Abstract
Transcripts of plant organelle genes are modified by cytidine-to-uridine (C-to-U) RNA editing, often changing the encoded amino acid predicted from the DNA sequence. Members of the PLS subclass of the pentatricopeptide repeat (PPR) motif-containing family are site-specific recognition factors for either chloroplast or mitochondrial C targets of editing. However, other than PPR proteins and the cis-elements on the organelle transcripts, no other components of the editing machinery in either organelle have previously been identified. The Arabidopsis chloroplast PPR protein Required for AccD RNA Editing 1 (RARE1) specifies editing of a C in the accD transcript. RARE1 was detected in a complex of >200 kDa. We immunoprecipitated epitope-tagged RARE1, and tandem MS/MS analysis identified a protein of unknown function lacking PPR motifs; we named it RNA-editing factor interacting protein 1 (RIP1). Yeast two-hybrid analysis confirmed RIP1 interaction with RARE1, and RIP1-GFP fusions were found in both chloroplasts and mitochondria. Editing assays for all 34 known Arabidopsis chloroplast targets in a rip1 mutant revealed altered efficiency of 14 editing events. In mitochondria, 266 editing events were found to have reduced efficiency, with major loss of editing at 108 C targets. Virus-induced gene silencing of RIP1 confirmed the altered editing efficiency. Transient introduction of a WT RIP1 allele into rip1 improved the defective RNA editing. The presence of RIP1 in a protein complex along with chloroplast editing factor RARE1 indicates that RIP1 is an important component of the RNA editing apparatus that acts on many chloroplast and mitochondrial C targets.
Keywords: nucleoid, RNA editosome, dual targeting
Posttranscriptional C-to-U RNA editing occurs in plastid and plant mitochondrial transcripts. In a typical vascular plant, ∼30 C targets in chloroplasts and over 500 C targets in mitochondria are targeted for editing (1, 2). The majority of the editing events results in encoding of a different amino acid than the one predicted from the genomic sequence. The editing-encoded amino acid is usually more conserved relative to residues present in homologous proteins in other organisms than the genomically encoded amino acid. Because there is presently no known case in which useful genetic variation results from partial editing of a transcript population, the current concept is that editing is a correction mechanism for thymidine-to-cytidine (T-to-C) mutations that have arisen in plant organelle genomes (1, 3, 4).
Little is known about the molecular apparatus that is responsible for recognizing the correct C target for editing and converting it to U, although plant mitochondrial RNA editing was discovered over 20 y ago (5–7). cis-Elements for recognition of editing sites have been identified proximal and 5′ to the nucleotide to be modified (8–10). As few as 22 nt in sequence surrounding the C target is sufficient to specify RNA editing (9). In 2005, a pentatricopeptide repeat (PPR) motif-containing protein termed CRR4 was discovered to be required for editing of the chloroplast ndhD start codon (11), and it binds to cis-elements on ndhD transcripts in vitro (12). Since that time, members of the PPR protein family have been identified as site-specific recognition factors for a number of C targets in either chloroplasts or mitochondria. PPR proteins consist of a tandem array of degenerate 35-aa repeats and can be divided into two major subfamilies based on the nature of their PPR motifs: the P and PLS subfamilies (13). The P subfamily contains a 35-aa motif, whereas the PLS subfamily exhibits longer or shorter variant PPR motifs within the tandem arrays. The PLS subfamily, which is specific to the plant kingdom, can be further separated into smaller subclasses based on two C-terminal motifs, the E and DYW motifs (14). All of the well-characterized organelle editing factors that are required for editing at specific sites are members of the PLS subfamily of PPR proteins (11, 15–29).
Other than the cis-elements and site-specific PPR proteins, the components of the editing machine are unknown. The enzymatic activity that converts C to U remains unidentified, although the DYW domain found in about one-half of the Arabidopsis PPR editing factors does contain a sequence similar to the conserved cytidine/deoxycytidylate deaminase motif (30). To identify additional components of the chloroplast editing apparatus in Arabidopsis, we immunoprecipitated an epitope-tagged PPR-DYW protein named RARE1, which is responsible for recognition of a C target in the chloroplast accD transcript (21). MS/MS analysis of the coimmunoprecipitated proteins resulted in the identification of a protein of unknown function lacking PPR motifs. Yeast two-hybrid analysis confirmed the interaction of RARE1 and the protein, which is named RNA-editing factor interacting protein 1 (RIP1). Although RIP1 was identified by its interaction with a chloroplast PPR protein, GFP localization experiments revealed its presence in both plastids and mitochondria. Virus-induced gene silencing of RIP1 resulted in defective editing of both chloroplast and mitochondrial C targets. A homozygous rip1 mutant line exhibited altered editing of 14 C targets in chloroplast transcripts and impaired editing of 266 of 368 mitochondrial editing sites that were assayed, with major loss of editing of 108 mitochondrial C targets. Transient introduction of a WT RIP1 allele into the mutant resulted in improvement in the defective RNA editing. Our findings indicate that RIP1, which belongs to a 10-member gene family, is required for efficient editing at most Arabidopsis mitochondrial editing sites and plays an important role in chloroplast editing as well. Identification of RIP1 is a significant step that will aid additional efforts to understand the mechanism of plant organelle RNA editing.
Results
Identification of RIP1 as an RARE1-Interacting Protein.
Our previous work reported the identification of RARE1, a plastid editing factor that controls the editing of accD-794 (21). We determined that RARE1 is present in a protein complex by performing size exclusion column chromatography on chloroplast stroma (SI Appendix, Fig. S1). To identify members of this complex, we produced transgenic plants that express RARE1 protein carrying a 3× FLAG tag (RARE1-3×F) (31) (SI Appendix, Fig. S2). Leaf protein extract from transgenic plants was incubated with α-FLAG agarose to isolate the RARE1 complex (SI Appendix, Fig. S3). The MS data indicated that the protein encoded by At3g15000 was the top candidate RARE1-interacting protein present in the immunoprepitate, because it had the largest number of matches of MS/MS spectra other than RARE1 (SI Appendix, Table S1). The gene encodes a member of the differentiation and greening (DAG) family; mutants in members of this gene family exhibit chloroplast biogenesis defects (32, 33). Yeast two-hybrid analysis confirmed the interaction between RARE1 and the protein encoded by At3g15000, which was, therefore, named RIP1 (SI Appendix, Fig. S4). Serial deletions of both RARE1 and RIP1 established the portions responsible for the interaction on the N termini of the proteins (SI Appendix, Fig. S5).
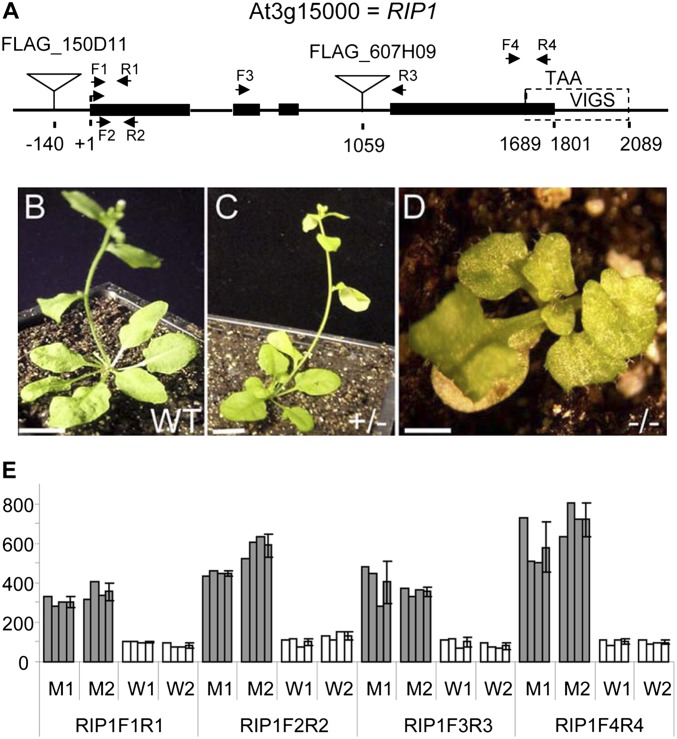
T-DNA Insertional rip1 Mutant Exhibits a Dwarf Phenotype and Altered Chloroplast RNA Editing.
