Abstract
Human malaria infection begins with a one-time asymptomatic liver stage followed by a cyclic symptomatic blood stage. All high-throughput malaria drug discovery efforts have focused on the cyclic blood stage, which has limited potential for the prophylaxis, transmission blocking, and eradication efforts that will be needed in the future. To address these unmet needs, a high-throughput phenotypic liver-stage Plasmodium parasite screen was developed to systematically identify molecules with liver-stage efficacy. The screen recapitulates liver-stage infection by isolating luciferase-expressing Plasmodium berghei parasites directly from the salivary glands of infected mosquitoes, adding them to confluent human liver cells in 384-well plates, and measuring luciferase activity after a suitable incubation period. Screening 5,375 known bioactive compounds identified 37 liver-stage malaria inhibitors with diverse modes of action, as shown by inhibition time course experiments. Further analysis of the hits in the Food and Drug Administration-approved drug subset revealed compounds that seem to act specifically on the liver stage of infection, suggesting that this phase of the parasite’s life cycle presents a promising area for new drug discovery. Notably, many active compounds in this screen have molecular structures and putative targets distinctly different from those of known antimalarial agents.
Despite sharply increased efforts to discover and develop new therapeutic agents and to implement improved preventive measures, malaria continues to burden large parts of the globe (1). The World Health Organization estimates malaria’s morbidity burden at over 200 million cases per year with a mortality burden of roughly a million deaths per year, primarily to children under 5 y and pregnant women in sub-Saharan Africa (2). Diminishing this burden will require a comprehensive attack on the causative agents, single-celled eukaryotic parasites belonging to the genus Plasmodium. These highly specialized parasites have multiple developmental stages that allow them to infect and move between their mosquito and human hosts encoded in their ∼23-Mb genome (3). In a human host, the overwhelming majority of parasites are either in the liver (infection initiated by sporozoites) or in the blood (cycles of red blood cell infection initiated by merozoites) developmental stage.
When an infected female mosquito takes her obligatory blood meal, Plasmodium sporozoites leave her salivary glands and enter the human host where they move quickly to the liver. After entering and then traversing, or migrating through, several liver cells, parasites invade a final cell, in which they replicate and expand their population by four orders of magnitude before exiting the liver as merozoites (4, 5). The two parasite forms, sporozoites and merozoites, must recognize, invade, and exploit completely different types of human cells, and this selectivity involves differences in everything from the extracellular proteins needed for invasion to the enzymes and pathways providing the energy and substrates for replication. Although the details of these differences are not well understood, transcript array and proteomic comparisons of liver and blood-stage parasites have shown profound qualitative and quantitative differences between the two stages (6–10). Unfortunately, the high percentage of genes with unknown function (>50%) within Plasmodium genomes makes it difficult to ascertain the functions of most stage-specific genes (3).
Whereas current therapies can clear blood-stage malaria, the parasite’s ability to develop drug resistance requires the continuous discovery and development of new therapeutic agents. These efforts have been hampered by the limited number of fully validated targets, the difficulty of predicting gene function from sequence analysis, and the restricted genetic tools available to probe the parasite’s cellular pathways. To date, high-throughput malaria screens have been limited to the parasite’s blood stage (11–14), and consequently relatively few drugs are known to inhibit malaria’s liver stages. Primaquine, for example, is a clinically used inhibitor of liver-stage malaria parasites (15). It is also the only drug used to clear Plasmodium vivax hypnozoites, a dormant hepatic stage that can cause relapsing malaria months or years after the original infection and that contributes substantially to malaria’s morbidity (16, 17). Despite this activity, primaquine’s other properties make it a poor choice for current therapy or even a useful starting point for analogs. It has a notably high IC50 (∼10 μM) in vitro (18) and causes hemolytic anemia in people with glucose-6-phosphate dehydrogenase deficiency (G6PD), the most common enzyme deficiency in malarious regions of Africa, South America, and Asia (19, 20). Atovaquone, another malaria drug, is a nanomolar inhibitor of liver-stage parasites, but it is not effective against the hypnozoite stage of P. vivax (17, 21).
Active compounds against the malaria parasite’s asymptomatic liver stage would enjoy strategic advantages, particularly the relatively small numbers of parasites, unavailable to currently used blood-stage drugs, but the technical difficulties involved in conducting high-throughput screens for this transient developmental stage have discouraged large-scale systematic searches for suitable drug candidates (22–24). Malaria’s blood stage is cyclic as merozoites invade red blood cells and proliferate until the cells rupture, whereupon they quickly reinvade uninfected red blood cells. Merozoites can be kept in continuous laboratory culture by the addition of fresh red blood cells (25, 26), and this culturability has allowed high-throughput phenotypic assays to be developed (11–14). Conversely, the liver stage occurs only once, and getting enough sporozoites, which must be obtained by appropriately timed microscopic dissection of the salivary glands of infected mosquitoes (27), to perform a high-throughput screen has been technically challenging. Here, we report the development of a high-throughput drug screen targeting the liver stage of malaria infection. The screen successfully identified liver-stage malaria inhibitors among a library of biologically active compounds. Analysis of these liver-stage inhibitors revealed several unique potential parasite targets and unique antimalarial chemotypes that could serve as the basis for further drug development and/or provide essential probes to explore the poorly understood biology of liver-stage Plasmodium parasites.
