Abstract
Nonribosomal peptide synthetases (NRPSs) usually catalyze the biosynthesis of peptide natural products by sequential selection, activation, and condensation of amino acid precursors. It was reported that some fatty acids, α-ketoacids, and α-hydroxyacids originating from amino acid metabolism as well as polyketide-derived units can also be used by NRPS assembly lines as an alternative to amino acids. Ecteinascidin 743 (ET-743), naphthyridinomycin (NDM), and quinocarcin (QNC) are three important antitumor natural products belonging to the tetrahydroisoquinoline family. Although ET-743 has been approved as an anticancer drug, the origin of an identical two-carbon (C2) fragment among these three antibiotics has not been elucidated despite much effort in the biosynthetic research in the past 30 y. Here we report that two unexpected two-component transketolases (TKases), NapB/NapD in the NDM biosynthetic pathway and QncN/QncL in QNC biosynthesis, catalyze the transfer of a glycolaldehyde unit from ketose to the lipoyl group to yield the glycolicacyl lipoic acid intermediate and then transfer the C2 unit to an acyl carrier protein (ACP) to form glycolicacyl-S-ACP as an extender unit for NRPS. Our results demonstrate a unique NRPS extender unit directly derived from ketose phosphates through (α,β-dihydroxyethyl)-thiamin diphosphate and a lipoyl group-tethered ester intermediate catalyzed by the TKase-ACP platform in the context of NDM and QNC biosynthesis, all of which also highlights the biosynthesis of ET-743. This hybrid system and precursor are distinct from the previously described universal modes involving the NRPS machinery. They exemplify an alternate strategy in hybrid NRPS biochemistry and enrich the diversity of precursors for NRPS combinatorial biosynthesis.
Keywords: primary metabolism, secondary metabolism, xylulose-5-phosphate, fructose-6-phosphate
Nonribosomal peptide natural products include many clinically important drugs such as vancomycin (antibacterial), bleomycin (anticancer), and cyclosporine (immunosuppression). They are biosynthesized from amino acid or relative precursors by nonribosomal peptide synthetases (NRPSs) by using the multienzyme thiotemplate model (1–4). Three enzymatic activities, which are organized into a condensation–adenylation–peptidyl carrier protein (C-A-PCP) module, are necessary for one complete elongation cycle of peptide synthesis. Generally, the A domain selects a cognate amino acid from the pool of available substrates and activates it as an aminoacyl adenylate. Subsequently, the activated amino acid is transferred onto the thiol group of the 4′-phosphopantetheinyl arm attached to the PCP domain. Next, the C domain catalyzes the peptide-bond formation between the upstream aminoacyl/peptidyl-S-PCP and the free amino group of the downstream aminoacyl-S-PCP, thus facilitating the translocation of the growing peptide chain onto the next module. The large diversity of precursors can be recognized and incorporated with positional specificity for elongation peptide chains by thioesters tethered to an assembly line, which create more structural diversity of final products and provide the molecular basis for combinatorial biosynthesis (2–4). As one of the most structurally diverse natural products, nonribosomal peptides contain not only nonproteogenic amino acids or aryl acids from amino acid metabolism but also fatty acids or polyketide-derived units (5). Accordingly, the hybrid NRPS/polyketide synthase (PKS) assembly lines carry out acyl transfer chemistry and tether monomers and growing polymers as pantheinyl thioesters to carrier protein domains (3, 6). Although current knowledge of NRPS and hybrid NRPS/PKS systems provides context for more efficient efforts in combinatorial biosynthesis and total biosynthesis, no doubt discovery of other hybrid systems and characterization of novel pathways that fashion different monomers for NRPS assembly line use will highlight new chemistry and strategies for NRPS engineering (3, 6, 7).
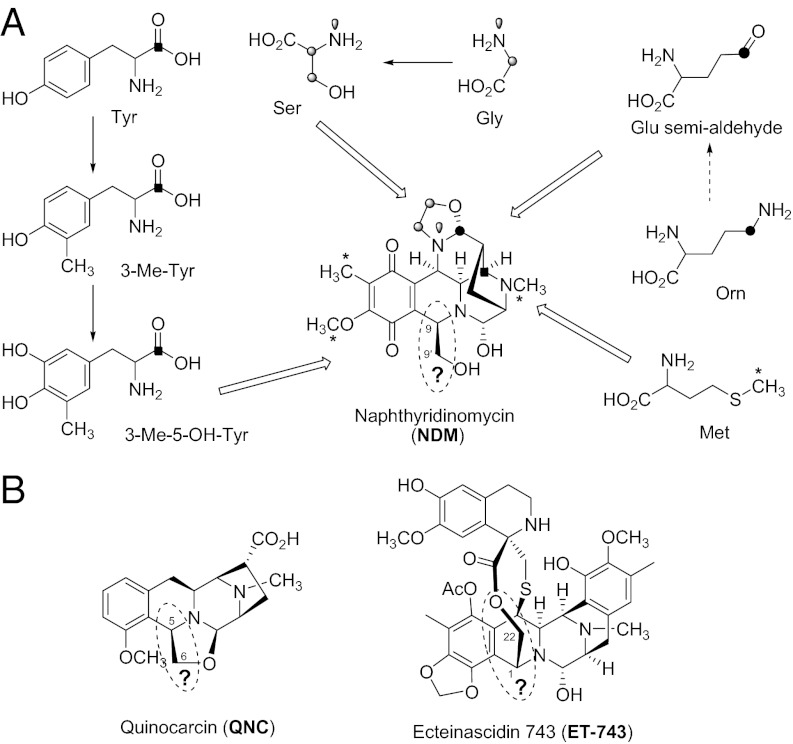
Naphthyridinomycin (NDM; Fig. 1A), an antitumor antibiotic, was isolated in 1974 from Streptomyces lusitanus as the first member of tetrahydroisoquinoline family of alkaloids, which includes ∼60 natural products classified into the saframycin, NDM, and quinocarcin (QNC; Fig. 1B) subfamilies (8). The great interest in this family of natural products stems from their novel structure, mode of action, and potent antibacterial and antitumor activities (8). NDM shows strong antimicrobial activities, extraordinarily effective against Gram-positive bacteria, even against methicillin-resistant Staphylococcus aureus (9). Of particular significance within this family is ecteinascidin 743 (ET-743, Yondelis; Fig. 1B), a marine natural product from Ecteinascidia turbinata, which has been approved in Europe since 2007 for treatment of advanced soft-tissue sarcoma (10, 11). Accordingly, the chemical synthesis of this family of alkaloids has been extensively pursued (8, 11–13); however, the deeply biosynthetic studies were limited to several members of saframycin group (14–18). Although labeled precursor feeding experiments showed that l-tyrosine, l-methionine, glycine, and d,l-ornithine could be incorporated into NDM (19–21), to date the origin and the molecular basis of the two carbon (C2) unit C9–C9′ for the biosynthesis of NDM, C5–C6 for QNC, or C1–C22 for ET-743 have remained unknown (Fig. 1).