Two mutant lines with insertions in the RIP1 locus (Fig. 1) were obtained from the INRA FLAGdb T-DNA collection (34). Homozygous mutants could not be recovered from the FLAG_607H09 line; possibly, the T-DNA insertion in FLAG_607H09 might be lethal because of the complete loss of expression. Homozygous FLAG_150D11 mutants, which have a T-DNA inserted 140 bp upstream of the RIP1 coding region, exhibit a dwarf phenotype (Fig. 1D). We measured the level of RIP1 transcript in the homozygous FLAG_150D11 mutant line and homozygous WT siblings by quantitative RT-PCR. The expression of the RIP1 ORF was found to be increased four- to sixfold in the T-DNA mutant compared with the WT (Fig. 1E). Nevertheless, the proximity of the T-DNA insertion to the ORF may result in impaired production of RIP1 protein; abnormal phenotypes have previously been reported in T-DNA insertional mutants that exhibited increased rather than reduced target gene transcript abundance (35).
Fig. 1.
A rip1 mutant exhibits dwarf phenotype and increases in RIP1 transcript. (A) Map of At3g15000 (RIP1) with exons shown as black rectangles, T-DNA insertions shown as triangles, the region used for VIGS indicated, and the location of primers used for quantitative RT-PCR shown as facing arrows. (B–D) WT, heterozygous, and homozygous progeny of a heterozygous plant carrying the FLAG_150D11 insertion. Plants are 32 d old. (Scale bars: B and C, 10 mm; D, 1 mm.) (E) The expression of RIP1 is increased four- to sixfold in the T-DNA mutant compared with WT. Quantitative RT-PCR measured the level of RIP1 transcript in two homozygous mutants (M1 and M2) and two homozygous WT siblings (W1 and W2). Quantitative RT-PCR assays were replicated three times for each plant. The expression level was arbitrarily set at 100 for W1. SDs are indicated (n = 3).
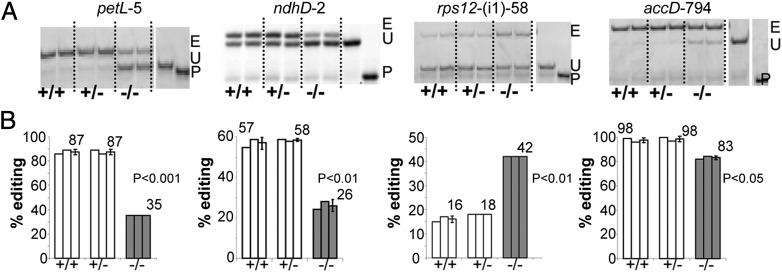
Because RIP1 coimmunoprecipitates and interacts in vivo with RARE1, a chloroplast editing factor, we surveyed the editing extent of all known Arabidopsis chloroplast editing sites in segregating progeny for the FLAG_150D11 T-DNA insertion. A poisoned primer extension (PPE) assay is shown in Fig. 2 for accD-794 and the three sites showing the most pronounced editing extent variation in the mutant relative to WT. PetL-5 and ndhD-2 exhibit a significant reduction of editing extent in the mutant (60% and 55%, respectively), whereas rps12-(i1)58, a site in the first intron of rps12, shows a significant increase of editing extent in the mutant (Fig. 2). PPE data for accD-794, the site under the control of RARE1, indicate that editing in the homozygous mutant is reduced relative to WT as observed for petL-5 and ndhD-2 but to a lesser extent (83% in mutant compared with 98% in WT or a 15% reduction). The mutation is clearly recessive, because the editing extent of the heterozygous plant for these sites is similar to the homozygous wild plants (Fig. 2).
Fig. 2.
Mutation in RIP1 affects the editing extent of plastid sites. (A) Acrylamide gels separate the PPE products obtained from sibling plants: two homozygous WT (+/+), two heterozygous (−/+) mutants, and two homozygous mutants (−/−). E, edited; P, primer; U, unedited. The name of the site assayed is given above each gel. (B) The quantification of editing extent derived from the measure of the band’s intensity is represented by a bar below each lane of the acrylamide gels. The average is given for each genotypic class with SD. The sites petL-5, ndhD-2, and accD-794 show a significant decrease of the editing extent in the mutant, representative of the majority of the plastid sites showing an effect of the RIP1 mutation. The site rps12-(i1)-58 shows a significant increase of editing extent in the mutant compared with the WT and heterozygous plants as observed only in two other plastid editing sites.
Of the 34 known chloroplast C targets of editing present in Arabidopsis, 14 C targets exhibited significant changes in RNA editing extent between the homozygous WT and the homozygous mutant plants (SI Appendix, Table S2); 11 of 14 sites exhibit a decrease in editing extent in the mutant, whereas an increase of editing extent in the mutant is observed for only 3 sites (SI Appendix, Table S2). The editing extent of the heterozygote was not significantly different from the homozygous WT at any of the chloroplast sites.
RIP1 Is Dual-Targeted to both Chloroplasts and Mitochondria.
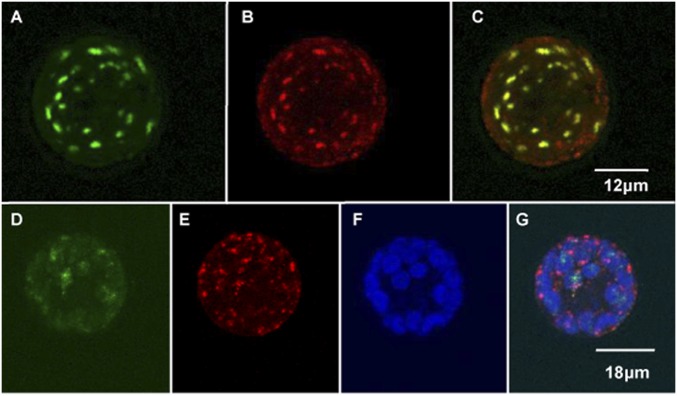
RIP1 has been previously reported to be located in mitochondria according to characterizations of the Arabidopsis mitochondrial proteome (36, 37). In addition, the dwarf phenotype of the FLAG_150D11 T-DNA insertional mutant could be indicative of mitochondrial dysfunction. Therefore, we determined the location of RIP1 by transiently expressing a construct encoding the full-length RIP1 attached to GFP under the control of a 35S promoter into Arabidopsis protoplasts (38). Our observations indicate that RIP1 is dually targeted to chloroplasts and mitochondria (Fig. 3). Most of the Arabidopsis protoplasts showed RIP1 to be localized in mitochondria (Fig. 3C). Occasionally, we observed RIP1 both in mitochondria and chloroplasts (Fig. 3G). This observation is reminiscent of a recent report on PPR2263, a maize PPR-DYW that is dually targeted to mitochondria and chloroplasts with a preference for mitochondria (39).
Fig. 3.
RIP1 is dual-targeted to mitochondria and chloroplasts. Protoplasts prepared from leaves of Arabidopsis accession Col-0 were transfected with a construct encoding an RIP1-GFP fusion protein under the control of a 35S promoter. Protoplasts were examined for fluorescence 16 h after incubation with the construct. (A and D) GFP signal is green. (B and E) Mitochondria (red) were labeled with Mitotracker Orange. (C) Merge of GFP and mitochondrial signal is yellow. (F) Chlorophyll autofluorescence is shown in blue. (G) Merge of D–F gives turquoise signals where GFP and chlorophyll overlap and yellow images where GFP and Mitotracker overlap.