Results and Discussion
Development of a High-Throughput Liver-Stage Malaria Screen.
To explore potential vulnerabilities of liver-stage sporozoites to small molecules, we developed a high-throughput screen using a luciferase-expressing sporozoite strain of Plasmodium berghei ANKA (28) harvested from mosquito salivary glands to infect human liver HepG2 cells (Fig. S1). The assay requires the sequential dissection of live mosquitoes (∼200 mosquitoes per plate) shortly before every screen, because parasite viability decreases within hours of their removal from the mosquito host. The assay was optimized for screening in 384-well plates with a Z-factor between 0.5 and 0.7. It was used to screen a small molecule library of known bioactive compounds with 5,375 members and included a 640-member small molecule library of Food and Drug Administration (FDA)-approved drugs. The most sensitive variable affecting the Z-factor was sporozoite density in the infected mosquitoes. Higher sporozoite density led to cleaner parasite preps and higher Z-factors.
Although the assay itself is robust and efficient, its scale is ultimately limited by the requirement for fresh parasites—a limitation imposed by the biology of the parasite’s developmental program. Currently, parasite harvesting effectively limits the screen described here to libraries in the low tens of thousands.
Compared with the blood stages, malaria’s transient liver stages are poorly understood and largely lack genetic and molecular tools. Even chemical approaches such as pull-down experiments with affinity reagents are troubled by the exceptionally high ratio of human liver proteins to parasite proteins. These limitations in biological understanding and technical approaches complicate target identification for screening positives that emerge in any phenotypic liver-stage screen, especially positives with liver-stage specificity. Target identification, which has remained challenging for blood-stage drug discovery and development, will be even more challenging for liver-stage screens. We addressed the dual limitations imposed by parasite harvesting and target identification by focusing this initial screen on relatively small libraries with a high percentage of molecules with known, or suspected, cellular targets.
Preliminary Analysis of Screening Hits.
Our screen tested well-characterized small molecules with known biological actions for inhibition of infection of HepG2 cells by luciferase-expressing P. berghei parasites. Screening positives were identified in the luminescence screen as compounds that reduced hepatic parasite load by ≥95% at ≥5 μg/mL without affecting HepG2 viability (Fig. 1A). These screening positives were retested first from the source plate and then from independent samples obtained from vendors. Inhibition of parasite load by the compounds at 10 μM was also confirmed by parasite imaging with antibody staining. The independently obtained compounds were then used to acquire 50% inhibitory concentrations (IC50 values), and screening positives that had IC50 values below 10 μM were considered confirmed hits (Fig. 1B).
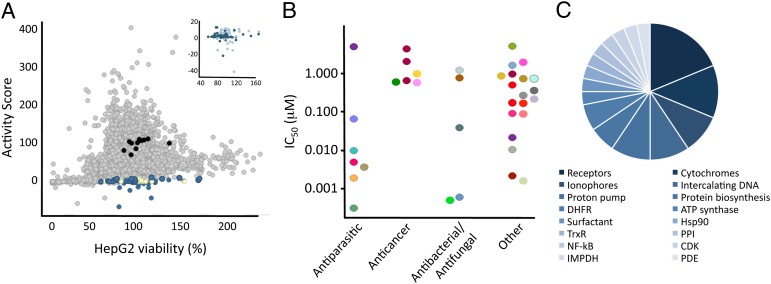
Fig. 1.
Analysis of screening results. (A) Negative controls (black), positive controls (yellow), 5,375 small molecules (gray), and screening actives (blue) are plotted as a function of their effect on HepG2 viability and inhibition of liver-stage malaria (activity score). Inset shows screening hits (dark blue) and actives that were eliminated as candidates with secondary assays (light blue). (B) Hits were grouped by function and plotted vs. their determined liver-stage Plasmodium IC50. Different compound classes of screening hits (31 total) are represented by different colors. (C) Distribution of hit function or human targets of screening hits.
This protocol identified 37 compounds as screening hits (0.7% hit rate). Functional analysis of the hits based on their known biological activities revealed a surprising number with unanticipated targets (Fig. 1B). In fact, only 20% of the active compounds were known antiparasitic agents, including coccidiostats like salinomycin and decoquinate, or compounds with known liver-stage antimalarial activity like atovaquone and primaquine. A significant fraction (18%) of the total hits were cytotoxic anticancer agents like shikonin, plicamycin, and daunorubicin, and a roughly equal number (15%) were antibacterial or antifungal agents including monensin, dequalinium, and cycloheximide. These hits may not represent a surprising extension of the compounds’ known biological activities, but for many, this screen is a unique report characterizing their activity against liver-stage Plasmodium. Almost half of the actives (47%) had diverse therapeutic activities (Fig. 1B) that included ulcer drugs, migraine medications, antihistamines, and a high blood pressure treatment.