Fig. 1.
The tetrahydroisoquinoline family natural products. (A) Isotope feeding experiment showed that l-tyrosine, l-methionine, glycine, and d,l-ornithine could be incorporated into NDM. (B) Structures of QNC and ET-743.
Here we report the NDM NRPS system from S. lusitanus NRRL 8034 and the QNC NRPS system from Streptomyces melanovinaceus NRRL 12388 that contain a two-component transketolase (TKase) that catalyzes the transfer of a C2 fragment, a glycolaldehyde unit, from ketose (donor substrate) to the lipoyl group to yield the glycolicacyl lipoic acid intermediate and then transfers the C2 unit to an acyl carrier protein (ACP) to form glycolicacyl-S-ACP as a extender unit for NRPS.
Results
Comparative Analysis of the Gene Cassettes in NDM and QNC Gene Cluster.
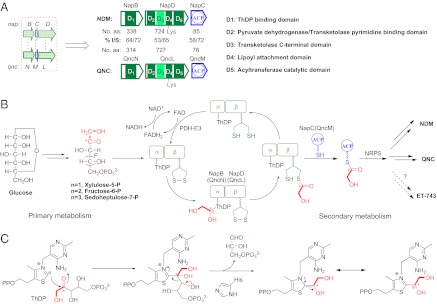
We have been interested in the biosynthesis of these natural products for several years (16, 22–24). Recently, comparative analysis of the NDM gene cluster in S. lusitanus NRRL 8034 and the QNC gene cluster in S. melanovinaceus NRRL 12388 revealed a three-gene cassette in each, napB–napC–napD and qncN–qncM–qncL, with significant overall identify to each other (Fig. 2A). NapB and QncN show high sequence similarity to the pyruvate dehydrogenase (PDH) E1α component from Streptomyces coelicolor A3 (2) (64% identity and 74% similarity) and moderate similarity to the thiamin diphosphate (ThDP) binding domain (D1) of TKase. Both NapD and QncL contain four domains—the pyrimidine binding domain (D2) of PDH E1β or TKase, the C-terminal domain (D3) of TKase, the lipoyl attachment domain (ETSKAVE) (D4) of the 2-oxoacid dehydrogenase E2 component, and the catalytic domain (D5) of acyltransferase or 2-oxoacid dehydrogenase—that are all characteristic of known 2-oxoacid dehydrogenases (25). NapC and QncM are homologs of ACP with a conserved Ser residue (AWTSL) as the active site for phosphopantetheinylation. Remarkably, the NapB/NapD-NapC three enzymes in NDM biosynthesis exhibit high sequence similarity (53–64% identity and 65–72% similarity) and have an identical domain organization to QncN/QncL-QncM, which is involved in the biosynthesis of QNC in S. melanovinaceus (Fig. 2A). Very recently, the gene cluster of ET-743 was reported by metagenomic sequencing of total-genome DNA from the tunicate/microbial consortium (18), which also includes an etuP1/etuP2 pair encoding two components (E1 and E2) of the PDH complex with moderate sequence homology (34–37% identity and 59–65% similarity) and an identical domain organization to NapB/NapD or QncN/QncL (Fig. S1). This finding suggests that these enzymes should be responsible for the biosynthesis of the common moiety among NDM, QNC, and ET-743, potentially being the hydroxyethyl C2 unit based on the mechanism of PDH complex.
Fig. 2.
Proposed mechanism of the TKase-ACP systems involved in the tetrahydroisoquinoline biosynthesis by diverting a hydroxyacyl unit from ketosugars into a nonribosomal peptide assembly line. (A) Comparison between the TKase-ACP systems in NDM and QNC biosynthesis. aa, amino acid; I/S, identity/similarity. (B) The proposed biosynthetic pathway and catalytic cycle by TKase-ACP system. (C) The ThDP-dependent reaction mechanism catalyzed by TKase.
NapB/NapD and QncN/QncL Play an Important Role in NDM and QNC Biosynthesis.