To confirm the dual localization of RIP1 to both organelles, we repeated the previous experiment by transfecting Nicotiana benthamiana protoplasts. In contrast to Arabidopsis protoplasts, all of the transfected N. benthamiana protoplasts showed a dual localization of RIP1 to both mitochondria and chloroplasts (SI Appendix, Fig. S6). DAPI staining of the N. benthamiana protoplasts showed that some of the small punctate structures targeted by RIP1-GFP colocalize with nucleoids (SI Appendix, Fig. S6H).
rip1 Mutant Exhibits Altered Mitochondrial Editing.
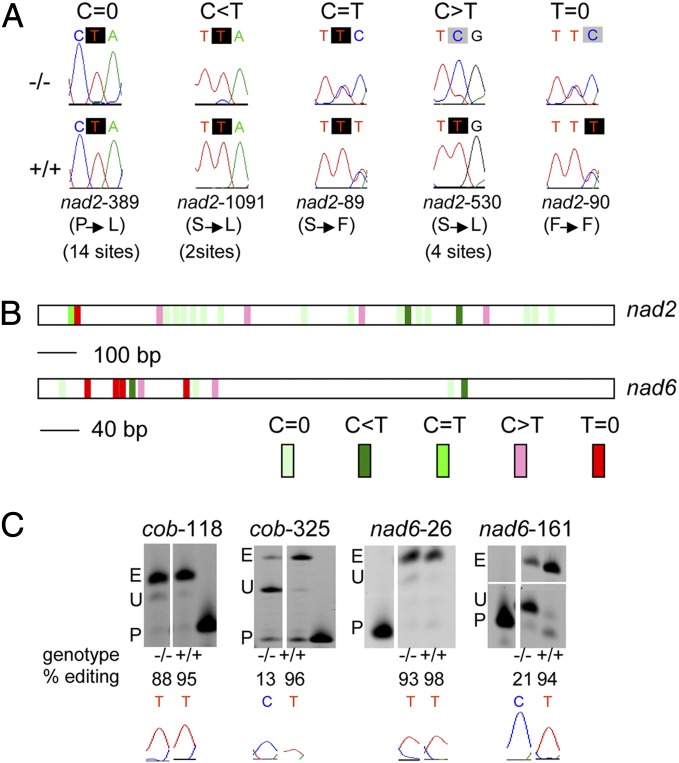
We conducted a bulk sequencing screen of the 33 mitochondrial protein-coding genes known to harbor editing sites by comparing the sequencing electrophoretograms of the RT-PCR products obtained from the homozygous T-DNA mutant with the homozygous WT line. A typical result is shown in Fig. 4A, where the editing extent in the nad2 transcript is not uniformly affected by the RIP1 mutation along the transcript. The majority of the nad2 sites, 14 of 22 sites, do not show any reduction in editing extent in the mutant compared with the WT (Table 1). However, editing of nad2-90 is not detectable in the mutant, because only a C peak is observed at that position (Fig. 4A). Between these two extremes are detected sites in which editing is reduced to less than one-half of WT, about one-half of WT, or more than one-half of WT as observed in nad2-1091, nad2-89, and nad2-530, respectively (Fig. 4A).
Fig. 4.
Editing extent is not uniformly affected along mitochondrial transcripts in rip1 mutants. (A) Portions of electrophoretograms from RT-PCR bulk sequencing of nad2 are shown for the homozygous T-DNA mutant (−/−) and homozygous WT (+/+). Below the electrophoretograms are given the position of the editing site in the nad2 transcript with the amino acid change on editing between parenthesis and the number of sites in nad2 sharing the same molecular phenotype. The editing phenotype of the mutant was classified in one of five categories above the electrophoretograms from C = 0 (no effect of the mutation on the editing extent) to T = 0 (total loss of editing in the mutant). The C target of editing is highlighted by a black shade for T and a gray shade for C, and it is shown according to its position in the codon. (B) Distribution of the effect of the RIP1 mutation on the editing extent of mitochondrial sites on nad2 and nad6 transcripts. Each site is represented by a block with background color that indicates the strength of the rip1 mutation’s effect on the editing extent as detected by bulk sequencing. (C) PPE assays confirm the reduction of editing extent of mitochondrial sites in cob and nad6 transcripts previously detected by bulk sequencing (Region of nad6-161 gel lacking signal removed for space considerations). The PPE products run on acrylamide gels are shown on top, with the name and position of the site being assayed above the gel. E, edited; P, primer; U, unedited. Below the gels are shown the electrophoretograms of the editing site.
Table 1.
Effect of FLAG_150D11 insertion on RNA editing of mitochondrial C targets
| Effect |
|||||
| Gene | C = 0 | C < T | C = T | C > T | T = 0 |
| nad1 | 14 | 4 | 1 | ||
| nad2 | 14 | 2 | 1 | 4 | 1 |
| nad3 | 3 | 4 | |||
| nad4 | 21 | 4 | 3 | 2 | |
| nad4L | 2 | 2 | 2 | 4 | |
| nad5 | 16 | 4 | 4 | 1 | |
| nad6 | 3 | 2 | 2 | 4 | |
| nad7 | 14 | 2 | 2 | 3 | 1 |
| nad9 | 4 | 2 | 1 | ||
| Complex I | 82 | 24 | 10 | 22 | 15 |
| cob—complex III | 3 | 3 | 2 | ||
| cox2 | 5 | 2 | 1 | 1 | 3 |
| cox3 | 5 | 1 | 1 | ||
| Complex IV | 5 | 7 | 2 | 2 | 3 |
| atp1 | 1 | 1 | 1 | ||
| atp4 (orf25) | 1 | 3 | 3 | 1 | |
| atp6-1 | 1 | ||||
| atp9 | 2 | 2 | |||
| Complex V | 5 | 6 | 1 | 3 | 1 |
| ccmB (ccb206) | 1 | 4 | 27 | ||
| ccmFn-2 (ccb203) | 1 | 5 | 4 | ||
| ccmC (ccb256) | 1 | 1 | 17 | 7 | |
| ccmFn-1 (ccb382) | 1 | 5 | 7 | ||
| ccmFc (ccb452) | 4 | 2 | 4 | 1 | |
| cytochrome c biogenesis | 5 | 2 | 8 | 32 | 45 |
| rpl2 | 1 | ||||
| rpl5 | 2 | 1 | 3 | 4 | |
| rpl16 | 2 | 2 | 1 | 1 | |
| rps3 | 2 | 2 | 1 | 2 | |
| rps4 | 3 | 1 | 1 | 5 | |
| rps7 | 1 | ||||
| rps12 | 5 | 3 | |||
| rps14 | 1 | ||||
| Ribosomal protein | 2 | 10 | 5 | 11 | 16 |
| matR | 2 | 2 | 5 | ||
| mttb (OrfX) | 6 | 21 | |||
| Total | 102 | 52 | 28 | 78 | 108 |
The five categories of RIP1 mutation effect on mitochondrial editing, from no effect (C = 0) to a total loss of editing (T = 0) extent, are presented in the text and Fig. 4.
Table 1 summarizes the results of the bulk sequencing screen by presenting the number of sites for each mitochondrial gene that falls into one of five categories described for nad2 transcript from no effect of the RIP1 mutation to an apparent absence of editing. Of the 33 mitochondrial genes surveyed, only atp6-1, which contains one reported editing site at position 475, does not show any dependence on a functional RIP1 for efficient editing. Overall, mutation in RIP1 affects the editing extent of a very high number of mitochondrial sites; 108 of 368 sites surveyed show a major loss of editing in the mutant (Table 1). A very similar number of sites (102 sites) do not show any variation in editing extent in the mutant. A complete list of all of the affected mitochondrial C targets of editing among the 368 sites assayed is shown in Dataset S1.