Analysis of Putative Targets.
As noted earlier, we hoped to counter the difficulties of liver-stage Plasmodium target identification by using known bioactive drug libraries, as every member has at least one putative target. These previously reported targets were used to formulate initial hypotheses for targets responsible for the antimalarial activity seen in the screen (Fig. 1C and Table 1). Identification of potential Plasmodium falciparum targets was completed with a standard protein BLAST [National Center for Biotechnology Information (NCBI) and PlasmoDB] for parasite proteins with homology to known human protein targets. In some cases multiple results had significant homology to the human target but only the sequence with the greatest identity is shown in Table 1. A similar analysis was used to identify putative targets of blood-stage screening hits (12).
Table 1.
Putative targets for liver-stage malaria hits
| Putative target | No. of inhibitors | Example | P. falciparum locus |
| Electron transport chain | 4 | Atovaquone | PlfaoMp3 |
| Metal regulation | 3 | Monensin | NA* |
| Protein biosynthesis | 2 | Cycloheximide | MAL1P3.03a |
| Intercalating DNA | 3 | Daunorubicin | NA |
| Surfactant | 1 | Nonoxynol-9 | NA |
| Histamine receptor | 2 | Clemastine | NA |
| Estrogen receptor | 1 | Clomiphene | NA |
| Angiotensin receptor | 1 | Telmisartan | NA |
| Serotonin receptor | 2 | Methysergide | NA |
| Dihydrofolate reductase | 2 | Pyrimethamine | PFD0830w |
| Thioredoxin reductase | 1 | Auranofin | PFI1170c |
| Inosine monophosphate dehydrogenase | 1 | Mycophenolic acid | PFI1020c |
| Phosphodiesterase | 1 | Zaprinast | MAL13P1.118 |
| Protein phosphatase | 1 | Cyclosporin A | PFC0975c |
| Cyclin-dependent kinase | 1 | Cdk1/2 inhibitor III | MAL13P1.279 |
| ATPase/proton pump | 4 | Esomeprazole | PFA0310c |
| Hsp90 | 1 | Gedunin | PF07_0029 |
| Nuclear factor κB | 1 | NF-κB activation inhibitor | NA |
Reported genes are identified with BLASTP against known human protein targets. Only one locus is shown but several targets have multiple homologs.
*No obvious homology to P. falciparum locus.
Comparison of the screening hits and their putative targets revealed that some compounds likely inhibited well-known blood-stage malaria targets, including the cyctochrome bc1 complex and dihydrofolate reductase (DHFR) (Fig. 1C). These findings are not surprising as the known bioactive libraries contain currently used malarial therapeutics like atovaquone, a cytochrome bc1 complex inhibitor, and pyrimethamine, a DHFR inhibitor (23). Heat shock protein 90 (Hsp90) is a proposed target of the screening hit gedunin (29) (Table 1). Hsp90 is a known blood-stage parasite target, and it is likely critical to liver-stage Plasmodium development as well. Identification of these known blood and liver-stage malaria targets provided additional validation of the assay. Other blood-stage inhibitors, such as artemisinin and quinine, were represented in the compound libraries but were not active in the screen. This lack of activity validates the existence of liver stage-specific inhibitors whose discovery was prevented by the lack of drug screening tools for liver-stage parasites.
Liver-stage Plasmodium infection was also inhibited by ionophores (9% of hits), molecules that intercalate DNA (9%), protein biosynthesis inhibitors (6%), and a surfactant (3%). Ionophores are a previously characterized class of blood-stage malaria inhibitors (30) and we found three molecules in this class that inhibit liver-stage malaria parasites with low nanomolar to picomolar potency (salinomycin, monensin, and lasalocid A; Table S1). Another hit with an IC50 value in the low nanomolar range, a quinazoline derivative, inhibits nuclear factor kappaB (NF-κB) activation. Inhibition of DNA transcription would be a likely target given the parasite’s tremendous proliferation in the liver.
The most abundantly targeted class of proteins included ligand-regulated receptors (19%) that bind a diverse array of ligands ranging from monoamines like serotonin and histamine to the steroid estrogen and the peptide hormone angiotensin. The histamine receptor antagonist astemizole was previously identified as a malaria inhibitor in a blood-stage parasite screen (31). Although astemizole was removed from the United States market due to potentially fatal side effects, we found that clemastine, an over-the-counter antihistamine used to treat or prevent allergy symptoms, along with astemizole, inhibits liver-stage Plasmodium. Subsequent testing revealed that both drugs inhibit liver and blood-stage parasites with submicromolar potency. However, no Plasmodium gene with significant homology to the mammalian histamine receptors has been annotated. Clemastine’s activity suggests that antihistamine scaffolds could be good starting points for inhibitors of both parasite stages, and previous work has shown efficacy in mouse models (31). Additionally, the inhibition of both blood and liver-stage parasites suggests a Plasmodium target active in both stages with binding specificity similar to that of known human histamine receptors.
