Abstract
Intron-containing genes are often transcribed more efficiently than nonintronic genes. The effect of introns on transcription of genes is an evolutionarily conserved feature, being exhibited by such diverse organisms as yeast, plants, flies, and mammals. The mechanism of intron-mediated transcriptional activation, however, is not entirely clear. To address this issue, we inserted an intron in INO1, which is a nonintronic gene, and deleted the intron from ASC1, which contains a natural intron. We then compared transcription of INO1 and ASC1 genes in the presence and absence of an intron. Transcription of both genes was significantly stimulated by the intron. The introns have a direct role in enhancing transcription of INO1 and ASC1 because there was a marked increase in nascent transcripts from these genes in the presence of an intron. Intron-mediated enhancement of transcription required a splicing competent intron. Interestingly, both INO1 and ASC1 were in a looped configuration when their genes contained an intron. Intron-dependent gene looping involved a physical interaction of the promoter and the terminator regions. In addition, the promoter region interacted with the 5′ splice site and the terminator with the 3′ splice site. Intron-mediated enhancement of transcription was completely abolished in the looping defective sua7-1 strain. No effect on splicing, however, was observed in sua7-1 strain. On the basis of these results, we propose a role for gene looping in intron-mediated transcriptional activation of genes in yeast.
Keywords: chromosome conformation capture, RNA polymerase II
The protein encoding genes in eukaryotes differ from their prokaryotic counterparts in having noncoding intervening regions called introns, which are removed by splicing to generate mature mRNA. Since their discovery in 1977, there has been considerable debate regarding the functional role of introns in eukaryotes (1). It is widely believed that introns increase proteomic complexity by facilitating expression of multiple proteins from a single gene by alternative splicing (2). In budding yeast, where more than 95% of genes are without introns and there are very few instances of alternative splicing, introns do not contribute significantly to the proteomic diversity (3). The presence of introns in all eukaryotes, despite the high cost of maintaining them and the existence of the elaborate splicing machinery needed to remove them, suggest that introns are playing a more fundamental and evolutionarily conserved role in eukaryotic cells.
One role of introns that has been remarkably conserved among diverse organisms, and which confers an additional advantage to eukaryotic genes, is their effect on efficiency of gene expression (4–6). Introns significantly enhance the transcriptional output of genes that harbor them. The expression level of intronless transgenes in mammalian cells is often 10–100 times lower than their intron-containing counterparts (5). The inclusion of just one intron near the 5′ end of the gene increases transcription of the gene many folds. A number of mammalian genes, including β-globin, growth hormone, thymidylate synthase, purine nucleoside phosphorylase, cathepsin L, and HIV-1 require introns for their normal expression (7–14). A similar intron-dependent stimulation of transcription has been observed in a variety of genes in plants (6, 15).
In yeast, less than 5% of genes contain introns, but these intron-containing genes produce approximately 27% of total cellular mRNA (4, 16). On average, intronic genes produce 3.9-fold more mRNA than their nonintronic counterparts in yeast (3). Removal of introns from yeast genes, like in plants and mammalian systems, decreases their mRNA output (3, 17). Deletion of introns from several yeast genes affected their transcription sufficiently to cause a phenotypic growth defect (3, 18). Introns also have been shown to stimulate transcription in flies. However, there are genes in all classes of organisms whose expression remains high irrespective of the presence or absence of introns, whereas a minority of genes are negatively regulated by introns (5, 6). In some cases, introns increase translational output without any increase in transcription of the gene by either affecting the stability of mRNA or by facilitating transport of mRNA out of the nucleus into the cytoplasm (5, 6).
The intron-mediated stimulation of transcription is physiologically relevant for several reasons: (i) A significant number of eukaryotic genes exhibit the phenomenon; (ii) it is an evolutionary conserved feature that is exhibited by such diverse organisms as yeast, nematodes, flies, plants, and mammals; and (iii) it often confers fitness to the organism by allowing tissue specific expression or developmentally regulated expression.
Intron-mediated transcriptional regulation can be broadly divided into two categories: (i) splicing-independent regulation and (ii) splicing-dependent regulation. Introns can often stimulate transcription because of the presence of an enhancer or a promoter element within their sequence (6). Such introns can influence transcription even if their orientation is reversed (19). In contrast, splicing-dependent regulation of transcription requires a functional, splicing-competent intron within the body of the gene. Such introns cannot affect transcription if their splicing is compromised by a mutation in the conserved sequences at the 5′ splice site, 3′ splice site, branchpoint or if they are inserted in an antisense orientation (8, 17). This direct effect of introns on transcription of genes is often referred to as “intron-mediated enhancement” (IME) and will be the focus of this investigation (6). IME requires the presence of an intron near the 5′ end of the gene. It has been proposed that a promoter proximal 5′ splice site facilitates recruitment of the transcription machinery to the promoter and, therefore, helps in initiation of transcription. The interaction of U1 snRNA with general transcription factor TFIIH and the effect of this interaction on reinitiation of transcription provided support to this hypothesis (20). The precise mechanism of intron-dependent enhancement of transcription, however, is not entirely clear.