Plant mitochondrial sites in the rip1 mutant analyzed can be divided into two categories: totally RIP1-dependent (108 of 368 sites or 29%) and totally RIP1-independent (102 of 368 sites or 28%). Although these categories are approximately equal in size in the entire population of genes analyzed, RIP1 seems to play a larger role in editing of transcripts for proteins of certain mitochondrial complexes than others. For example, transcripts of complex 1 genes exhibit 10% (15/153) C targets affected by the RIP1 mutation and 45% (82/153) unaffected. In contrast, the cytochrome c biogenesis complex exhibits 49% C targets (45/92) affected and only 5% (5/92) sites with editing extent that is unaffected (Table 1). The effect of RIP1 mutation on mitochondrial extent does not seem to be related to the location of the C target on the transcript, because there is no apparent pattern in the distribution of the RIP1-dependent and -independent sites along the transcript (Fig. 4B).
Editing events can be divided into two classes: nonsilent (when editing changes the encoded amino acid) or silent (when the amino acid is unchanged). Nonsilent sites are predominant in the population of sites surveyed (335 nonsilent sites or 91%) (Dataset S1). There are somewhat fewer nonsilent sites in the group of sites that are strongly affected in the rip1 mutant than there are in the entire population of surveyed sites [83% (90 nonsilent sites to 108 sites) vs. 91% respectively] (Dataset S1).
We also examined a small selection of editing sites by the PPE assay, which is more precise and sensitive than the RT-PCR/bulk sequencing method that we used to survey the 368 sites in the rip1 mutant (40). We chose some mitochondrial editing sites that exhibited either no or complete dependence on functional RIP1 (Fig. 4A). Although no editing of the C targets in cob-325 and nad6-161 was detected by the less-sensitive bulk sequencing method, we found that both exhibit a residual editing extent detectable by PPE (13% and 21%, respectively) (Fig. 4C). The negative effect of the rip1 mutation on cob-325 and nad6-161 is greater than its effect on any chloroplast C targets (SI Appendix, Table S2). When the editing extent of these two sites was assayed by PPE in homozygotes, heterozygotes, and WT, we found no difference between heterozygotes and WT, indicating the mutation is completely recessive with respect to editing efficiency at these two C editing targets as well as other mitochondrial sites (SI Appendix, Fig. S7).
rip1 Mutation Affects Transcript Abundance.
We examined the level of a selection of mitochondrial transcripts in the mutant to investigate the possibility of a link between the steady-state level of mitochondrial transcripts and the editing extent of their targeted C sites. Among the 10 mitochondrial transcripts assayed by quantitative RT-PCR, 5 transcripts showed a significant increase (approximately four- to sixfold) in their abundance compared with the WT, 3 transcripts showed a moderate increase (1.3- to 1.5-fold) in the mutant, and 2 transcripts were in similar amounts in both the mutant and the WT (SI Appendix, Fig. S8). Although ccmB, the transcript harboring the highest number of sites with editing extents that are severely affected by rip1 mutation (Table 1), shows the highest increase of transcript abundance in the mutant (SI Appendix, Fig. S8A), there is no obvious correlation between the steady-state level of transcript and the incidence of rip1 mutation on the editing extent. For example, nad9, which exhibits a similar increase of its transcript abundance in the mutant as ccmB (SI Appendix, Fig. S8A), does not harbor any site with editing extent that is greatly impaired in the mutant (Table 1). Conversely, ccmFn-2, with 4 of 10 targeted sites showing an apparent total loss of editing in rip1 (Table 1), experiences only a slight increase of its transcript abundance in the mutant relative to the WT (SI Appendix, Fig. S8C). Eight of the sites on nad7 transcript show a reduced editing extent in rip1, whereas nad7 abundance is similar to the WT (Table 1 and SI Appendix, Fig. S8D).
A model can be proposed in which a dosage effect is transcript-specific, and therefore, a slight increase of ccmB transcript abundance is sufficient to have an effect on the editing extent of some of its sites. In this model, some of the recognition transfactors directing the specific editing site of targeted C sites are in limiting amounts, and therefore, even a slight increase of the transcript abundance might deplete these recognition factors, resulting in an apparent reduction in editing efficiency. However, this possibility is refuted by a close examination of the nad4, ccmFn-2, and cox3 transcript abundances, which show a very similar and slight increase in the rip1 mutant (SI Appendix, Fig. S8C). These three transcripts each possess a site, cox3-422, nad4-124, and ccb203 (ccmFn-2)-344, recognized by the same recognition factor, MEF11 (27). In the rip1mutant, the editing extent of ccmFn-2-344 is severely reduced, whereas the editing extent of cox3-422 is only slightly reduced; nad4-124 does not show any detectable difference in editing extent between rip1 and the WT (Dataset S1). In conclusion, our mutant analysis data clearly indicate independence of the editing extent of the sites carried by a mitochondrial transcript and its abundance.
Unlike the mitochondrial transcripts, the three plastid transcripts assayed by quantitative RT-PCR all show a reduction of steady-state level in rip1 compared with WT (SI Appendix, Fig. S8E). Similar to editing of mitochondrial transcripts, there is no clear connection between the editing extent of plastid sites and the amount of transcript that carries these sites. The ndhD and petL sites show a decrease of editing extent in rip1, whereas rpoC1-488 editing extent is significantly increased in the mutant (SI Appendix, Table S2).
Editing Defects in the rip1 Mutant Differ from the Minor Defects Seen in Other Types of Mutants.
We investigated organelle editing in two mutants that mimic the growth phenotype of the rip1 mutant and are compromised in some aspects of organelle RNA metabolism or organelle biogenesis. Tissue was available from a mutant in the chloroplast polynucleotide phosphorylase, which has a major role in maturing mRNA and rRNA 3′ ends but also participates in RNA degradation through exonucleolytic digestion and polyadenylation (41). We obtained a second mutant that was affected in the gene encoding a chloroplast envelope membrane protein containing a putative LrgB domain, which has been reported recently to play an important role in A. thaliana chloroplast development (42). Examination of the editing extent in two null mutants of the genes encoding these plastid proteins shows no difference from the WT in the editing extent of nad6-161 and cob-325, two mitochondrial sites that show a drastic reduction of editing extent in rip1 (SI Appendix, Fig. S9). Among the five plastid sites with editing extent that showed the largest variation in the rip1 mutant, we observed only an increase of editing extent in the null pnp and lrgB mutants for certain sites. NdhD-2, with an editing extent in the rip1 mutant that is about one-half the amount observed in WT (SI Appendix, Table S2), shows an increase of editing extent in both mutants (110% and 40% in pnp and lrgB mutants, respectively) (SI Appendix, Fig. S9). AccD-794 and petL-5 exhibit an increase of editing extent only in the pnp mutant, whereas rpoC1-488 editing extent is markedly increased only in the lrgB mutant. Rps12-158 editing extent is invariant in both the pnp and lrgB plants.
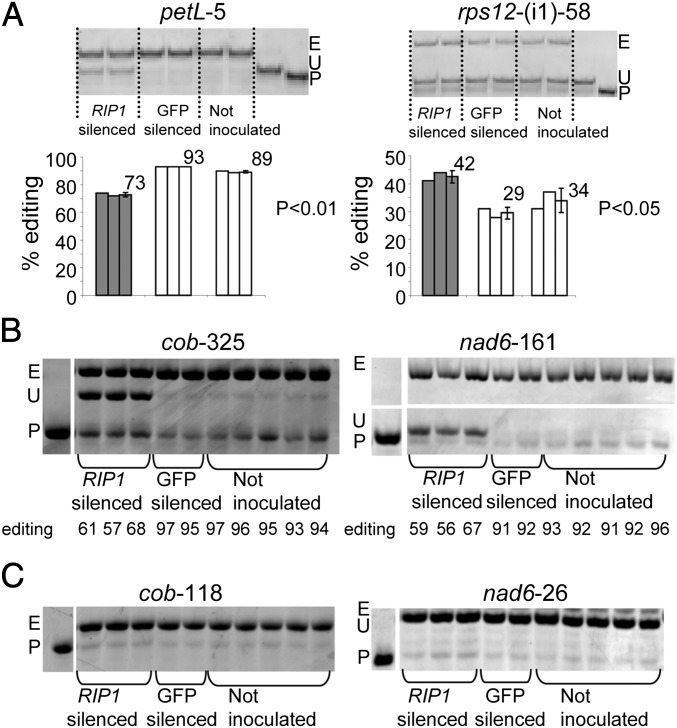
Virus-Induced Gene Silencing of RIP1 Affects Chloroplast and Mitochondrial Editing Efficiency.