Two screening hits, methysergide and ketanserin, target serotonin receptors but there is no parasite gene with obvious sequence homology to known serotonin-binding sequences. A previous blood-stage screen also proposed that several of their hits target the mammalian serotonin receptor (12). Another surprising liver-stage inhibitor with nanomolar potency was telmisartan (25 nM), a blood pressure lowering agent (32). The telmisartan analogs valsartan and olmesartan, tested at 0.5 μM, do not inhibit liver-stage Plasmodium growth. All three compounds lower blood pressure by binding the human angiotensin II receptor, and valsartan and olmesartan bind more tightly than telmisartan. Their lack of activity may be due to their shorter biological half-lives (6 and 13 h, respectively, vs. 24 h for telmisartan) (32), or telmisartan may have an alternative Plasmodium target.
The second most frequently targeted class of proteins was ATP synthases/proton pumps (13%). Three hits within this class are compounds used to treat ulcers: esomeprazole (Nexium); its over-the-counter racemic form, omeprazole (Prilosec); and tenatoprazole (reviewed in ref. 33). Esomeprazole, the S-enantiomer of omeprazole, was the third-highest–grossing drug sold in 2009 and has high bioavailability and limited toxicity (34). Esomeprazole had an IC50 that was twofold lower than that of racemic omeprazole (290 nM vs. 680 nM, Table S1) and it was more active than R-omeprazole at 1 μM, which indicates a stereoselective interaction with a specific target. Esomeprazole, omeprazole, and tenatoprazole target mammalian proton pumps and their stereoselective inhibitory activity in the liver-stage screen is consistent with a similar mechanism (35). The Plasmodium genome contains at least two conserved predicted ATP synthases (PFA0310c and PFL0590c) with homology to the mammalian proton ATPase subunit. PFA0310c has greater identity than PFL0590c (also known as PfATP4) to the human gastric hydrogen potassium ATPase. Importantly PfATP4 was recently identified as the target of spiroindolones, a potent class of antimalarials (36). Thus, it is possible that inhibition of this parasite ATP synthase and/or PFA0310c is responsible for the observed activity of esomeprazole and its derivatives; however, these putative targets await experimental verification (discussed further below).
Other Plasmodium proteins including peptidylprolyl isomerases (PPIase), cyclin-dependent kinases (CDK), inosine monophosphate dehydrogenase (IMPDH), thioredoxin reductase (TrxR), and phosphodiesterases (PDE) were identified as putative parasite targets from our analysis of the liver-stage screening hits (Table 1). Taken together, this screen’s hits identify several Plasmodium proteins that may function as drug targets for the development of future antimalarials, pinpoint over-the-counter drugs with potential liver-stage utility, and describe several unique antimalarial chemotypes (Fig. 1B).
Plasmodium Infection Time-Course Studies.
In a little over 3 d, a single P. berghei sporozoite introduced into its mammalian host progresses through several stages into mature schizonts containing as many as 10,000 infectious merozoites. In the 2 h following sporozoite introduction to the hepatocytes, the parasites first migrate through several liver cells and then invade a single hepatocyte (4, 5). In the next 24 h, the parasite’s apicoplast and mitochondrion elongate, and then extremely rapid nuclear division occurs. At ∼52 h postinfection, the apicoplast and then the mitochondrion divide, the plasma membrane invaginates, and finally merozoite formation is complete (37).
To see what, if any, developmental stage specificity screening hits might possess, a time course was generated by treating Plasmodium-infected HepG2 cells with compounds at varying times (0, 3, 8 and 24 h) postsporozoite infection. Parasite development was assessed at 48 h. On the basis of this analysis, the majority of the hits (62%) inhibited processes that are necessary even after 24 h postinfection (Fig. 2A), during the proliferative burst. Electron transport and transcriptional inhibitors were in this category, as were the antihistamines (Fig. 2B) and ionophores. Whereas the vulnerability of the parasite during this stage might be expected, this activity illustrates the ability of some small molecules to clear parasites from the liver after infection occurs. Other hits inhibited processes that occur only between 0 and 24 h postinfection (26% of hits). For example, gedunin, the Hsp90 inhibitor, effectively inhibits liver-stage malaria parasites in this period (Fig. 2B), which could indicate that Hsp90 is less critical to the later stages of liver Plasmodium development. Molecules that target PPIases and the angiotensin receptor (Fig. 2B) were also effective only in the initial 24 h postinfection. Other hits influenced critical processes that occur only between 0 and 3 h (zaprinast, a PDE inhibitor) or 3 and 8 h (methysergide, a serotonin antagonist) after infection.