Here, we show that inclusion of an intron in INO1 resulted in constitutive activation of the gene. The intron-mediated activation was due to an increase in nascent transcription of INO1, required splicing, and was independent of the transcription activator Ino2. In the presence of the intron, the promoter of INO1 interacted with its terminator region to form a gene loop. Similar results were obtained with ASC1. Intron-mediated transcriptional activation was abolished in the looping defective sua7-1 strain. These results suggest a role for gene looping in intron-mediated enhancement of transcription in budding yeast.
Results
Intron-Mediated Enhancement of Transcription of INO1 and ASC1.
To investigate intron-dependent enhancement of transcription in budding yeast, we inserted an intron into an intronless gene and deleted the intron from a natural intron-containing gene. Transcription of the genes was then compared under the intron-plus and intron-minus conditions.
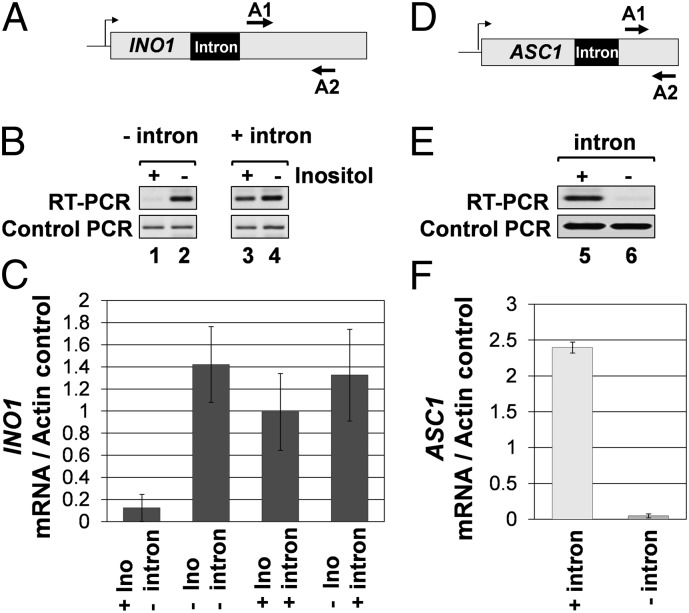
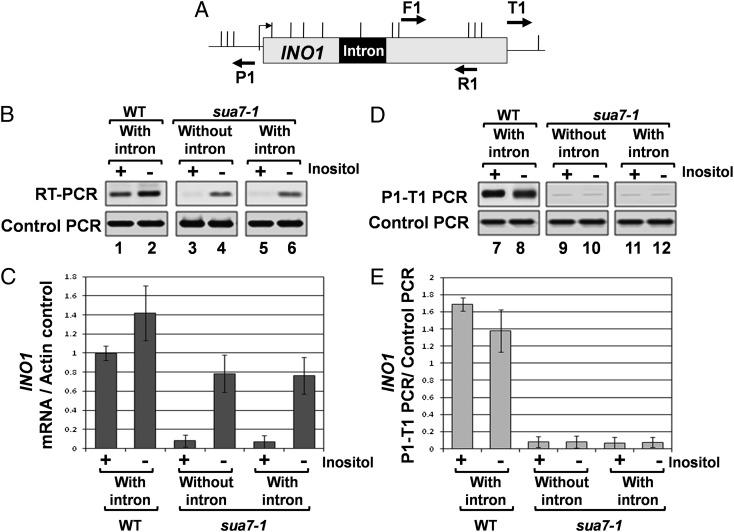
INO1 is an intronless gene whose transcription is regulated by inositol. The gene is repressed in the presence of inositol and is transcriptionally activated upon depletion of inositol from the growth medium (21, 22). To examine the effect of an intron on transcription of INO1, a 308-nt-long ACT1 intron was inserted in the gene 500 bp downstream of the initiator codon (SI Appendix, Fig. 1). The inserted intron was splicing competent because it was efficiently removed from the INO1 transcripts (SI Appendix, Fig. 2C, lanes 5 and 6). Transcription of INO1 was monitored in cells grown in the presence or absence of inositol by RT-PCR. The intron had little effect on the transcript level of INO1 under inducing conditions (Fig. 1 B, lanes 2 and 4 and C). However, there was a 10-fold increase in steady-state level of INO1 mRNA in the presence of inositol in the medium under intron-plus condition (Fig. 1 B, lanes 1 and 3 and C). Thus, the presence of intron caused an increased accumulation of INO1 transcripts under noninducing conditions.
