes in the bacterial world, and the genome sequences and related findings in this study should help us get further insights in many aspects of the bacterial evolution.
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
The draft sequences of 26 test strains were produced by using Illumina paired-end sequencing technology at the BGI–Shenzhen (Table S2). Assembly was done with SOAPdenovo (38). Gene prediction was done using Glimmer v3.0 (39). Annotation of protein coding sequence was performed by using Basic Local Alignment Search Tool (BLAST) against COG and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The draft genomes of 26 test strains have been deposited in GenBank under the project accession no. PRJNA77219 (Table S1).
Comparative Genomics.
The BBH approach (4) was used to identify all of the orthologous pairs between test rhizobial genomes. All pairwise BBH shared genes were compared, and the common dataset of shared genes among test strains was defined as their core genome. The total set of genes within test genomes was defined as the pan genome. Documented symbiosis or nonsymbiosis functional genes (Table S3) were mapped across rhizobial genomes, and the resulting table was used to calculate the Average Linkage dendrogram based on Euclidean distance with SPSS package. The bootstrap tests were performed using the R environment package Pvclust (40). The ANI values between genomes were calculated by using the NUCmer algorithm integrated in Jspecies (18).
Phylogenetic Analyses.
Single gene alignments were aligned with molecular evolutionary genetics analysis (MEGA) (41). The neighbor-joining trees were constructed by using the same software, and 1,000 bootstraps were done. The maximum likelihood trees were constructed with PhyML or PAUP* 4.0b10, when necessary 100 bootstrap analysis was performed by using PHYLIP (31, 42, 43). The best nucleotide substitution model for each alignment was obtained by MODELTEST analysis (44). The supertree of 295 core proteins was constructed by using a pipeline described in the legends of the figures in SI Text. The strict consensus tree of 295 core genes was reconstructed by using Clann (45).
Analyses of Recombination Including LGT.
The effect of intergenic recombination was investigated by testing whether the gene tree is congruent to the species tree in a ML schema with the SH test (33) implemented in PAUP* 4.0b10 (31). The presence or absence of intragenic recombination was assessed by the single break point (SBP) method (34) and the Фw statistic (36) (*P < 0.05). To calculate r/m and ρ/θ, multiple genome alignments (the 100 longest blocks obtained by using Mauve and stripSubsetLCBs) were subject to ClonalFrame analysis (36, 46). To identify putatively recently transferred genes of biased codon use, the χ2 goodness-of-fit test was used; those genes with similar codon frequencies to putatively highly expressed genes (PHEG) of S. meliloti 1021 and B. japonicum USDA110 (47) were identified by calculating the codon adaptation index (48).
Supplementary Material
Acknowledgments
We thank C. Masson-Boivin, J. Peter W. Young, and three anonymous reviewers for their valuable comments on the manuscript. This work was supported by National Basic Research Program of China (973 Program) Grant 2010CB126500, National Natural Science Foundation of China Grant 31170002, Innovative Project of State Key Laboratory of Agrobiotechnology (SKLAB) Grants 2010SKLAB01-1 and 2012SKLAB01-9, Chinese Universities Scientific Fund Grant 2010TD001, and China Postdoctoral Science Foundation Grant 20100480377. Genome sequencing was partially supported by BGI–Shenzhen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences of 26 test strains reported in this paper have been deposited in the GenBank database (BioProject no. PRJNA77219).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120436109/-/DCSupplemental.