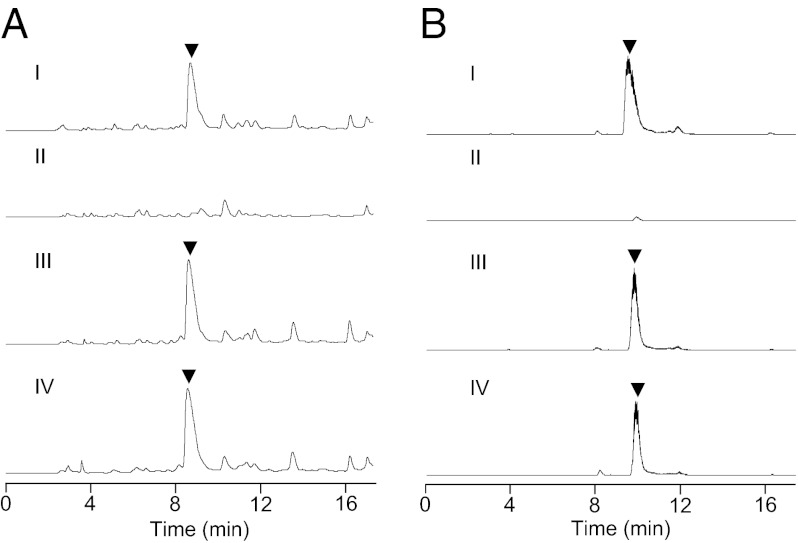
To validate this hypothesis, we first inactivated napB and napD by replacing them with a mutant copy in which the respective gene was disrupted by the spectinomycin resistance gene aadA (Fig. S2). The resultant mutant strains S. lusitanus TG3001 (ΔnapB) and TG3002 (ΔnapD) completely lost the ability to produce NDM (Fig. 3 A, II and B, II). Introduction of pTG3012, in which the expression of the napB–napC–napD gene cassette is under the control of the constitutive PermE* promoter, into S. lusitanus TG3001 or TG3002 restored the production of NDM, which was confirmed by HPLC and liquid chromatography/MS (LC-MS) analysis (Fig. 3 A, III and B, III). Additionally, heterologous complementation by using the qncN–qncM–qncL gene cassette from S. melanovinaceus under the control of PermE* promoter (pTG3013) also resumed the production of NDM with similar yield (Fig. 3 A, IV and B, IV). Altogether, these findings unambiguously established that the two gene cassettes share the same function, which is necessary for NDM and QNC biosynthesis.
Fig. 3.
Verification of the necessary role of TKase in the biosynthesis of the tetrahydroisoquinoline family. (A) HPLC analysis (UV at 270 nm). I, WT S. lusitanus NRRL 8034. II, mutant S. lusitanus TG3001 (ΔnapB); III, mutant S. lusitanus TG3003 (TG3001 harboring the napB–napC–napD expression plasmid pTG1012); IV, mutant S. lusitanus TG3005 (TG3001 harboring the qncN–qncM–qncL expression plasmid pTG1013). (B) LC-MS analysis. I, WT S. lusitanus NRRL 8034; II, mutant S. lusitanus TG3002 (ΔnapD); III, mutant S. lusitanus TG3004 (TG3002 harboring the napB–napC–napD expression plasmid pTG1012); IV, mutant S. lusitanus TG3006 (TG3002 harboring the qncN–qncM–qncL expression plasmid pTG1013). ▼, NDM.
Isotope Labeling Indicates That the Substrate of QncN/QncL Is a Ketose.
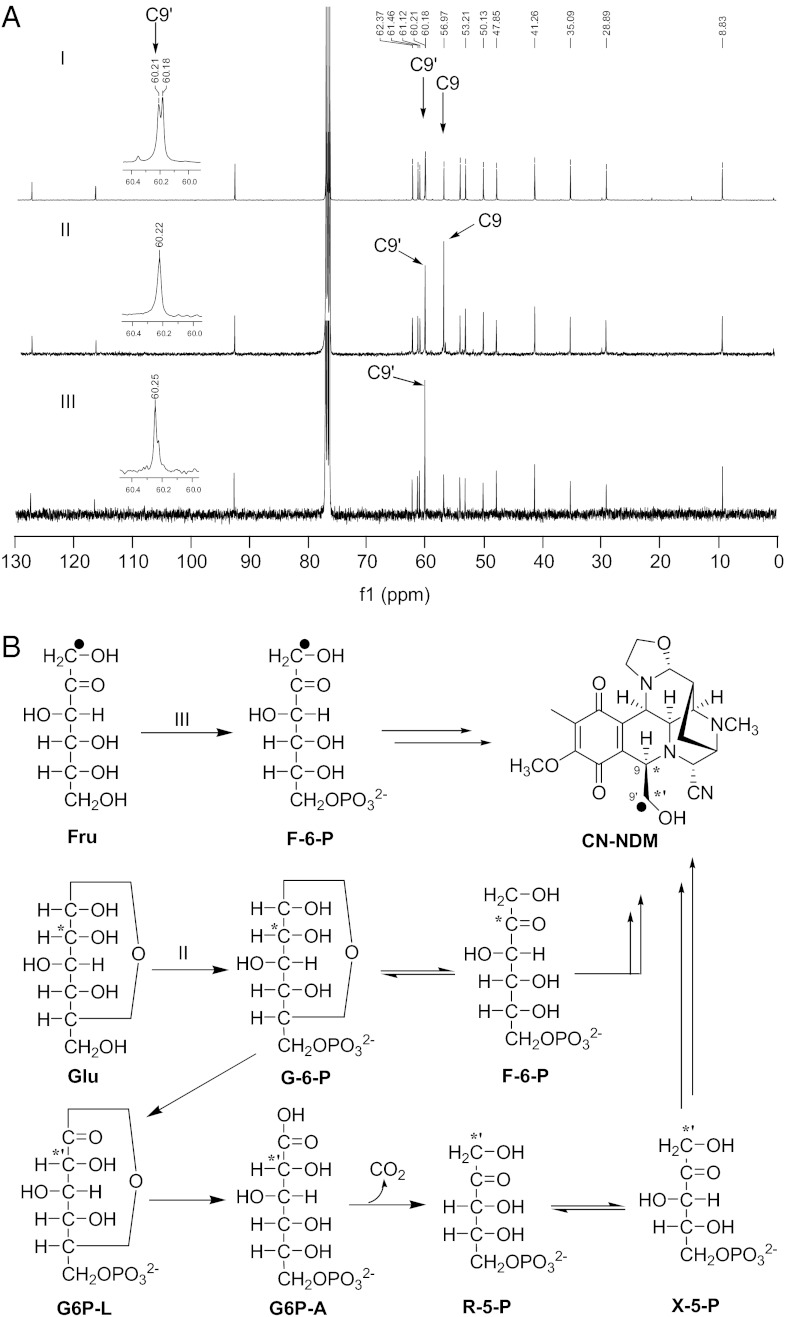
On the basis of the homology to PDH and the biochemical characterization of YerE, a ThDP-dependent enzyme that catalyzed the attachment of the C2 branched chain onto sugar from pyruvate (26), we reasoned that the unknown C2 unit of NDM would be derived from pyruvate by a ThDP-dependent decarboxylation process. We first explored this assumption by feeding [2-13C]pyruvate into the fermentation