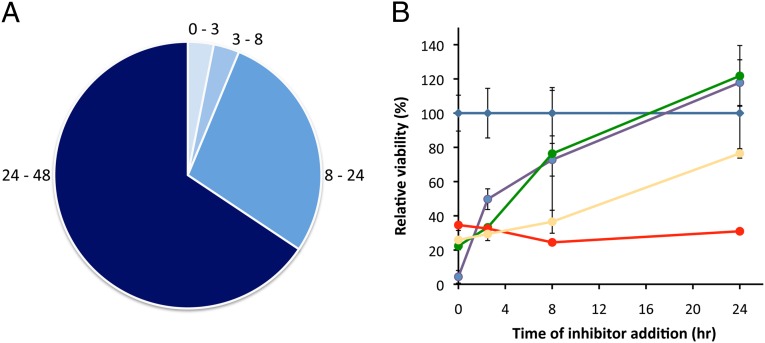
Fig. 2.
Evaluation of the efficacy of screening hits (10 μM) at various stages of liver infection. (A) Hits were evaluated for inhibition of parasite processes 0–3 h, 3–8 h, 8–24 h, and 24–48 h after infection. (B) Most hits (61%) inhibit parasite processes that occur 24–48 h postinfection. Representative time course data are shown for DMSO (blue), astemizole (red), gedunin (yellow), dequalinium (purple), and telmisartan (green). All parasite levels were measured 48 h postinfection.
A more detailed analysis of the data was provided by a comparison of the rate of change in the time-course plots. Fig. 2B shows that dequalinium and telmisartan inhibit liver-stage malaria parasites relative to the control between 0 and 24 h postinfection, but they are not equally effective at inhibiting parasite processes during the first 3 h. The different slopes of the regression lines indicate that dequalinium’s ability to inhibit the parasite diminishes rapidly within the first few hours of infection, whereas telmisartan’s target is more critical between 0 and 3 h than between 3 and 8 h. Further analysis of Plasmodium infection time courses could provide a new method for evaluating the mode of action of liver-stage inhibitors. In addition to their potential therapeutic value, inhibitors that specifically affect early or late stage infection could function as chemical tools for probing the complex biology of the liver-stage malaria parasite.
FDA-Approved Drugs.
Within the 5,375 compounds screened, at least 640 are FDA-approved drugs, and these drugs were of special interest as their chemistry and biology have been extensively investigated and each drug typically has numerous analogs and significant human safety data. Fifteen of these compounds were identified as inhibitors of liver-stage malaria parasites, and 2 are over-the-counter drugs. The majority of the FDA-approved hits (60%) had a diverse range of functions that included treatments for ulcers, migraine, and blood pressure, functions that would not normally be associated with antiparasitic activity. Two-thirds of them selectively inhibit liver-stage parasites over blood-stage parasites by at least a factor of 10 and therefore would not have been identified in blood-stage screens.
A previous report found that ulcer medications (proton pump inhibitors), including omeprazole, inhibited blood-stage P. falciparum D6, W2, and TM91C235 with IC50 values between 7 and 70 μM (38), but in our blood-stage assay omeprazole or esomeprazole did not inhibit P. falciparum Dd2 or 3D7 isolates even at 50 μM (Fig. 3A). Unfortunately this weak activity eliminates the option of raising resistant blood-stage parasites as a means for proton pump inhibitor target identification.
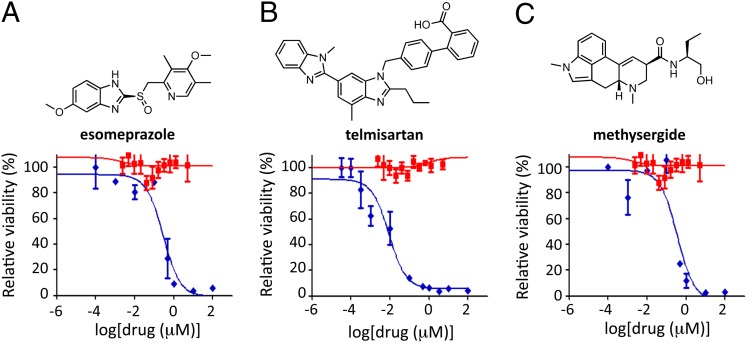
Fig. 3.
Characterization of liver-stage malaria inhibitors within the FDA-approved drug library. Representative dose–response curves for P. berghei ANKA liver stage (blue curve) and P. falciparum 3D7 blood stage (red curve) are shown for esomeprazole (A, Lower), telmisartan (B, Lower), and methysergide (C, Lower). (A–C, Upper) Structures are shown.
Methysergide and telmisartan do not inhibit blood-stage parasites at 5 μM (Fig. 3 B and C). These drugs were of special interest as RNA interference (RNAi) knockdown of the human serotonin and angiotensin receptors indicated that these genes are not critical to Plasmodium infection (Fig. S2). Taken at face value, these results argue that telmisartan and methysergide are unlikely to be inhibiting parasites through their known human targets.
Metabolic Activation.