Fig. 1.
The presence of an intron leads to increased accumulation of INO1 and ASC1 mRNA. (A and D) Schematic depiction of intron-containing INO1 and ASC1 genes indicating A1 and A2 primers used in RT-PCR analysis. (B and E) RT-PCR analysis of INO1 and ASC1 genes with and without an intron. The reverse transcription was performed by using oligo dT, and the cDNA obtained was PCR amplified by A1 and A2 primers. The control PCR represent the ACT1 RNA levels, which is used as a loading control indicating that equal amounts of template were present in each RT-PCR. (C and F) Quantification of transcript levels of the intron-less and intron-containing INO1 and ASC1.
An increase in transcript level of a gene can be attributed either to enhanced transcription of the gene or to an increase in mRNA stability (23). To address this issue, transcription run-on (TRO) analysis of INO1 was performed in intron-plus and intron-minus states of the gene. The TRO assay measures the level of nascent transcripts that are still attached to the elongating polymerase on the template. In the native intron-less INO1, only a weak TRO signal could be detected on the gene when the cells were grown in inositol-containing medium (SI Appendix, Fig. 3 B, lanes 1–8 and C). In the presence of an intron, however, there was a marked increase in TRO signal throughout the gene under noninducing conditions (SI Appendix, Fig. 3 E, lanes 25–29 and F). These results clearly indicate that the intron-dependent increase in RNA level of INO1 under noninducing conditions is due to transcriptional activation of the gene. Furthermore, the presence of the intron conferred constitutive activation of INO1 (Fig. 1C and SI Appendix, Fig. 3F).
We next asked whether deletion of the native intron from ASC1 had a reciprocal effect on gene expression. ASC1 has a single 272-nt-long intron positioned 583 bp downstream of the start codon (Fig. 1D). The intron was precisely removed from the chromosomal copy of the gene (SI Appendix, Fig. 4). Deletion of the intron resulted in a 25-fold decrease in the transcript level of ASC1 (Fig. 1 E, lanes 5 and 6 and F). TRO analysis revealed at least a 10-fold decrease in nascent transcription of ASC1 after removal of the intron (SI Appendix, Fig. 5 B, lanes 2–4 and 9–11 and C). Similar results were reported earlier with a plasmid borne copy of ASC1 (17). An increase in TRO signal at the 3′ end of ASC1 was observed in the absence of intron (SI Appendix, Fig. 5B, lane 12). This result could be due to the presence of a promoter for transcription of anti-sense RNA at the 3′ end of ASC1. To check this possibility, we performed RT-PCR analysis for the ASC1 anti-sense transcript near the 3′ end of the gene. We could not detect any signal for anti-sense RNA at the 3′ end of intron-less ASC1 (SI Appendix, Fig. 6B, lane 1), but we could observe the signal for sense transcript (SI Appendix, Fig. 6B, lane 3). These results suggest that TRO signal at the 3′ end of intron-less ASC1 is not due to anti-sense transcription but due to the accumulation of transcribing RNAP II molecules near the terminator region of ASC1. The overall conclusion of these experiments is that both INO1 and ASC1 exhibit intron-dependent activation of transcription in yeast cells.
Intron-Mediated Enhancement Depends on Splicing.
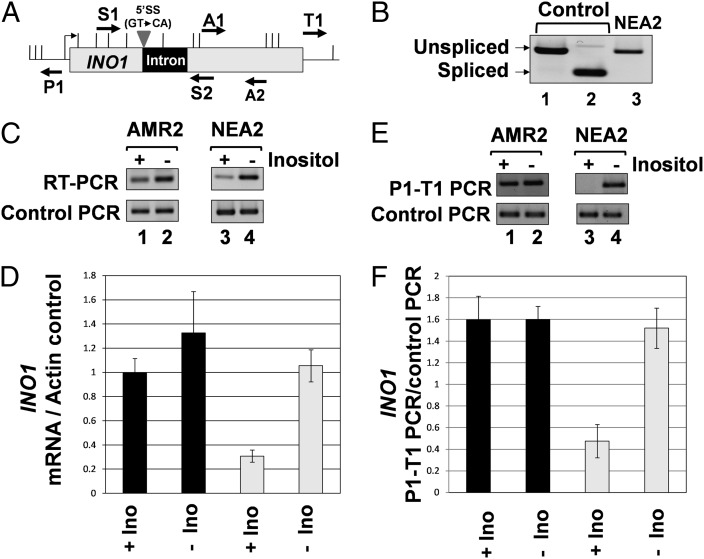
Introns located in the 5′ UTR of several plant and mammalian genes function as enhancer elements (6). Such introns bring about transcriptional activation of genes in a splicing-independent manner. To determine whether intron-mediated enhancement of INO1 transcription was due to the splicing function of intron, the 5′ splice site of ACT1 intron was mutated from GT to CA as described in Furger et al. (17). The mutant intron was inserted in INO1 gene as described. As expected, the intron with a mutated 5′ splice site could not be spliced out of INO1 precursor mRNA, and a longer transcript was produced (Fig. 2B, lane 3). Results from RT-PCR analysis revealed that the intron-mediated transcriptional stimulation of INO1 under noninducing conditions decreased by approximately 60–70% in the presence of a splicing defective intron (Fig. 2C, lane 3). Thus, the intron-mediated increase in transcription of INO1 under noninducing conditions depends on its splicing function.