Because additional mutant lines with a second independent T-DNA insertion in RIP1 were not available, we silenced RIP1 by virus-induced gene silencing (VIGS) to confirm that the effect on RNA editing was specifically attributable to a defective RIP1 gene. Two types of control plants were used in this experiment: uninoculated plants and plants inoculated with a silencing vector containing only GFP. Quantitative RT-PCR showed that the level of RIP1 transcript in RIP1-silenced plants was reduced to 38% of the level detected in uninoculated plants (SI Appendix, Fig. S10). Unexpectedly, the level of RIP1 transcript in GFP-silenced plants was increased to about two times the level in uninoculated plants. Both RIP1- and GFP-silenced plants show a significant reduction of GFP transcript compared with the uninoculated plants (87% and 95%, respectively) (SI Appendix, Fig. S10).
PPE assays on transcripts from uninoculated and silenced plants showed that silencing of RIP1 results in an average 18% decrease in petL-5 editing extent and a 24% increase in rps12-(i1)-58 editing extent compared with the uninoculated plants (P < 0.01 and P < 0.05, respectively) (Fig. 5A). A decrease in petL-5 editing and an increase in rps12-(i1)-58 editing was likewise observed in the T-DNA mutant (SI Appendix, Table S2), although as expected, the effect of rip1 knockdown in silenced plants is less than in the mutant. No significant difference in editing extent was detected between the GFP-silenced and uninoculated plants (Fig. 5A). The editing extent for rps12-(i1)-58 in the uninoculated WT siblings was 16% (Fig. 2), whereas the uninoculated GFP plants exhibited a 34% editing efficiency (Fig. 5A). This discrepancy results from the fact that the WT siblings of the mutant are in Wassilewskija background, whereas our Arabidopsis GFP line used for VIGS is in Columbia background. PPE assay on mitochondrial transcripts from uninoculated and silenced plants also confirms the variation in mitochondrial editing extent observed in the T-DNA mutant. cob-325 and nad6-161 exhibit a very significant reduction of editing extent in RIP1-silenced plants compared with uninoculated control plants [35% (P < 10−4) and 34% (P < 10−4), respectively] (Fig. 5B). As expected, cob-118 and nad6-26, two C targets with editing efficiencies that are not affected in the rip1 mutant, do not show any decrease of editing extent in RIP1-silenced plants (Fig. 5C). Not all C targets with editing that was greatly impaired in the rip1 mutant exhibited detectable reduction in the silenced plants. For example, no effect on nad6-88 and nad6-89 editing extent was detected in knockdown plants, although they were strongly affected in the mutant (SI Appendix, Fig. S11). There may be sufficient RIP1 present in the silenced plants to allow editing of some sites to proceed normally.
Fig. 5.
RIP1 silencing recapitulates the effect of rip1 mutation on editing extent of organelle sites. (A) PPE assays on plastid sites with quantification of the editing extent represented by a bar below each lane. The average is given with SDs on the right of each group of plants: RIP1-silenced and two sets of controls (GFP-silenced and uninoculated plants). petL-5 and rps12-intron C targets were chosen for assay, because they exhibit reduction and increase, respectively, in the rip1 T-DNA mutants. (B) PPE assays on mitochondrial sites cob-325 and nad6-161 in RIP1-silenced plants compared with the two sets of controls. cob-325 and nad6-161 are C targets that also show a very strong reduction of editing extent in rip1 mutants. (C) RIP1 silencing does not induce any change in the editing extent of cob-118 and nad6-26, two sites with editing extent that was also not affected in the RIP1 mutant. E, edited; P, primer; U, unedited. (The unedited band is not detectable on the cob-118 PPE gel).
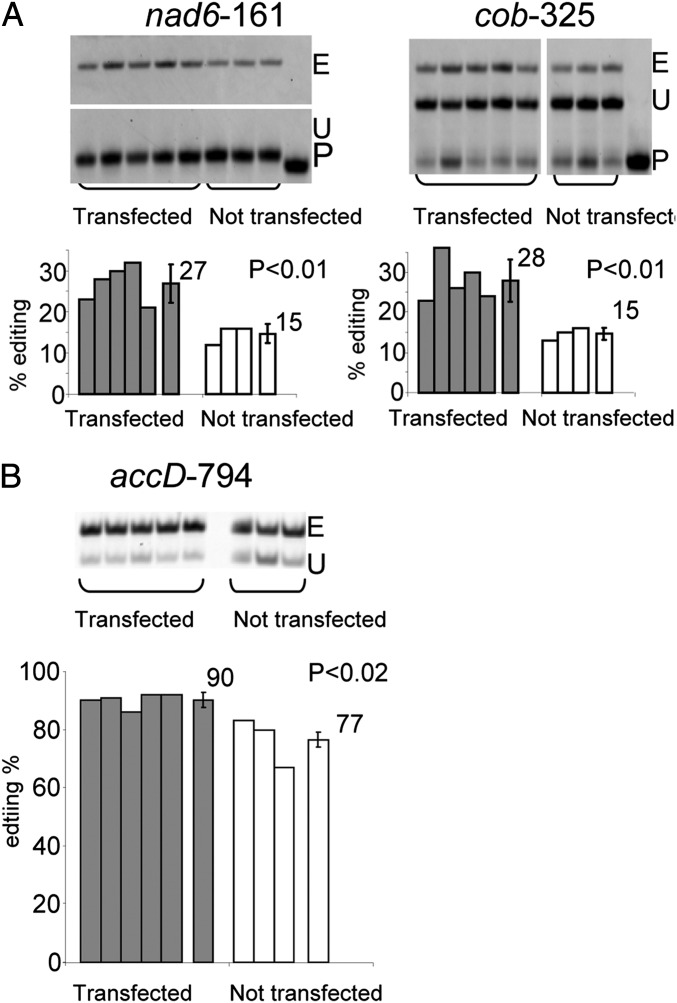
Transient Expression of RIP1 in the rip1 Mutant Seedling Partially Complements the Defect in Mitochondrial and Chloroplast RNA Editing.
We used a transient transformation system in which an Agrobacterium strain is vacuum infiltrated directly into young Arabidopsis seedlings (43). rip1 homozygous mutant seedlings were infiltrated with an Agrobacterium strain carrying a binary vector containing a 35S-RIP1 construct. Three days postinfiltration, PPE analysis showed that editing extent in the mitochondrial nad6-161 and cob-325 and plastid accD-794 sites was significantly increased in transfected vs. not transfected seedlings [80%, 86% (P < 0.01), and 17% (P < 0.02), respectively] (Fig. 6). This experiment provides additional evidence for the necessity of functional RIP1 for efficient mitochondrial and plastid RNA editing.
Fig. 6.
Transfection of rip1 mutant with a WT version of RIP1 partially complements the editing defect in both organelles. (A) Transfection increases editing extent of mitochondrial nad6-161 and cob-325. (B) Transfection increases editing extent of plastid accD-794. (Upper) The PPE products obtained from plants either transfected or not with a construct containing a functional copy of RIP1 under the control of a 35S promoter. (Lower) Graphs depicting the quantification of editing extent for each lane; on the right of each group, the average is shown with SD. E, edited; P, primer; U, unedited.