References
- 1.Masson-Boivin C, Giraud E, Perret X, Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Young JPW, et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraud E, et al. Legumes symbioses: Absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- 4.Amadou C, et al. Genome sequence of the beta-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 2008;18:1472–1483. doi: 10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao C, Qiu J, Wang C, Charles TC, Sobral BW. NodMutDB: A database for genes and mutants involved in symbiosis. Bioinformatics. 2005;21:2927–2929. doi: 10.1093/bioinformatics/bti427. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor Appl Genet. 2008;117:857–871. doi: 10.1007/s00122-008-0825-0. [DOI] [PubMed] [Google Scholar]
- 7.Man CX, et al. Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil. 2008;310:77–87. [Google Scholar]
- 8.Vinuesa P, et al. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the asiatic continent. Appl Environ Microbiol. 2008;74:6987–6996. doi: 10.1128/AEM.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han LL, et al. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil. 2009;324:291–305. [Google Scholar]
- 10.Li QQ, et al. Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei Province, China. Microb Ecol. 2011;61:917–931. doi: 10.1007/s00248-011-9820-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YM, et al. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China Plain. Appl Environ Microbiol. 2011;77:6331–6342. doi: 10.1128/AEM.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li QQ, et al. Ensifer sojae sp. nov., isolated from root nodules of Glycine max grown in saline-alkaline soils. Int J Syst Evol Microbiol. 2011;61:1981–1988. doi: 10.1099/ijs.0.025049-0. [DOI] [PubMed] [Google Scholar]
- 13.Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeve WG, et al. The Sinorhizobium medicae WSM419 lpiA gene is transcriptionally activated by FsrR and required to enhance survival in lethal acid conditions. Microbiology. 2006;152:3049–3059. doi: 10.1099/mic.0.28764-0. [DOI] [PubMed] [Google Scholar]
- 15.Camacho M, et al. Soils of the Chinese Hubei province show a very high diversity of Sinorhizobium fredii strains. Syst Appl Microbiol. 2002;25:592–602. doi: 10.1078/07232020260517733. [DOI] [PubMed] [Google Scholar]
- 16.Yang LF, et al. The pha2 gene cluster involved in Na+ resistance and adaption to alkaline pH in Sinorhizobium fredii RT19 encodes a monovalent cation/proton antiporter. FEMS Microbiol Lett. 2006;262:172–177. doi: 10.1111/j.1574-6968.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 17.Pocard JA, et al. Molecular characterization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34. Microbiology. 1997;143:1369–1379. doi: 10.1099/00221287-143-4-1369. [DOI] [PubMed] [Google Scholar]
- 18.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saldaña G, et al. Genetic diversity of fast-growing rhizobia that nodulate soybean (Glycine max L. Merr) Arch Microbiol. 2003;180:45–52. doi: 10.1007/s00203-003-0559-y. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miché L, et al. Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that nod-independent stem nodulation is not restricted to photosynthetic bradyrhizobia. Environ Microbiol. 2010;12:2152–2164. doi: 10.1111/j.1462-2920.2009.02090.x. [DOI] [PubMed] [Google Scholar]
- 22.Bontemps C, et al. Burkholderia species are ancient symbionts of legumes. Mol Ecol. 2010;19:44–52. doi: 10.1111/j.1365-294X.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- 23.Tian CF, Young JP, Wang ET, Tamimi SM, Chen WX. Population mixing of Rhizobium leguminosarum bv. viciae nodulating Vicia faba: The role of recombination and lateral gene transfer. FEMS Microbiol Ecol. 2010;73:563–576. doi: 10.1111/j.1574-6941.2010.00909.x. [DOI] [PubMed] [Google Scholar]
- 24.Marchetti M, et al. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol. 2010;8:e1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner SL, Young JPW. The glutamine synthetases of rhizobia: Phylogenetics and evolutionary implications. Mol Biol Evol. 2000;17:309–319. doi: 10.1093/oxfordjournals.molbev.a026311. [DOI] [PubMed] [Google Scholar]
- 26.Sprent JI. Evolving ideas of legume evolution and diversity: A taxonomic perspective on the occurrence of nodulation. New Phytol. 2007;174:11–25. doi: 10.1111/j.1469-8137.2007.02015.x. [DOI] [PubMed] [Google Scholar]
- 27.Goldenfeld N, Woese C. Biology’s next revolution. Nature. 2007;445:369. doi: 10.1038/445369a. [DOI] [PubMed] [Google Scholar]
- 28.Doolittle WF, Nesbo CL, Bapteste E, Zhaxybayeva O. Lateral gene transfer. In: Pagel M, Pomiankowski A, editors. Evolutionary Genomics and Proteomics. Sunderland, MA: Sinauer; 2008. pp. 45–79. [Google Scholar]
- 29.Boto L. Horizontal gene transfer in evolution: Facts and challenges. Proc Biol Sci. 2010;277:819–827. doi: 10.1098/rspb.2009.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 31.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 32.Kuo CH, Wares JP, Kissinger JC. The Apicomplexan whole-genome phylogeny: An analysis of incongruence among gene trees. Mol Biol Evol. 2008;25:2689–2698. doi: 10.1093/molbev/msn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 34.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 35.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Retief JD. Phylogenetic analysis using PHYLIP. Methods Mol Biol. 2000;132:243–258. doi: 10.1385/1-59259-192-2:243. [DOI] [PubMed] [Google Scholar]
- 43.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 44.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 45.Creevey CJ, McInerney JO. Trees from trees: Construction of phylogenetic supertrees using clann. Methods Mol Biol. 2009;537:139–161. doi: 10.1007/978-1-59745-251-9_7. [DOI] [PubMed] [Google Scholar]
- 46.Darling AE, Mau B, Perna NT. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puigbò P, Romeu A, Garcia-Vallvé S. HEG-DB: A database of predicted highly expressed genes in prokaryotic complete genomes under translational selection. Nucleic Acids Res. 2008;36(Database issue):D524–D527. doi: 10.1093/nar/gkm831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.