The methysergide and esomeprazole results given above did not resolve the difference in potency of these molecules in liver and blood-stage assays, and a possible explanation for the discrepancy could be differing degrees of host cell metabolic activity in the two assays. Esomeprazole, its derivatives, and methysergide are prodrugs (39, 40). Methysergide requires metabolic activation by cytochrome P450 enzymes in the liver (41) and esomeprazole requires an acidic environment. Liver cells, unlike human red blood cells, are metabolically active. To test this possibility we evaluated compound efficacy for all of the screening hits at two different cell densities (5,000 cells per well and 20,000 cells per well) with the expectation that compounds requiring metabolic activation will be more potent at higher liver cell concentrations. Most of the 37 screening hits are not influenced by liver cell density; however, esomeprazole, omeprazole, tenatoprazole, and methysergide are more potent at a higher concentration of liver cells (Fig. S3). This experiment indicates that liver-stage malaria assays can reveal metabolically activated drugs that would be missed by blood-stage parasite screening.
In humans, proton pump inhibitors are activated by low pH in gastric parietal cells (33). This protonation leads to the formation of the active sulfonamide that irreversibly binds proton pumps. The inhibitors also interact with cytochrome P450s in the liver and it is known that omeprazole is oxidized to the sulphone and metabolized to 5-hydroxy and 5-O-desmethyl omeprazole (39, 42). Thus, existing studies on omeprazole suggest two potential routes of parasite inhibition: a pH-dependent mechanism or liver cell metabolism. Both blood and liver-stage parasites contain acidic vacuoles and it is possible that currently unknown differences between these vacuoles in the two parasite stages allow for omeprazole’s observed stage specificity. We also have not eliminated the possibility that the proton pump inhibitors influence parasite development through a host ATPase mechanism. However, if this were the case, we would not have expected the dependence of activity on liver cell density (Fig. S3). The elucidation of Plasmodium inhibition by proton pump inhibitors is an active area of our research.
In Vivo Efficacy of Salinomycin.
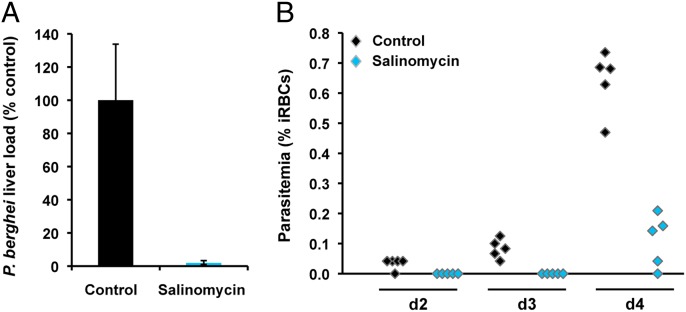
Within the FDA-approved drug set salinomycin was one of the most potent screening hits that had not been previously identified as a potential liver-stage antimalarial. We sought to evaluate the potential of the compound to inhibit malaria infection in vivo, using an established rodent model of malaria. Mice received a single treatment with 30 mg per kg of body weight administered orally immediately after i.v. injection of 10,000 luciferase-expressing P. berghei sporozoites. Liver parasite burdens were determined by bioluminescence of P. berghei; in parallel, blood parasitemias were monitored daily starting day 2 postinfection. Salinomycin was able to reduce parasite liver load by 98% relative to the vehicle-treated controls (Fig. 4A). Moreover, our results show that administration of the drug led to a 2-d delay in the appearance of blood parasitemia. (Fig. 4B).
Fig. 4.
In vivo efficacy of salinomycin in mice. (A) P. berghei parasite load in mouse livers 44 h after infection with luciferase-expressing sporozoites, determined by luminescence in vehicle-treated (black) and salinomycin-treated (blue) animals. (B) Plots of blood parasitemia at days 2–4 after infection for vehicle-treated (black) and salinomycin-treated (blue) animals. Mice received a single treatment by oral administration of 30 mg/kgb.w. salinomycin or an equivalent amount of vehicle (controls).
In summary, we have developed a phenotypic liver-stage malaria screen in a high-throughput format and used it to evaluate a library of well-characterized bioactive molecules with putative targets. The screen identifies several molecules that could become candidates for further development, including one with high in vivo efficacy. The discovery that at least two over-the-counter drugs have activity against liver-stage malaria could become significant, as combating global infectious disease requires inexpensive drugs with minimal or no side effects. The screen’s results also demonstrate that screening of liver-stage inhibitors can expose parasite vulnerabilities to drugs that require metabolic activation, prodrugs, that are not apparent with blood-stage parasite screens. Finally, the diverse drug scaffolds with their disparate associated targets identify malaria’s liver-stage sporozoites as a target-rich area for drug discovery, and identifying and exploiting these targets could lead to a new generation of antimalarial agents for prophylaxis, transmission blocking, and possibly eradication.
Materials and Methods
High-Throughput Screen.