Discussion
Unlike other proteins known to affect editing efficiency at specific C targets of organelle transcripts, RIP1 is not a member of the PPR family. RIP1 affects editing efficiency of hundreds of C targets, whereas known PPR proteins specify editing at one to seven C targets on either chloroplast or mitochondrial transcripts. The high level of RIP1 transcript expression in both green and nongreen tissues and across many developmental stages is consistent with an essential function for RIP1 in a variety of plastid types as well as mitochondria (SI Appendix, Fig. S12). The presence of homologs to the RIP1 family members in monocots, such as rice and maize, shows its evolution before the split between eudicot and monocots and emphasizes the importance of this family in plant biology. The disparity in level of transcript expression of RIP1 vs. a typical PPR protein may be explained if RIP1 is required in many editing complexes, whereas PPR proteins are needed only for recognition of a small number of target transcripts.
RIP1 has been observed to affect both chloroplast and mitochondrial editing, whereas PPR protein editing factors have been shown to affect editing in either chloroplasts or mitochondria but not both organelles. Recently, OTP87, a PPR-E editing factor, was shown to be dual-targeted to mitochondria and chloroplasts; however, although mutation of OTP87 was shown to affect mitochondrial editing, no effect on chloroplast editing extent in the mutant was reported (29). RIP1 is among about 50 dual-targeted proteins in plants reported to date; 16 of these proteins are amino acyl-tRNA synthetases (reviewed in ref. 44). One theory holds that dual targeting may comprise a means of interorganelle communication; sending the same protein to both organelles may ensure that the protein’s activity occurs in a coordinated manner (44).
RIP1 belongs to a small family of Arabidopsis proteins that contains 10 members, all of which are predicted to be targeted to chloroplasts or mitochondria. RIP1 has been annotated as similar to DAG protein, which refers to the Antirhinnum majus protein DAG; it was shown to affect expression of the rpoB, encoding a subunit of the chloroplast RNA polymerase, to affect accumulation of plastid-targeted nuclear gene products targeted to plastids, and to control chloroplast development at a very early stage (32). The molecular function of DAG was not investigated in the report of its discovery. The only member of the Arabidopsis protein family characterized to date is DAG-LIKE 1, AT2G33430.1, mutants of which have a yellow phenotype and have been shown to have defects in chloroplast rrn operon processing (33). The possible involvement of other members of the RIP1 family in RNA editing obviously merits experimental investigation.
RIP1 Is a Positive and Negative Regulator of Plastid RNA Editing.
RIP1 controls RNA editing of 14 of 34 Arabidopsis editing sites in the chloroplast sites (SI Appendix, Table S2). Editing extent of these 14 sites was significantly altered in the rip1 T-DNA insertional mutant (Fig. 2 and SI Appendix, Table S2), but editing of most chloroplast sites was unaffected. Because there are fewer plastid sites than mitochondrial sites, the amount of RIP1 in the mutant may still be sufficient to accomplish much of its editing function in the plastid, especially if RIP1 import into chloroplasts is more efficient than entry into mitochondria.
Three plastid sites exhibited an increase of editing extent in the rip1 T-DNA insertional mutant (SI Appendix, Table S2). This unexpected effect of the rip1 mutation was also confirmed for rps12-(i1)-58 in RIP1-silenced plants (Fig. 5A). The increased rate of editing of rps12-(i1)-58 in RIP1-silenced plants was less pronounced than in rip1 mutant plants. The expression of RIP1 is expected to be less affected in a silenced plant than a homozygous mutant. In addition to RIP1, mutations in two PPR protein-encoding genes have been shown to increase editing extent. Mutation of REME1, which encodes a PPR-DYW protein, negatively affects editing of both nad2-558 and orfX-552 but also increases editing extent in at least two sites, matR-1771 and rpl5-92 (23). A null mutant of PPR596, which encodes a PPR-P protein, showed an increase of editing extent in several sites in the rpS4 transcript (45). Site-specific inhibition of editing by protein factors has previously been reported in the apolipoprotein B (apoB) RNA editing system in mammals. Antisense inhibition of expression of the proteins GRY-RBP or CUGBP2 in McA cells led to a two- to threefold increase in endogenous apoB RNA editing, suggesting that both these factors may participate in the apoB editing complex as negative regulators in vivo (46, 47). In contrast to these apoB factors, RIP1 is able to promote editing at many sites while inhibiting editing at a few other sites.
Examination of editing extent in two mutants impaired in either plastid RNA metabolism or plastid biogenesis suggests that the increase in editing observed for some plastid sites in rip1 might be an indirect effect of the mutation (SI Appendix, Fig. S9 and SI Appendix, Table S2). Another indication that plastid editing activity might be indirectly compromised by RIP1 mutation is the observation that plastid-encoded polymerase (PEP) transcripts (e.g., petL and ndhB) show decreased editing, whereas most nucleus-encoded polymerase (NEP) transcripts (e.g., rpoA, rpoB, and rpoC) show increased editing (SI Appendix, Table S2). However, if the plastid editing effect observed in the rip1 mutant was indirectly caused by PEP dysfunction, we would expect to observe increase in transcript abundance generated by NEP, which is generally observed for mutants impaired in PEP activity (48). Our data clearly disprove this model, because the rpoC1 (NEP-generated) transcript level is reduced in rip1 as well as levels of PEP transcripts ndhD and petL (SI Appendix, Fig. S8). In addition, the accD (NEP-generated) transcript exhibits a reduction of editing extent at site 794 in rip1, which is unlike other NEP transcripts (SI Appendix, Table S2). More importantly, the current view that genes of photosystems I and II are completely dependent on PEP transcription and a few housekeeping genes, including the rpoB operon, are transcribed exclusively from NEP promoters has been recently challenged in a study of the barley chloroplast transcriptome (49). In this study, which included a PEP-lacking plastid mutant, Zhelyazkova et al. (49) observed that most genes, including genes coding for photosynthesis proteins, have both NEP and PEP promoters.
It remains possible that many of the minor alterations in editing extent of plastid and mitochondrial sites could possibly be caused by indirect effects on transcript or transfactor abundance. For example, altered organelle metabolism could potentially affect the abundance of particular PPR protein editing factors and thus, result in minor differences in editing extent at specific sites. However, we have also shown, in the case of the editing transfactor MEF11, that the sites on which it operates are differentially affected in the rip1 mutant.
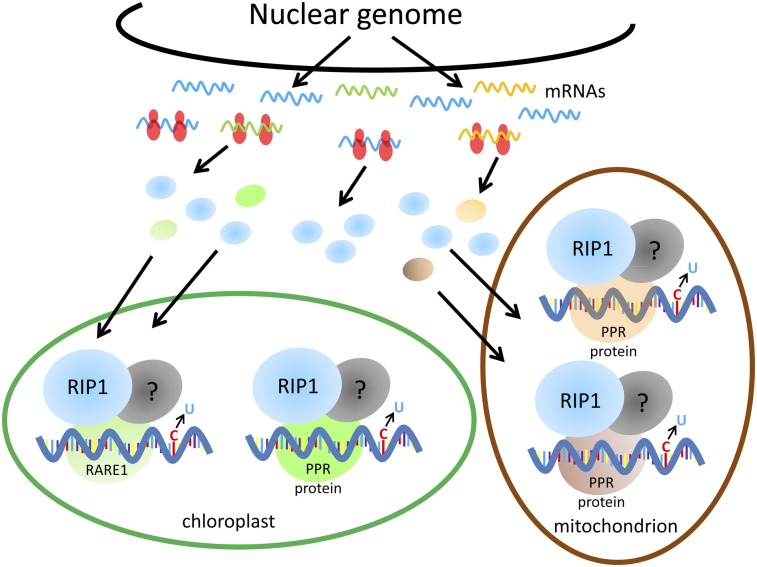
RIP1 and RARE1 Function in an Editing Complex.