Fig. 2.
Splicing is required for intron-dependent enhancement of INO1 transcription and gene looping. (A) Schematic depiction of INO1 with a 5′ splice site mutated intron indicating the position of AluI and EcoRV restriction sites (vertical lines) and PCR primers used in RT-PCR and CCC analysis. The strain with the mutated 5′ splice site is NEA2, whereas the strain with the wild-type splice sites is AMR2 (B) ACT1 intron with a mutated 5′ splice site is not removed from INO1 pre-mRNA as indicated by the position of RT-PCR product obtained by using primers S1 and S2 flanking the intronic region of INO1 (lane 3). Control PCRs with and without intron were obtained by using genomic DNA from AMR2 and BY4733 as template using primers S1 and S2. (C) RT-PCR analysis of INO1 harboring a 5′ splice site mutated intron. The reverse transcription was performed by using oligo dT, and the cDNA obtained was PCR amplified by A1 and A2 primers. The control PCR represent the ACT1 mRNA levels that are used as a loading control, indicating that equal amount of template was present in each of the RT-PCRs. (D) Quantification of the data shown in C. (E) CCC analysis of INO1 containing a 5′ splice site mutated intron. The PCR products are indicated by the primers used for amplification. A P1-T1 PCR product indicates a looped configuration. The A1-A2 PCR represent the loading control, indicating that equal amounts of template DNA were present in each CCC PCR. (F) Quantification of the data shown in E.
Activated transcription of INO1 requires the transcription activator Ino2, which is constitutively bound to the UAS element of INO1 but brings about stimulation of transcription only in the absence of inositol (21, 24). To determine whether intron-mediated enhancement of INO1 requires Ino2, we repeated RT-PCR in an ino2− strain containing INO1with an intron. The intron-mediated constitutive activation of INO1 was maintained in ino2− strain (SI Appendix, Fig. 7B, lanes 1 and 2). These data indicate that intron-dependent enhancement of INO1 transcription does not require the activator protein.
Gene Looping Accompanies Intron-Mediated Enhancement of Transcription.
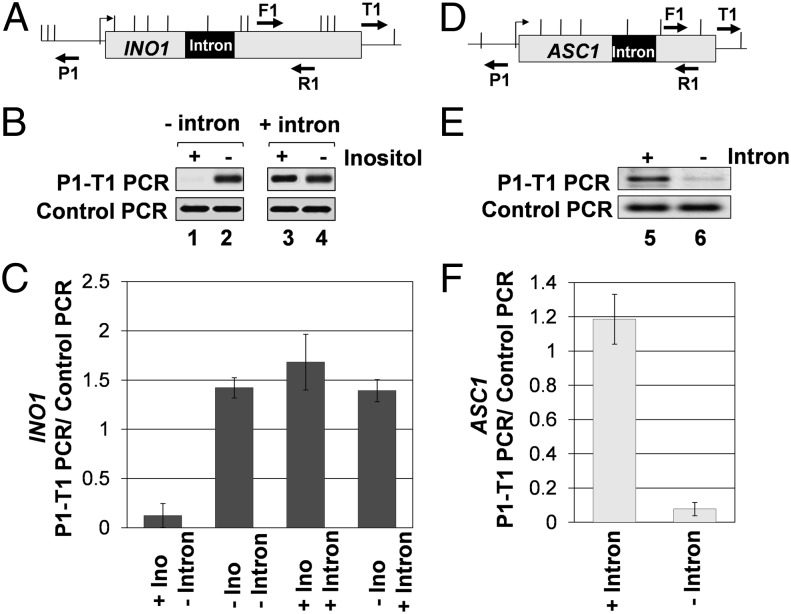
Gene looping is defined as the physical interaction of the promoter and terminator regions of a gene in a transcription-dependent manner (25). We earlier demonstrated that Ino2-mediated transcriptional activation of INO1 is accompanied by formation of a looped gene configuration (21). We also showed that gene looping accompanies activator-dependent increase in transcription of other yeast genes as well. We therefore asked whether intron-mediated enhancement of INO1 transcription also results in the formation of a looped structure. To detect gene loops, the chromosome conformation capture (CCC) assay was used. We have used this assay to show transcription-dependent looping of several yeast genes (21, 25, 26). A PCR product obtained by using divergent primers P1 and T1 was taken as a measure of the interaction of the promoter and terminator regions in these experiments (Fig. 3 A and D). To find out whether intron-mediated enhancement is accompanied by gene looping, CCC analysis of intron-containing INO1 was carried out in the cells grown in the presence and absence of inositol. A robust P1-T1 looping signal was obtained for intronic INO1 both in the presence and absence of inositol (Fig. 3B, lanes 3 and 4). This result is in contrast to nonintronic INO1, which exhibited gene looping only in the absence of inositol (Fig. 3B, lane 2). The extent of gene looping in the presence of the intron was almost the same under inducing and noninducing conditions (Fig. 3 B and C). The insertion of intron with the mutated 5′ splice site did not result in a looped gene configuration of INO1 under noninducing conditions (Fig. 2E, lane 3). Similar results were obtained with ASC1. In the presence of the intron, CCC analysis of ASC1 yielded a distinct P1-T1 PCR product (Fig. 3E, lane 5). This signal was reduced more than 10-fold after removal of the intron from the gene (Fig. 3E, lane 6 and F). These analyses reveal that both INO1 and ASC1 are in a looped configuration during intron-mediated enhancement of transcription.