broth of S. lusitanus NRRL 8034. Characterization of the resultant cyanonaphthyridinomycin (CN-NDM) by NMR revealed only weak 13C enrichment at C9′ (Fig. S3) but unexpected specific 13C enrichment at C9, thereby ruling out the possibility of the origin from pyruvate after the ThDP-dependent decarboxylation process. We next turned our attention to ketoses, which are the substrates of TKases (27). Similarly, CN-NDM was isolated from the [1-13C]fructose or [2-13C]glucose feeding experiments, and NMR analysis confirmed 13C enrichment at C9′ or C9′ and C9, respectively (Fig. 4A), thereby suggesting an origin of the C2 unit derived from ketoses, which are metabolized from glucose (Fig. 4B).
Fig. 4.
Investigation of the possible precursors of C9–C9′ by feeding experiments with 13C-labeled sugars. (A) 13C NMR spectra of CN-NDM without feeding (I), with feeding of [2-13C]glucose (II), and with feeding of [1-13C]fructose (III). (B) Proposed mechanism of relative sugar metabolism and incorporation into NDM. Fru, fructose; Glu, glucose; G-6-P, glucose 6-phosphate; G6P-L, 6-phosphoglucono-δ-lactone; G6P-A, 6-phosphogluconate; R-5-P, ribulose-5- phosphate.
Given the fact that TKases usually use phosphorylated ketose as donor substrates (27), we proposed that both of the two-component TKases, NapB/NapD and QncN/QncL, serve as the key roles to divert hydroxyethyl units from phosphorylated ketoses into nonribosomal peptide assembly line in NDM and QNC biosynthetic pathways, respectively. The first half-reaction of these two unique TKase catalytic cycles is formation of (α,β-dihydroxyethyl)-ThDP as the C2 unit donor by splitting the C2–C3 bond of xylulose-5-phosphate (X-5-P), fructose-6-phosphate (F-6-P), or sedoheptulose-7-phosphate (S-7-P) (Fig. 2C); the second half-reaction is capture of the hydroxyethyl C2 unit by the bound lipoic acid cofactor to form a thiolate intermediate, which should be similar to the reaction catalyzed by PDH (Fig. 2B). Then, the hydroxyethyl C2 unit is subsequently transferred to ACP, yielding glycolicacyl-S-ACP, which is further used as a carrier protein-tethered monomer for NRPS extension in the biosynthesis of NDM, QNC, and even ET-743 (Fig. 2B).
QncN/QncL also Transfers the Glycolicacyl C2 Unit onto QncM to Form Glycolicacyl-S-ACP in Vitro.