The current model for RNA editing holds that PPR proteins recognize cis-elements near C targets of editing, serving as molecular adaptors that recruit an editing activity to specific transcripts (50). CRR4, a PPR-E protein necessary for the editing of the plastid ndhD transcript, directly interacts with the transcript in the area surrounding the targeted C for editing (12). Two other PPR editing trans-factors, the Physcomitrella patens PpPPR_71 and A. thaliana OTP87, have been shown to bind the RNA sequence surrounding their target editing sites (29, 51). However, the identity of all the proteins that act in conjunction with PPR proteins to convert C targets to U targets is unknown.
Deamination is the favored process to explain the base modification, because the sugar phosphate backbone and the nucleotide base are retained during C-to-U conversion (52). The DYW domain found in about one-half of the Arabidopsis PPR editing factors has been suggested to be catalyzing the editing activity based on the similarity of one of its motifs to the conserved cytidine deaminase motif (30). In addition, the phylogenetic distribution of the DYW domain in plant taxa is strictly correlated with RNA editing (30). However, about one-half of the Arabidopsis editing factors lack the DYW domain; moreover, mutant complementation experiments with a truncated DYW protein have proved that the DYW domain is dispensable for editing (53). More importantly, in vitro experiments with recombinant DYW proteins failed to detect any RNA editing activity (53, 54). Examination of the RIP1 sequence for motifs with a web-based tool (http://www.genome.jp/tools/motif/) did not detect any known motif; in particular, the conserved cytidine deaminase motif is absent from RIP1. However, it remains possible that a complex of RIP1 and one or more PPR proteins could constitute an editing activity.
The coimmunoprecipitation and yeast two-hybrid interaction of RIP1 and RARE1 indicate that they are present in the same protein complex in chloroplasts. The C-terminal PPR repeats are not needed for interaction with RIP1, and the C-terminal portion of RIP1 is dispensable for interaction with RARE1 in yeast two-hybrid analysis. Perhaps the C-terminal PPR repeats are involved in interaction with RNA cis-elements, whereas the N-terminal repeats mediate interaction with RIP1. Supporting this model is the recent finding that two PPR motifs are sufficient to bind to an RNA target in vitro (55).
RIP1 Controls the Editing Extent of Many More Mitochondrial C Targets than Any Identified PPR Protein Editing Factor.
The editing extent of 33 mitochondrial genes in the RIP1 T-DNA homozygous mutant exhibited a variable decrease, from undetectable editing for 108 sites by bulk sequencing to a mild decrease in 52 sites (Table 1). The intermediate level of decrease in editing in the mutant ranges from severe in 78 sites to moderate in 28 sites (Table 1). Thus, the number of mitochondrial sites with editing that is affected by RIP1 equals 266 and represents roughly 70% of the sites assayed. A residual editing extent was detected by the sensitive PPE method at two sites, cob-325 and nad6-161, although no editing was detected by bulk sequencing (Fig. 4C). To study the function of RIP1, we have used a hypomorphic allele with an upstream insertion that likely allows accumulation of some active RIP1 protein. A low level of editing at most affected C targets may explain why the rip1 homozygous mutant is viable.
Examination of transcript level by quantitative RT-PCR clearly shows that the changes in mitochondrial editing observed in rip1 cannot be trivially explained by changes in RNA abundance. Mutation in RIP1 has a generally positive effect on mitochondrial transcript levels as previously observed in respiratory mutants (24, 39).
All of the currently identified Arabidopsis mitochondrial PPR protein editing factors belong to the 152-member PLS subfamily, of which 65 members contain only the C-terminal E domain and 87 members exhibit both the E and DYW domains (14). Approximately two-thirds or about 100 of these proteins are predicted to be targeted to mitochondria. Among the 13 Arabidopsis members of the PLS subfamily reported to be mitochondrial editing factors (22–29), only 5 members have been observed to control the editing extent at more than one site. MEF1 and MEF11 control the editing of three sites (22, 27), whereas REME1, SLO1, and OTP87 control two sites each (23, 24, 29). Whether 100 PPR proteins are sufficient to recognize the over 500 C targets of editing in Arabidopsis mitochondria is presently unknown. Some PPR proteins are known to have roles in other aspects of RNA metabolism instead of editing (56–60).
Although mutation of RIP1 negatively affects the editing extent of a large number of mitochondrial C targets, editing of some C targets was not affected at all. It is possible that RIP1 interacts with only a subset of PPR proteins that interact with target RNAs, whereas other members of the RIP1 family interact with a different subset of PPR proteins to stimulate editing at other C targets. The discovery of the important role of RIP1 in mitochondrial editing will open new inquiry into the functions of its 10-member gene family.
Materials and Methods
Genotyping.
All genotyping was done by PCR with BioMix Red (Bioline). For amplification of RARE1 in transgenic plants, primer Rare1_+1933F and the 3xFLAG-StrepIIR primer were used. For genotyping of the FLAG_150D11 line, the WT allele and T-DNA alleles were amplified with primer pairs At3g15000_-442F with At3g15000_+99R or At3g15000_-442F with FLAG_LB4, respectively (SI Appendix, Table S3). Likewise, for the FLAG_607H09 line, the primer pairs were At3g15000_+856F with At3g15000_+1334R and FLAG_Tag3 with At3g15000_+1334R. Both lines were obtained from the INRA FLAGdb T-DNA collection (34).
VIGS.
VIGS of At3g15000 using a GFP cosilencing marker as in refs. 21 and 61 was performed with Complete Arabidopsis Transcriptome MicroArray (CATMA) primers (62) At3g15000_VF and At3g15000_VR (SI Appendix, Table S3). Tissue was collected 18 d postinoculation.
Analysis of RNA Editing by PPE.
All 34 known Arabidopsis chloroplast RNA editing C targets (63, 64) were assayed as in ref. 21. Mitochondrial RNA editing sites were assayed by RT-PCR bulk sequencing using primers described in refs. 65 and 66. PPE analysis on mitochondrial sites cob-118, cob-325, nad6-26, and nad6-161 was conducted as in ref. 23 with primers cob-118, cob325, nad6-26, and nad6-161 (SI Appendix, Table S3).
Transient Transformation of rip1 Seedlings.
Production of the binary vector.
RIP1 ORF was transferred from the gateway entry clone G67651 (ABRC; Ohio State University) into the binary vector pH7RWG2.0 (67) by recombination using LR Clonase II (Invitrogen) After sequence verification, the plasmid was transformed into Agrobacterium tumefaciens GV3101.
Transformation of rip1 seedlings.
Sterile seeds from RIP1-FLAG_150D11-T-DNA heterozygous plant were germinated on MS plates. After 2 wk, the homozygous mutant plants, which were distinguishable from the other progeny because of their dwarf phenotype, were collected onto new MS plates and subjected to Agrobacterium infiltration according to the protocol in ref. 44. DNA and RNA were collected 3 d postinfiltration. DNA genotyping confirmed the visual assignment of the mutant seedlings based on the dwarf phenotype.
Subcellular Localization of RIP1.
The RIP1 ORF minus the stop codon was amplified from the clone G67651 with primers At3g15000_+1F and At3g15000_+1185R (SI Appendix, Table S3) and cloned into pCR8/GW/TOPO (Invitrogen). After sequence verification, the insert was transferred into the pEarleyGate 103 vector (68) by recombination using LR Clonase II (Invitrogen), creating an RIP1-GFP fusion driven by the 35S promoter. Protoplasts from Arabidopsis Col-0 accession or N. benthamiana were transfected with the plasmid using the protocol in ref. 39. Protoplasts were checked for fluorescence under the confocal microscope 16 h after incubation with the plasmid. Protoplasts were incubated with MitoTracker Orange CM-H2TMRos (Invitrogen) at a final concentration of 500 nM for 30 min, centrifuged, and resuspended in dye-free medium. Images were acquired using a Leica SP2 confocal microscope. For chloroplast nucleoid staining, N. benthamiana protoplasts were incubated with 3 μg/mL DAPI (Sigma) for 5 min before visualization using a Zeiss 710 confocal microscope.