HepG2 cells (ATCC) were maintained in DMEM (Invitrogen), 10% (vol/vol) FBS (Sigma), and 1% (vol/vol) antibiotic–antimycotic (Invitrogen) in a standard tissue culture incubator (37 °C, 5% CO2). Plasmodium-infected Anopheles stephensi mosquitoes were obtained from the New York University Langone Medical Center Insectary (New York). For assays, ∼17,500 HepG2 cells per well were added to a 384-well microtiter plate in duplicate, using a multichannel pipette (Rainin). After 18–24 h at 37 °C the media were exchanged and compounds (100 nL) and DMSO controls were added to the plates with a pin array on a robot arm. The final concentration of DMSO was 0.3% (vol/vol) and compounds were at 6–10 μg/mL. Atovaquone and halofuginone (1 μM) were used as positive controls for parasite inhibition (43). After 1 h, parasites obtained from freshly dissected mosquitoes were added to the plates (4,000 parasites per well), the plates were spun for 10 min at 1,000 rpm, and then the plates were incubated at 37 °C. The final assay volume was 30 μL. After a 48-h incubation at 37 °C, Bright-Glo (Promega) was added to the parasite plate to measure relative luminescence and CellTiter-Glo (Promega) was added to the duplicate plate to measure relative liver cell viability. The relative signal intensity of each plate was evaluated with an EnVision (PerkinElmer) system. There was a crosstalk correction of 0.27. This protocol routinely yielded Z-factors between 0.5 and 0.7.
For the purpose of screening the known bioactive libraries the protocol above was slightly modified. At 24 h postcompound transfer the plate media were exchanged and compounds and DMSO were again transferred to the plates. This step reduced the probability of culture contamination from addition of nonsterile parasites and provided an additional opportunity for the compounds to function before liver cell metabolism. These plates were incubated at 37 °C another 20 h before assessing parasite load and HepG2 viability in the presence of the drugs. The parasite plate and the HepG2 viability plate were measured in duplicate as described above. The percentage of HepG2 viability was determined relative to the DMSO control and the activity score for each compound was determined by standardizing the parasite signal in the presence of the compound to the positive and negative controls. Screening data have been deposited in the Chembl-NTD database (http://www.ebi.ac.uk/chemblntd).
Confirmation of Screening Positives and IC50 Determinations.
Compounds determined as hits (95% inhibition of parasite signal in both replicates) were purchased from Sigma, Toronto Research Chemicals, or Santa Cruz Biotechnology for secondary assays. Activity was confirmed first by retesting the hit from the source plate and then with the purchased compounds. Dose–response curves were generated by evaluating parasite growth in the presence of varying concentrations of the drugs (0–10 μM in DMSO) in triplicate. Data analysis was carried out using GraphPad Prism and curves were fit with a standard inhibition dose–response curve to generate an IC50 value. All statistical results are the mean IC50 value averaged from two to three independent experiments.
Blood-Stage Malaria Assays.
P. falciparum 3D7 and Dd2 blood-stage assays were performed following a previously published procedure (11). Data analysis was carried out using GraphPad Prism and curves were fit with a standard inhibition dose–response curve to generate an IC50 value.
Time-Course Studies.
Liver-stage malaria assays were performed as described above but with varying times of compound addition. Liver-stage hits (10 μM final) were added to HepG2 cells 2 h before or 3, 8, and 24 h after sporozoite addition. Parasite load was determined as described above.
Supplementary Material
Acknowledgments
We thank the Institute of Chemistry and Cell Biology Screening Facility where the screen was performed; Roger Wiegand (Harvard School of Public Health) for blood-stage assays; and Joshua Blodgett (Harvard Medical School), Nathaniel Fernhoff (Stanford University), and Ralph Mazitschek (Massachusetts General Hospital) for useful discussions. We thank Andreia Magalhães and Inês Albuquerque for technical assistance and useful discussions. This work was supported by the Harvard Medical School–Portugal Program in Translational Research and Information for laboratory support (M.M.M. and J.C.) and by National Institutes of Health Grant F32GM093510 for fellowship support (to E.R.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The screening data reported in this paper have been deposited in the Chembl-NTD database (http://www.ebi.ac.uk/chemblntd).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118370109/-/DCSupplemental.