Fig. 3.
Gene looping accompanies intron-mediated stimulation of transcription of INO1 and ASC1. (A and D) Schematic depiction of the intron-containing INO1 and ASC1 genes, indicating the position of restriction sites (vertical lines) and PCR primers used in CCC analysis. (B and E) CCC analysis of intronless and intron-containing INO1 and ASC1. The PCR products are indicated by the primers used for amplification. A P1-T1 PCR product indicates a looped configuration. The F1-R1 PCR represent the loading control indicating that equal amounts of template DNA were present in each CCC PCR. (C and F) Quantification of the CCC results shown in B and E.
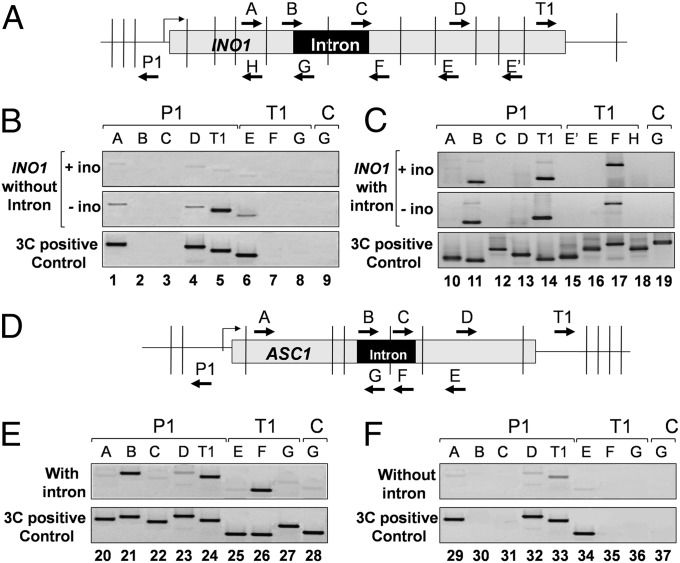
CCC mapping of looping interactions across the INO1 and ASC1 genes revealed that the promoter, in addition to contacting the terminator (Fig. 4 C, lane 14 Top and Middle and E, lane 24), also interacted with the region near 5′ splice site (Fig. 4 C, lane 11 Top and Middle and E, lane 21). Furthermore, the terminator region made contact with the 3′ splice site (Fig. 4C, lane 17 Top and Middle and E, lane 26). We were, however, unable to detect a direct interaction of the 5′ splice site with the 3′ splice site (Fig. 4 C, lane 19 Top and Middle and E, lane 28), and we did not detect interaction of the promoter or terminator with any other internal region of either genes (Fig. 4 C, lanes 10, 12, 13, 15, 16, and 18 Top and Middle and E, lanes 20, 22, 23, 25, and 27). A similar interaction of the intron with the promoter and terminator regions was observed during gene looping of human BRCA1 (27).
Fig. 4.
Interaction of splice sites with the promoter and terminator regions during gene looping. (A and D) Schematic depiction of intron-containing INO1 and ASC1, indicating the position of restriction sites (vertical lines) and PCR primers used in the high-resolution CCC mapping. (B, C, E, and F) PCR products derived from the indicated primer pairs by CCC analysis show that the promoter interacts with the 5′ splice site and the terminator with the 3′ splice site in the intron-containing INO1 and ASC1. The CCC positive control PCR products were generated to check the amplification efficiency of the primer pairs in the PCR.
Gene Looping Has a Possible Role in Intron-Mediated Enhancement of Transcription.