Real-Time Quantitative RT-PCR Conditions and Analysis.
Quantitative RT-PCR was performed as described in ref. 69. All of the primers used for quantitative RT-PCR are given in SI Appendix, Table S3. The results of the quantitative RT-PCR analysis were normalized using the gene At2g28390, which has been shown to be a superior reference gene for transcript normalization in Arabidopsis (70).
Supplementary Material
Acknowledgments
We thank Carol Bayles for instruction in confocal microscopy and Lin Lin for excellent technical assistance. We also thank Benoit Castandet for providing us with RNA from pnp and its WT sibling. This work was supported by the National Science Foundation from the Molecular and Cellular Biosciences, Gene and Genome Systems Grants 1020636 (to S.B.), 0716888 (to M.R.H.), and 0929423 (to M.R.H.) and Integrative Organismal Systems Grant 0922560 (to K.J.v.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 8372 (volume 109, number 22).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121465109/-/DCSupplemental.
References
- 1.Shikanai T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu Rev Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 3.Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- 4.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 5.Covello PS, Gray MW. RNA editing in plant mitochondria. Nature. 1989;341:662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- 6.Gualberto JM, Lamattina L, Bonnard G, Weil JH, Grienenberger JM. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989;341:660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- 7.Hiesel R, Wissinger B, Schuster W, Brennicke A. RNA editing in plant mitochondria. Science. 1989;246:1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- 8.Bock R, Hermann M, Kössel H. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 1996;15:5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri S, Maliga P. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 1996;15:5958–5964. [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes ML, Reed ML, Hegeman CE, Hanson MR. Sequence elements critical for efficient RNA editing of a tobacco chloroplast transcript in vivo and in vitro. Nucleic Acids Res. 2006;34:3742–3754. doi: 10.1093/nar/gkl490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 12.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem. 2006;281:37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 13.Small ID, Peeters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 14.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA. 2007;104:8178–8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chateigner-Boutin AL, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, et al. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 2009;58:82–96. doi: 10.1111/j.1365-313X.2008.03766.x. [DOI] [PubMed] [Google Scholar]
- 18.Cai W, et al. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 2009;150:1260–1271. doi: 10.1104/pp.109.136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammani K, et al. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21:3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda K, et al. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 2010;61:339–349. doi: 10.1111/j.1365-313X.2009.04059.x. [DOI] [PubMed] [Google Scholar]
- 21.Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA. 2009;15:1142–1153. doi: 10.1261/rna.1533909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentolila S, Knight WE, Hanson MR. Natural variation in Arabidopsis leads to the identification of REME1, a pentatricopeptide repeat-DYW protein controlling the editing of mitochondrial transcripts. Plant Physiol. 2010;154:1966–1982. doi: 10.1104/pp.110.165969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung TY, Tseng CC, Hsieh MH. The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J. 2010;63:499–511. doi: 10.1111/j.1365-313X.2010.04258.x. [DOI] [PubMed] [Google Scholar]
- 25.Takenaka M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in the nad7 transcript in mitochondria of Arabidopsis. Plant Physiol. 2010;152:939–947. doi: 10.1104/pp.109.151175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takenaka M, Verbitskiy D, Zehrmann A, Brennicke A. Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J Biol Chem. 2010;285:27122–27129. doi: 10.1074/jbc.M110.128611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbitskiy D, Zehrmann A, van der Merwe JA, Brennicke A, Takenaka M. The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J. 2010;61:446–455. doi: 10.1111/j.1365-313X.2009.04076.x. [DOI] [PubMed] [Google Scholar]
- 28.Verbitskiy D, Härtel B, Zehrmann A, Brennicke A, Takenaka M. The DYW-E-PPR protein MEF14 is required for RNA editing at site matR-1895 in mitochondria of Arabidopsis thaliana. FEBS Lett. 2011;585:700–704. doi: 10.1016/j.febslet.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 29.Hammani K, et al. The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J Biol Chem. 2011;286:21361–21371. doi: 10.1074/jbc.M111.230516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salone V, et al. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581:4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 31.Gillman JD, Bentolila S, Hanson MR. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007;49:217–227. doi: 10.1111/j.1365-313X.2006.02953.x. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee M, et al. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996;15:4194–4207. [PMC free article] [PubMed] [Google Scholar]
- 33.Bisanz C, et al. The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol Biol. 2003;51:651–663. doi: 10.1023/a:1022557825768. [DOI] [PubMed] [Google Scholar]
- 34.Samson F, et al. FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang YH. How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Tech. 2008;1:11–20. [Google Scholar]
- 36.Kruft V, Eubel H, Jänsch L, Werhahn W, Braun HP. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 2001;127:1694–1710. [PMC free article] [PubMed] [Google Scholar]
- 37.Heazlewood JL, et al. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 39.Sosso D, et al. PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell. 2012;24:676–691. doi: 10.1105/tpc.111.091074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peeters NM, Hanson MR. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA. 2002;8:497–511. doi: 10.1017/s1355838202029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Germain A, et al. Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J. 2011;67:381–394. doi: 10.1111/j.1365-313X.2011.04601.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, et al. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2012;193:81–95. doi: 10.1111/j.1469-8137.2011.03867.x. [DOI] [PubMed] [Google Scholar]
- 43.Marion J, et al. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 2008;56:169–179. doi: 10.1111/j.1365-313X.2008.03596.x. [DOI] [PubMed] [Google Scholar]
- 44.Carrie C, Giraud E, Whelan J. Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- 45.Doniwa Y, et al. The involvement of a PPR protein of the P subfamily in partial RNA editing of an Arabidopsis mitochondrial transcript. Gene. 2010;454:39–46. doi: 10.1016/j.gene.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Anant S, et al. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 47.Blanc V, et al. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J Biol Chem. 2001;276:10272–10283. doi: 10.1074/jbc.M006435200. [DOI] [PubMed] [Google Scholar]
- 48.Kindgren P, et al. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 2011;68:28–39. doi: 10.1111/j.1365-313X.2011.04865.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhelyazkova P, et al. The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008;13:663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Tasaki E, Hattori M, Sugita M. The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 2010;62:560–570. doi: 10.1111/j.1365-313X.2010.04175.x. [DOI] [PubMed] [Google Scholar]
- 52.Rajasekhar VK, Mulligan RM. RNA editing in plant mitochondria: [alpha]-Phosphate is retained during C-to-U conversion in mRNAs. Plant Cell. 1993;5:1843–1852. doi: 10.1105/tpc.5.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuda K, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21:146–156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura T, Sugita M. A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 2008;582:4163–4168. doi: 10.1016/j.febslet.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi K, et al. Identification and characterization of the RNA binding surface of the pentatricopeptide repeat protein. Nucleic Acids Res. 2012;40:2712–2723. doi: 10.1093/nar/gkr1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz-Linneweber C, et al. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Longevialle AF, et al. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell. 2007;19:3256–3265. doi: 10.1105/tpc.107.054841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008;55:619–628. doi: 10.1111/j.1365-313X.2008.03529.x. [DOI] [PubMed] [Google Scholar]
- 60.Chateigner-Boutin AL, et al. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J. 2011;65:532–542. doi: 10.1111/j.1365-313X.2010.04441.x. [DOI] [PubMed] [Google Scholar]
- 61.Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006;142:21–27. doi: 10.1104/pp.106.084624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crowe ML, et al. CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 2003;31:156–158. doi: 10.1093/nar/gkg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tillich M, et al. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005;43:708–715. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- 64.Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35:e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bentolila S, Chateigner-Boutin AL, Hanson MR. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol. 2005;139:2006–2016. doi: 10.1104/pp.105.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bentolila S, Elliott LE, Hanson MR. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics. 2008;178:1693–1708. doi: 10.1534/genetics.107.073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 68.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 69.Bentolila S, Stefanov S. A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes. Plant Physiol. 2012;158:996–1017. doi: 10.1104/pp.111.190231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]