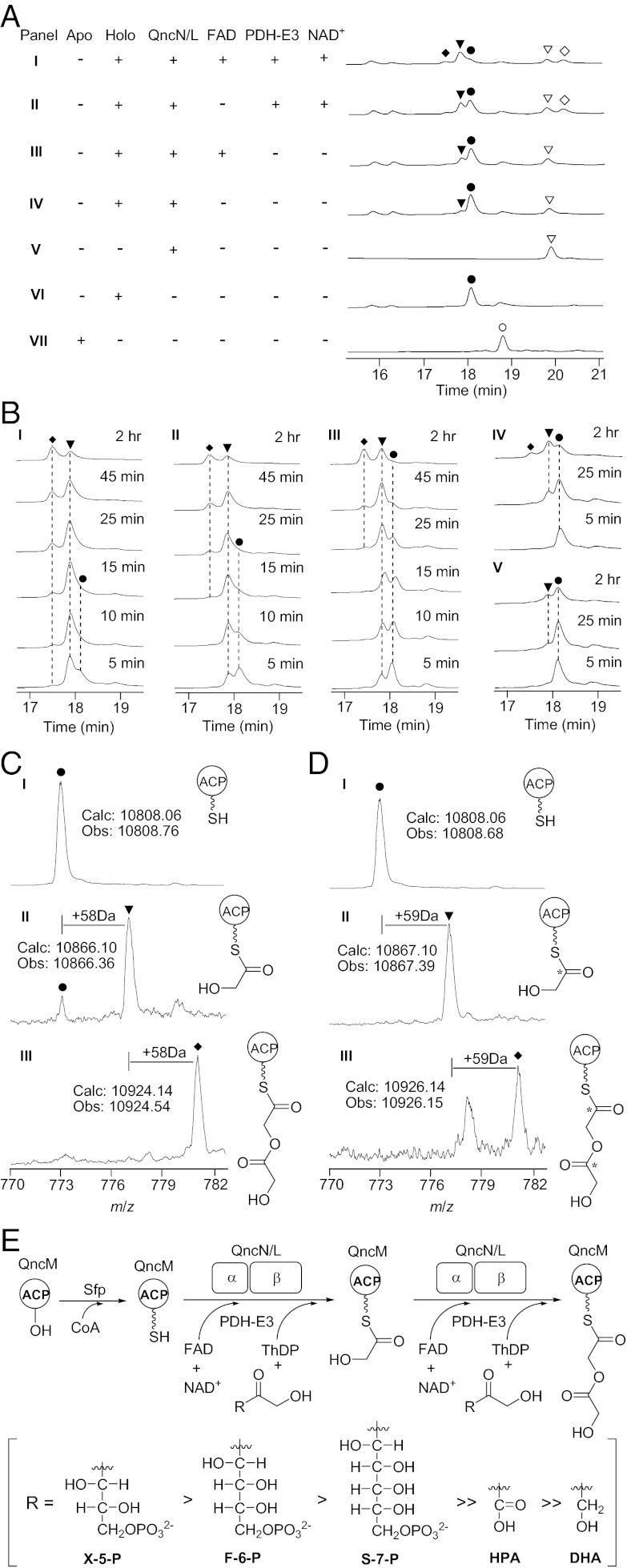
Seeking an understanding of the chemical underlying C2 derivation, we characterized the two-component TKases in vitro. First attempts to express NapB and NapD involved in NDM biosynthesis from S. lusitanus were unsuccessful because they are insoluble in Escherichia coli. We therefore turned to qncN and qncL in the QNC gene cluster from S. melanovinaceus. We successfully overexpressed QncN and QncL by coexpression of qncN and qncL using pCDFDuet vector as well as ACP encoded by qncM in E. coli and purified the resultant enzymes as His6-tagged fusion proteins to homogeneity (Fig. S4 A and B). Because QncM overexpressed in E. coli is the nonfunctional apo form, we coexpressed it in E. coli BL21 (DE3) with Sfp, which catalyzes the phosphopantetheinylation to ensure that all ACPs are converted into the functional holo forms (Fig. 5A). We incubated the holo-ACP (QncM) with X-5-P, ThDP, Mg2+, and TKase (QncN/QncL) to directly test hydroxyacyl loading by subjecting the reaction mixtures to HPLC analysis. By HPLC analysis, a unique peak appeared that seemed to depend on the TKase, and the production increased by addition of coenzyme FAD (Fig. 5A). The complete conversion of holo-ACP to the unique product was observed when PDH E3 component [which is purified from E. coli BL21 (DE3) by expressing its gene from Streptomyces lividans (Fig. S4C)] and NAD+ were also incubated with the reaction together (Fig. 5A). The unique product was subjected to quadrupole–time of flight/MS (Q-TOF-MS) analysis (Fig. 5C); the addition of 58 Da to holo-ACP is in agreement with the mass increase for the glycolicacyl C2 unit. Further investigation of the reaction time course led to the discovery that the produced glycolicacyl-S-ACP could be converted into another unique deriviate (Fig. 5B), which was identified by Q-TOF-MS analysis as glycolicacyl-O-glycolicacyl-S-ACP, the secondary loading of the glycolicacyl C2 unit (Fig. 5 C and E). To further confirm this conclusion, [2-13C]F-6-P, which is generated in situ from [2-13C]glucose by glucokinase and phosphoglucose isomerase, was used as substrate to perform the same assay and resulted in the [M + 1] and [M + 2] products as expected (Fig. 5D).
Fig. 5.
Biochemical characterization of the TKase-ACP systems in vitro. (A) HPLC analysis of enzymatic reaction. ○, apo-ACP; ●, holo-ACP; ▼, glycolicacyl-S-ACP; ◆, glycolicacyl-O-glycolicacyl-S-ACP; ▽, QncN or QncL; ◇, PDH E3. (B) Time course of enzymatic reaction using X-5-P (I), F-6-P (II), S-7-P (III), HPA (IV), and DHA (V) as donor substrates, respectively. The concentration of substrate used in the assay is 10 mM for DHA and 2 mM for others. (C) Q-TOF-MS analysis of holo-ACP (I), glycolicacyl-S-ACP (II), and glycolicacyl-O-glycolicacyl-S-ACP (III) using X-5-P as substrate. (D) Q-TOF-MS analysis of holo-ACP (I), glycolicacyl-S-ACP (II), and glycolicacyl-O-glycolicacyl-S-ACP (III) using [2-13C]F-6-P as substrate. (E) Proposed mechanism of the in vitro enzymatic reaction catalyzed by the TKase-ACP system.