References
- 1.Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- 2.WHO . World Malaria Report 2010. Geneva: World Health Organization; 2010. [Google Scholar]
- 3.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 5.Prudêncio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: The Plasmodium liver stage. Nat Rev Microbiol. 2006;4:849–856. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- 6.Bozdech Z, et al. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 8.Silvestrini F, et al. Genome-wide identification of genes upregulated at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem Parasitol. 2005;143:100–110. doi: 10.1016/j.molbiopara.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Westenberger SJ, et al. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis. 2010;4:e653. doi: 10.1371/journal.pntd.0000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams CT, Azad AF. Transcriptional analysis of the pre-erythrocytic stages of the rodent malaria parasite, Plasmodium yoelii. PLoS ONE. 2010;5:e10267. doi: 10.1371/journal.pone.0010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baniecki ML, Wirth DF, Clardy J. High-throughput Plasmodium falciparum growth assay for malaria drug discovery. Antimicrob Agents Chemother. 2007;51:716–723. doi: 10.1128/AAC.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamo FJ, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 13.Guiguemde WA, et al. Chemical genetics of Plasmodium falciparum. Nature. 2010;465:311–315. doi: 10.1038/nature09099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plouffe D, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci USA. 2008;105:9059–9064. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baird JK, Hoffman SL. Primaquine therapy for malaria. Clin Infect Dis. 2004;39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- 16.Bassat Q, Alonso PL. Defying malaria: Fathoming severe Plasmodium vivax disease. Nat Med. 2011;17:48–49. doi: 10.1038/nm0111-48. [DOI] [PubMed] [Google Scholar]
- 17.Price RN, et al. Vivax malaria: Neglected and not benign. Am J Trop Med Hyg. 2007;77(6 Suppl):79–87. [PMC free article] [PubMed] [Google Scholar]
- 18.Carraz M, et al. A plant-derived morphinan as a novel lead compound active against malaria liver stages. PLoS Med. 2006;3:e513. doi: 10.1371/journal.pmed.0030513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgoine KL, Bancone G, Nosten F. The reality of using primaquine. Malar J. 2010;9:376. doi: 10.1186/1475-2875-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371:64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- 21.Baird JK. Effectiveness of antimalarial drugs. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- 22.Derbyshire ER, Mota MM, Clardy J. The next opportunity in anti-malaria drug discovery: The liver stage. PLoS Pathog. 2011;7:e1002178. doi: 10.1371/journal.ppat.1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazier D, Rénia L, Snounou G. A pre-emptive strike against malaria’s stealthy hepatic forms. Nat Rev Drug Discov. 2009;8:854–864. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- 24.Prudêncio M, Mota MM, Mendes AM. A toolbox to study liver stage malaria. Trends Parasitol. 2011;27:565–574. doi: 10.1016/j.pt.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Golenda CF, Li J, Rosenberg R. Continuous in vitro propagation of the malaria parasite Plasmodium vivax. Proc Natl Acad Sci USA. 1997;94:6786–6791. doi: 10.1073/pnas.94.13.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 27.Kappe SH, Duffy PE. Malaria liver stage culture: in vitro veritas? Am J Trop Med Hyg. 2006;74:706–707. [PubMed] [Google Scholar]
- 28.Ploemen IH, et al. Visualisation and quantitative analysis of the rodent malaria liver stage by real time imaging. PLoS ONE. 2009;4:e7881. doi: 10.1371/journal.pone.0007881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J, et al. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ., Jr A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 32.Battershill AJ, Scott LJ. Telmisartan: A review of its use in the management of hypertension. Drugs. 2006;66:51–83. doi: 10.2165/00003495-200666010-00004. [DOI] [PubMed] [Google Scholar]
- 33.Roche VF. The chemically elegant proton pump inhibitors. Am J Pharm Educ. 2006;70:101. doi: 10.5688/aj7005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullah MA, et al. Relative bioavailability and pharmacokinetic properties of two different enteric formulations of esomeprazole in healthy Bangladeshi male volunteers: An open-label, single-dose, randomized-sequence, two-way crossover study. Clin Ther. 2010;32:1419–1426. doi: 10.1016/j.clinthera.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Andersson T, Weidolf L. Stereoselective disposition of proton pump inhibitors. Clin Drug Investig. 2008;28:263–279. doi: 10.2165/00044011-200828050-00001. [DOI] [PubMed] [Google Scholar]
- 36.Rottmann M, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanway RR, et al. Organelle segregation into Plasmodium liver stage merozoites. Cell Microbiol. 2011;13:1768–1782. doi: 10.1111/j.1462-5822.2011.01657.x. [DOI] [PubMed] [Google Scholar]
- 38.Riel MA, Kyle DE, Bhattacharjee AK, Milhous WK. Efficacy of proton pump inhibitor drugs against Plasmodium falciparum in vitro and their probable pharmacophores. Antimicrob Agents Chemother. 2002;46:2627–2632. doi: 10.1128/AAC.46.8.2627-2632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson T, et al. Identification of human liver cytochrome P450 isoforms mediating omeprazole metabolism. Br J Clin Pharmacol. 1993;36:521–530. doi: 10.1111/j.1365-2125.1993.tb00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koehler PJ, Tfelt-Hansen PC. History of methysergide in migraine. Cephalalgia. 2008;28:1126–1135. doi: 10.1111/j.1468-2982.2008.01648.x. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 42.Andersson T, Hassan-Alin M, Hasselgren G, Röhss K, Weidolf L. Pharmacokinetic studies with esomeprazole, the (S)-isomer of omeprazole. Clin Pharmacokinet. 2001;40:411–426. doi: 10.2165/00003088-200140060-00003. [DOI] [PubMed] [Google Scholar]
- 43.Derbyshire ER, Mazitschek R, Clardy J. Characterization of Plasmodium liver stage inhibition by halofuginone. ChemMedChem. 2012;7:844–849. doi: 10.1002/cmdc.201200045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.