The results described above demonstrate looping of INO1 and ASC1 during intron-mediated enhancement of transcription. It was, however, not clear from these experiments whether gene looping was responsible for intron-mediated stimulation of transcription. To address the issue, we analyzed transcription of intron-containing INO1 in the looping-defective TFIIB mutant, sua7-1 (28). TFIIB is an important determinant of gene looping in budding yeast (21, 29). The sua7-1 mutation abolishes gene looping (29). This mutant provides a convenient way to determine whether a cellular process depends on gene looping. Using this mutant, it was demonstrated that gene looping is required for maintenance of “transcriptional memory” in yeast (30, 31). Thus, to determine whether gene looping is required for intron-mediated enhancement, we assayed transcription of INO1 with an intron in sua7-1 cells by the RT-PCR approach. We did not observe any intron-mediated increase in the RNA level of INO1 under noninducing conditions in sua7-1 cells (Fig. 5 B, lane 5 and C). In a parallel experiment, CCC analysis showed the loss of gene looping of intronic INO1 in sua7-1 cells (Fig. 5 D, lanes 11 and 12 and E). These results demonstrate that intron-mediated constitutive enhancement of INO1 transcription was abolished in the looping defective sua7-1 mutant. Because splicing is required for intron-mediated stimulation of INO1 (Fig. 2), gene looping may directly influence intron-dependent enhancement of transcription or it may do so indirectly by affecting splicing. To clarify the issue, we checked for a possible splicing defect in sua7-1 strain by RT-PCR (SI Appendix, Fig. 2D). The primers were designed so that a spliced RNA will give a shorter PCR product, whereas an unspliced RNA will give a longer PCR product because of the presence of intron (SI Appendix, Fig. 2A). We found that the intron-containing INO1 precursor mRNAs were spliced as efficiently in sua7-1 cells as in wild-type cells (SI Appendix, Fig. 2 C, lanes 5 and 6 and D, lane 9). This observation ruled out the possibility of gene looping indirectly influencing intron-dependent transcription through splicing. These results suggested a more direct role for gene looping in intron-mediated enhancement of transcription. The possibility of the TFIIB mutation (sua7-1) affecting intron-mediated transcriptional activation through an aspect of transcription other than gene looping, however, still could not be ruled out.
Fig. 5.
Intron-dependent enhancement of transcription and gene looping are abolished in sua7-1 strain. (A) Schematic depiction of intron-containing INO1, indicating the position of AluI and EcoRV restriction sites (vertical lines) and PCR primers used in CCC analysis. (B) RT-PCR analysis of intronless and intron-containing INO1 in wild-type and sua7-1 strains. The reverse transcription was performed by using oligo dT, and the cDNA obtained was then PCR amplified by A1 and A2 primers shown in Fig. 1. The control PCR represents the ACT1 mRNA levels, which are used as a loading control, indicating that equal amounts of template was present in each of the RT-PCRs. (C) Quantification of RT-PCR results shown above in B. (D) CCC analysis of intronless and intron-containing INO1 in wild-type and sua7-1 strains after 3 h of induction. The PCR products are indicated by the primers used for amplification. A P1-T1 PCR product indicates a looped configuration. The F1-R1 PCR represents the loading control, indicating that equal amounts of template DNA were present in each CCC PCRs. (E) Quantification of the CCC results shown above in D.
Because gene looping of intronic INO1 involved juxtaposition of the promoter and terminator regions with each other, and with the ends of the intron, we next examined the interaction of 5′ and 3′ splice site with the promoter and terminator regions, respectively, in sua7-1 strain. We found that not only was the promoter-terminator contact abolished in the looping defective strain, but interaction of promoter with the 5′ splice site and the terminator with the 3′ splice site was also abrogated in sua7-1 cells (SI Appendix, Fig. 8B). Thus, there was a complete loss of intron-dependent looped gene architecture in looping defective cells.
Discussion
Here, we provide evidence that intron-mediated enhancement of transcription in yeast requires a splicing-competent intron and occurs when the gene is in a looped configuration. The presence of an intron may facilitate juxtaposition of the promoter and the terminator regions of a gene resulting in a looped gene structure. In the looping-defective sua7-1 mutant, although splicing was normal, there was no enhancement of transcription. The intron-mediated enhancement of transcription is therefore not due to splicing per se but may be due to the formation of a splicing-dependent looped architecture of the gene.