X-5-P Is the Best Donor Substrate for QncN/QncL.
On the basis of the studies described before that the conserved Lys is the active site for lipoyl attachment of PDH (25) and that the key His residue is necessary for stabilization of the active glycolaldehyde intermediate of TKase (27, 28), we therefore constructed QncN H245A, QncL H134A, and QncL K416A mutants (Fig. S5A), respectively. As expected, all of the mutants lost the biological activity (Fig. S5B). Further substrate specificity assays showed that X-5-P, F-6-P, or S-7-P could serve as the donor substrates, whereas xylulose without phosphorylation could not be recognized; hydroxypyruvate (HPA) and dihydroxyacetone (DHA) also could be used as substrates, but the efficiency is poor (Fig. 5 B and E). These in vitro biochemical results are also consistent with the observation of weak 13C enrichment at C9′ in the in vivo feeding experiment using [2-13C]pyruvate as precursor. Advanced kinetic analysis gave the apparent parameters of holo-QncM glycolicacylation by QncN/QncL using X-5-P, F-6-P, or S-7-P, which indicated that X-5-P is the best donor substrate, relatively (Fig. S6).
Discussion
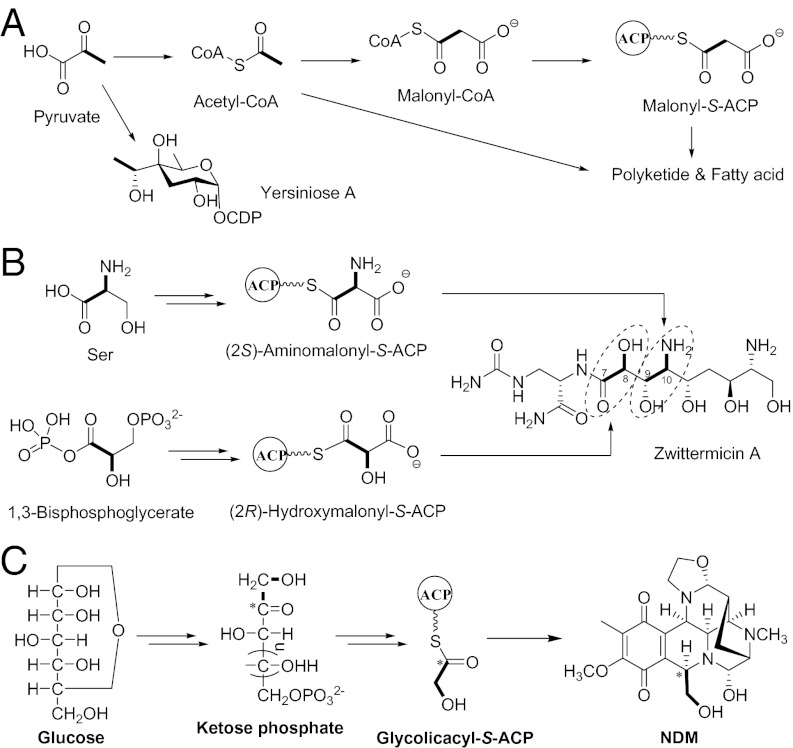
Nonribosomal peptides are a class of peptide secondary metabolites usually produced by microorganisms like bacteria and fungi. The backbone of nonribosomal peptides are usually synthesized by NRPSs, which, unlike ribosomes, are independent of messenger RNA. NRPSs consist of repeating modules that are responsible for sequential selection, activation, and condensation of amino acid precursors. However, the most popular two-carbon donor in the secondary metabolism, acetyl-CoA, which is first converted into the more reactive malonyl-CoA before its incorporation by PKS or fatty acid synthase (Fig. 6A), could also be incorporated into the peptides synthesis in the NRPS/PKS hybrid systems as a two-carbon donor. To date, more and more building blocks, such as fatty acids, α-ketoacids, and α-hydroxyacids originating from amino acid metabolism as well as polyketide-derived units can also be used by NRPS assembly lines in the hybrid systems (4, 5), which has greatly enriched the diversity of the peptide natural products.
Fig. 6.
Summary of the origins of the two-carbon unit in the biosynthesis. (A) YerE catalyzed the attachment of the C2 branched chain onto sugar from pyruvate by a ThDP-dependent decarboxylation process. (B) A hydroxymalonyl-ACP derived from the glycolytic pathway as an extender unit for PKS involved in the biosynthesis of zwittermicin. (C) A unique NRPS extender unit derived from ketose phosphates through (α,β-dihydroxyethyl)-ThDP and a lipoyl group-tethered ester intermediate catalyzed by the TKase-ACP platform in the context of NDM and QNC biosynthesis.
It has been shown previously that there are two strategies for the biosynthesis of α-hydroxyacyl units in nonribosomal peptides: either α-hydroxyacids were directly adenylated by the A domain (29) or α-ketoacids were activated and reduced by ketoreductase to the corresponding α-hydroxyacids while tethered to a PCP domain on the NRPS assembly line (30, 31). We have now characterized a unique NRPS extender unit derived from ketose phosphates through (α,β-dihydroxyethyl)-ThDP and a lipoyl group-tethered ester intermediate catalyzed by the TKase-ACP platform in the context of NDM and QNC biosynthesis (Fig. 6C). This strategy is totally different from that mentioned before about the biosynthesis of α-hydroxyacyl units in nonribosomal peptides. The recently published gene cluster of ET-743 also includes an etuP1/etuP2 pair encoding two components of the PDH complex with moderate sequence similarity and an identical domain organization to NapB/NapD or QncN/QncL, while the function of these two enzymes remains unclear (18). Our findings indicated that the etuP1/etuP2 pair should also play a similar role in the biosynthetic pathway of ET-743.