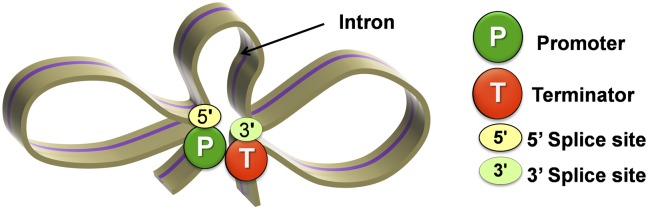
It was demonstrated that a 5′ splice site alone can bring about an increase in transcription of HIV-1 and β-globin genes (10). The enhancement of transcription elicited by a 5′ splice site, however, was much lower (75% less) compared with that brought about by a full-length intron. Our results show that in a looping defective sua7-1 strain, even a splicing competent intron cannot evoke a transcription activation response. The results presented here suggest that it is not merely the presence of a 5′ splice site or an intron, but an intron-facilitated looped gene configuration, that confers transcriptional enhancement of a gene. We further show that during intron-dependent gene looping, there are additional contacts between the promoter and 5′ splice site and the terminator and 3′ splice site (Fig. 6). The promoter–5′ splice site interaction explains why the 5′ splice site alone is able to bring about a modest increase in transcription of a gene on its own. However, to achieve the intron-mediated enhancement to the fullest possible extent, further contacts of the promoter region with the terminator are essential. How the presence of an intron facilitates gene loop formation is not yet clear. We propose that it is the interaction of 5′ splice site with the promoter and 3′ splice site with terminator that bring the two ends of a gene in close physical proximity and facilitate the promoter-terminator contact.
Fig. 6.
A model of intron-dependent gene looping.
Because looping-defective sua7-1 strain has a mutated TFIIB, it is possible that the loss of intron-mediated enhancement of transcription in sua7-1 mutant is not due to impairment of gene loop formation, but due to some other transcription defect associated with this mutation. The sua7-1 codes for a mutated form of TFIIB with glutamic acid at position 62 being replaced by lysine (TFIIB-E62K). The mutant is cold sensitive and exhibits altered transcription start site selection (32). The activator-dependent transcription that requires gene looping exhibited a kinetic lag in the sua7-1 mutant (21, 30). The binding affinity of TFIIB-E62K for the promoter region and its interactions with TBP and RNAP II are comparable to that of wild-type TFIIB (33). The recruitment of general transcription factors and RNAP II onto the promoter during assembly of preinitiation complex is also normal in the presence of TFIIB-E62K (33). The cross-linking of mutated TFIIB to the terminator region of actively transcribed genes, however, is severely compromised in sua7-1 cells (29). The localization of wild-type TFIIB to the 3′ end of genes is also abolished in the looping-defective mutants of Ssu72 and Rna15, which are 3′ end processing/termination factors (21, 25, 29). Because TFIIB has been shown to physically interact with Ssu72 and Rna15 (21, 34, 35), it has been proposed that TFIIB interaction with the terminator-bound factors is the molecular basis of gene looping (21, 25, 29). Accordingly, we recently purified a complex of TFIIB with a number of terminator-bound factors from yeast cells (36). This TFIIB–termination factor complex, which has been proposed to facilitate gene loop formation by bridging the promoter and the terminator regions, was not observed in looping-defective sua7-1 cells. The overall conclusion of these results is that TFIIB-E62K (sua7-1) is defective in its interaction with the terminator-bound factors and, consequently, transcriptionally activated genes are no longer in a looped configuration. Taken together, these results suggest a role for gene looping in intron-mediated enhancement of transcription in yeast cells.
A critical issue is how intron-dependent gene looping brings about enhancement of transcription. It has been proposed that when a gene is in a looped configuration, the proximity of the terminator to the promoter region facilitates release of polymerase from the terminator region. The polymerase is then recycled back to the juxtaposed promoter for reinitiation of transcription. Such a coupling of termination to reinitiation, with a concomitant increase in the transcriptional activity, has been demonstrated for RNAP III, RNAP I, mitochondrial polymerase, and archaeal polymerase (37–41). Recently, the terminator-promoter cross-talk also was shown during transcription by RNAP II (42). Several possible mechanisms have been proposed to explain transfer of polymerase from the terminator to the promoter for reinitiation (43). Gene looping is by far the most attractive of these proposed mechanisms. However, rigorous experimental evidence is needed to show that gene looping facilitates the release and transfer of RNAP II from the 3′ end to the 5′ end of a gene after termination of transcription. Nevertheless, this study provides an insight into the mechanism of intron-mediated enhancement of transcription in yeast that also may help understand the phenomenon in higher eukaryotes.
Materials and Methods
Yeast Strains.
The yeast strains used in this study are listed in SI Appendix, Table S1. Strains BY4733, AMR2, AMR6, and AMR15 are isogenic. Strain AMR2 was derived from BY4733 by introducing an intron in the INO1 gene at the 500 bp position of the ORF as described in Cheng et al. (44). Strain AMR6 was derived from AMR2 by inserting the kanamycin resistance gene (kanMX6) between INO1 and the downstream SNA3 genes. Strain AMR15 was derived from AMR2 by replacing the entire ORF of INO2 by KanMX6 as described in Wach et al. (45).