Thomas and coworkers reported another strategy for the biosynthesis of a unique α-hydroxyacid originating from 1,3-bisphosphateglycerate in the biosynthetic pathway of zwittermicin (32): A proposed glycolytic pathway intermediate is converted to a glyceryl-ACP intermediate as a PKS extender unit (Fig. 6B); while tethered to the ACP, the 3-hydroxyl of the glyceryl moiety is converted to an acid by the concerted actions of enzymes similar to 3-hydroxybutyryl-CoA dehydrogenases and acyl-CoA dehydrogenases. This mechanism is totally different from that of QncN/QncL. Studies on the biosynthetic pathway of Yersiniose A (26) showed that YerE (Fig. 6A) is also thiamine pyrophosphate-dependent, involving the formation of hydroxyethyl-ThDP as the nucleophilic two-carbon donor; however, the origin of the two carbons and the cleavage method for leaving the group are different from QncN/QncL: the two-carbon donor was formed by decarboxylation of thiamine pyrophosphate-bound intermediate originating from pyruvate and directly transferred to the sugar by the formation of a C—C bond.
It is totally unexpected for NRPS to incorporate hydroxyethyl C2 precursors directed from ketose, and the TKase-ACP/NRPS system is also rare for natural products biosynthesis. To our knowledge, the only similar enzyme is NasB, which, containing D1 to D4 domains and a β-ketoacyl-ACP synthase III domain, was very recently genetically characterized to convert branched-chain α-ketoacids into branched-chain acyl-ACPs involved in the biosynthesis of branched long-chain N-acyl amino acids (33). Although the domain organization from D1 to D4 is similar, the substrate and enzymatic process are totally different. In parallel to the interests in novel extender units for polyketide biosynthesis (34, 35), our finding exemplifies an alternate strategy in hybrid NRPS biochemistry and also set the stage for further combinatorial biosynthesis toward ET-743.
Materials and Methods
General.
NDM producer S. lusitanus NRRL 8034 was purchased from American Agricultural Research Service Culture Collection and was cultured as described previously (19–21). E. coli DH5α competent cells were used for routine subcloning and plasmid preparations. pCC1FOS vector (Epicentre) was used for fosmid library preparations. E. coli cells were grown in LB medium with appropriate antibiotics, when necessary. A complete list of the strains and plasmids used in this study is presented in Table S1. All other common biochemicals and chemicals were from standard commercial sources. PCR amplification was carried out by using either Taq DNA polymerase or PfuUltra DNA polymerase with genomic DNA or fosmid as a template and degenerate or specific primers, as listed in Table S2. Primer synthesis was performed at Invitrogen Shanghai Center. DNA sequencing was performed at Shanghai GeneCore Biotechnology. The genetic manipulations of S. lusitanus, fermentation, analysis of production, protein expression, and purification were performed by following the methods detailed in SI Materials and Methods.
Biochemical Assays of Glycolicacylation of QncM Catalyzed by QncN/QncL.
Standard assays were carried out in 50 μL of reaction mixture containing 50 mM Mops (pH 7.5), 2.5 mM MgCl2, 400 μM ThDP, 100 μM holo-ACP, 50 μg (1 mg/mL) QncN/QncL, 5 μM PDH E3, 250 μM FAD, and 1 mM NAD. The reaction was initiated by addition of donor substrate F-6-P (2 mM) and incubated at 30 °C. The reactions were quenched by the addition of trifluoroacetic acid (TFA) to a final concentration of 10%; after centrifugation, the clarified supernatant was subjected to HPLC analysis. The details for analysis, assay of the time courses, and the kinetics for the donor substrates are all provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Zixin Deng’s laboratory at Shanghai JiaoTong University for support in obtaining MS data of proteins. This work was supported in part by National Basic Research Program of China (973 Program) Grants 2010CB833200 and 2009CB118901; National Natural Science Foundation of China Grants 90913005, 21072214, 20832009, and 20921091; and the Chinese Academy of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences for and reported in this paper have been deposited in the GenBank database [accession nos. JQ765000 (napB–napC–napD) and JQ764999 (qncN–qncM–qncL)].
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204232109/-/DCSupplemental.
References
- 1.Felnagle EA, et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sieber SAM, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson B, Micklefield J. Chapter 14. Biosynthesis of nonribosomal peptide precursors. Methods Enzymol. 2009;458:353–378. doi: 10.1016/S0076-6879(09)04814-9. [DOI] [PubMed] [Google Scholar]
- 6.Walsh CT, Fischbach MA. Natural products version 2.0: Connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: In vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 8.Scott JD, Williams RM. Chemistry and biology of the tetrahydroisoquinoline antitumor antibiotics. Chem Rev. 2002;102:1669–1730. doi: 10.1021/cr010212u. [DOI] [PubMed] [Google Scholar]
- 9.Bernan VS, et al. Bioxalomycins, new antibiotics produced by the marine Streptomyces sp. LL-31F508: Taxonomy and fermentation. J Antibiot (Tokyo) 1994;47:1417–1424. doi: 10.7164/antibiotics.47.1417. [DOI] [PubMed] [Google Scholar]
- 10.Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Drug development from marine natural products. Nat Rev Drug Discov. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 11.Cuevas C, Francesch A. Development of Yondelis (trabectedin, ET-743). A semisynthetic process solves the supply problem. Nat Prod Rep. 2009;26:322–337. doi: 10.1039/b808331m. [DOI] [PubMed] [Google Scholar]
- 12.Siengalewicz P, Rinner U, Mulzer J. Recent progress in the total synthesis of naphthyridinomycin and lemonomycin tetrahydroisoquinoline antitumor antibiotics (TAAs) Chem Soc Rev. 2008;37:2676–2690. doi: 10.1039/b804167a. [DOI] [PubMed] [Google Scholar]
- 13.Liao XW, et al. Synthetic progress of the tetrahydroisoquinoline antitumor alkaloids. Chinese J Org Chem. 2010;30:317–329. [Google Scholar]
- 14.Pospiech A, Cluzel B, Bietenhader J, Schupp T. A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology. 1995;141:1793–1803. doi: 10.1099/13500872-141-8-1793. [DOI] [PubMed] [Google Scholar]
- 15.Velasco A, et al. Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2-2: Designing new cytotoxic compounds. Mol Microbiol. 2005;56:144–154. doi: 10.1111/j.1365-2958.2004.04433.x. [DOI] [PubMed] [Google Scholar]
- 16.Li L, et al. Characterization of the saframycin A gene cluster from Streptomyces lavendulae NRRL 11002 revealing a nonribosomal peptide synthetase system for assembling the unusual tetrapeptidyl skeleton in an iterative manner. J Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koketsu K, Watanabe K, Suda H, Oguri H, Oikawa H. Reconstruction of the saframycin core scaffold defines dual Pictet-Spengler mechanisms. Nat Chem Biol. 2010;6:408–410. doi: 10.1038/nchembio.365. [DOI] [PubMed] [Google Scholar]
- 18.Rath CM, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zmijewski M, Jr, Mikolajczak M, Viswanatha V, Hruby VJ. Biosynthesis of the antitumor antibiotic naphthyridinomycin. J Am Chem Soc. 1982;104:4969–4971. [Google Scholar]
- 20.Zmijewski M, Jr, Palaniswamy VA, Gould SJ. Naphthyridinomycin biosynthesis. The involvement of ornithine and the origin of the oxazolidine nitrogen. J Chem Soc Chem Commun. 1985;18:1261–1262. [Google Scholar]
- 21.Palaniswamy VA, Gould SJ. The incorporation of 3′-methyltyrosine and 5′-methyl DOPA into naphthyridinomycin. J Am Chem Soc. 1986;108:5651–5652. [Google Scholar]
- 22.Fu C-Y, et al. Biosynthesis of 3-hydroxy-5-methyl-O-methyltyrosine in the saframycin/safracin biosynthetic pathway. J Microbiol Biotechnol. 2009;19:439–446. doi: 10.4014/jmb.0808.484. [DOI] [PubMed] [Google Scholar]
- 23.Peng C, et al. In vivo investigation of the role of SfmO2 in saframycin A biosynthesis by structural characterization of the analogue saframycin O. Sci China Chem. 2012;55:90–97. [Google Scholar]
- 24.Tang M-C, Fu C-Y, Tang G-L. Characterization of SfmD as a Heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis. J Biol Chem. 2012;287:5112–5121. doi: 10.1074/jbc.M111.306316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Kok A, Hengeveld AF, Martin A, Westphal AH. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim Biophys Acta. 1998;1385:353–366. doi: 10.1016/s0167-4838(98)00079-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Guo Z, Liu HW. Biosynthesis of Yersiniose: Attachment of the two-carbon branched-chain is catalyzed by a thiamine pyrophosphate-dependent flavoprotein. J Am Chem Soc. 1998;120:11796–11797. [Google Scholar]
- 27.Kochetov GA, Sevostyanova IA. Binding of the coenzyme and formation of the transketolase active center. IUBMB Life. 2005;57:491–497. doi: 10.1080/15216540500167203. [DOI] [PubMed] [Google Scholar]
- 28.Fiedler E, et al. Examination of donor substrate conversion in yeast transketolase. J Biol Chem. 2001;276:16051–16058. doi: 10.1074/jbc.M007936200. [DOI] [PubMed] [Google Scholar]
- 29.Feifel SC, et al. In vitro synthesis of new enniatins: Probing the α-d-hydroxy carboxylic acid binding pocket of the multienzyme enniatin synthetase. ChemBioChem. 2007;8:1767–1770. doi: 10.1002/cbic.200700377. [DOI] [PubMed] [Google Scholar]
- 30.Magarvey NA, Ehling-Schulz M, Walsh CT. Characterization of the cereulide NRPS α-hydroxy acid specifying modules: Activation of α-keto acids and chiral reduction on the assembly line. J Am Chem Soc. 2006;128:10698–10699. doi: 10.1021/ja0640187. [DOI] [PubMed] [Google Scholar]
- 31.Calderone CT, Bumpus SB, Kelleher NL, Walsh CT, Magarvey NA. A ketoreductase domain in the PksJ protein of the bacillaene assembly line carries out both α- and β-ketone reduction during chain growth. Proc Natl Acad Sci USA. 2008;105:12809–12814. doi: 10.1073/pnas.0806305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan YA, et al. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc Natl Acad Sci USA. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig JW, Brady SF. Discovery of a metagenome-derived enzyme that produces branched-chain acyl-(acyl-carrier-protein)s from branched-chain α-keto acids. ChemBioChem. 2011;12:1849–1853. doi: 10.1002/cbic.201100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: The functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep. 2012;29:72–86. doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.