AMR14 was derived from W303-1A by first replacing the entire ORF of ASC1 by KanMX6 as described in Wach et al. (45). Second, an intron-less ASC1 was obtained by PCR amplifying ASC1 cDNA using primers that contained adaptor sequences homologous to the regions upstream and downstream of ASC1 ORF. Third, the intron-less ASC1 was inserted back into the yeast genome by homologous recombination. Replica plating was performed to make sure that the KanMX6 marker was lost, and the correct insertion of the intronless ASC1 gene was verified by PCR.
Strain MHA1 was derived from YMH124 by introducing an intron in the INO1 gene at the 500 bp position of the ORF as described earlier. NEA2 is identical to AMR2 except that the 5′ splice site of the intron integrated into INO1 was mutated from GT to CA.
Cell Culture.
Cells for RT-PCR and CCC analyses of INO1 were grown as described in El Kaderi et al. (21). Induction of INO1 in all experiments was performed by growing cells for 3 h in inositol-depleted medium. For ASC1, cells were grown in YP-dextrose till A600 reached 0.4. Cells were then harvested and processed for RT-PCR or CCC.
RT-PCR.
Isolation of RNA and RT-PCR analyses of INO1 and ASC1 was performed as described in El Kaderi et al. (21).
CCC.
CCC analyses for INO1 and ASC1 was performed essentially as described in El Kaderi et al. (21), except that chromatin was solubilized with 1% SDS before restriction digestion. For CCC analysis of INO1, chromatin was digested with AluI and EcoRV, whereas restriction digestion was performed with AluI, DraI, and NlaIV for CCC analysis of ASC1. Position of P1, T1 and other primers, and F1 and R1 control primers is indicated in figures. The CCC-positive control PCR products in Fig. 4 were generated as described in El Kaderi et al. (21).
TRO.
TRO assay was performed by the modification of protocols described in Birse et al. and Hirayoshi and Lis (46, 47). All TRO signals were quantified by using GEL LOGIC 200 (Kodak) system and normalized with respect to ACT1 control.
Quantification.
All quantifications and data analyses were performed as described in El Kaderi et al. (21).
Supplementary Material
Acknowledgments
We thank Dr. Michael Hampsey (University of Medicine and Dentistry of New Jersey), Dr. Beate Schwer (Cornell University), Dr. Victoria Meller (Wayne State University), and Dr. Lori Pile (Wayne State University) for critical reading of the manuscript. The plasmid pRKO for insertion of intron was a generous gift from Dr. Marc Gartenberg. We thank Dr. Shiv Grewal (National Institutes of Health) for helpful tips about TRO assay; laboratory members Scott Medler, Banupriya Mukundan, and Nadra Alhusini for helpful discussions; and S. Kiran Koya for help with Fig. 6. This work was supported by the start-up funds from Wayne State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112400109/-/DCSupplemental.
References
- 1.Mattick JS. Introns: Evolution and function. Curr Opin Genet Dev. 1994;4:823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ares M, Jr, Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 6.Rose AB. Intron-mediated regulation of gene expression. Curr Top Microbiol Immunol. 2008;326:277–290. doi: 10.1007/978-3-540-76776-3_15. [DOI] [PubMed] [Google Scholar]
- 7.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charron M, Chern JY, Wright WW. The cathepsin L first intron stimulates gene expression in rat sertoli cells. Biol Reprod. 2007;76:813–824. doi: 10.1095/biolreprod.106.057851. [DOI] [PubMed] [Google Scholar]
- 9.Choi T, Huang M, Gorman C, Jaenisch R. A generic intron increases gene expression in transgenic mice. Mol Cell Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damgaard CK, et al. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 11.Deng TL, Li Y, Johnson LF. Thymidylate synthase gene expression is stimulated by some (but not all) introns. Nucleic Acids Res. 1989;17:645–658. doi: 10.1093/nar/17.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonsson JJ, Foresman MD, Wilson N, McIvor RS. Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res. 1992;20:3191–3198. doi: 10.1093/nar/20.12.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacy-Hulbert A, et al. Interruption of coding sequences by heterologous introns can enhance the functional expression of recombinant genes. Gene Ther. 2001;8:649–653. doi: 10.1038/sj.gt.3301440. [DOI] [PubMed] [Google Scholar]
- 14.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 16.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parenteau J, et al. Deletion of many yeast introns reveals a minority of genes that require splicing for function. Mol Biol Cell. 2008;19:1932–1941. doi: 10.1091/mbc.E07-12-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bianchi M, Crinelli R, Giacomini E, Carloni E, Magnani M. A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene. 2009;448:88–101. doi: 10.1016/j.gene.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Kwek KY, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 21.El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Martínez J, Aranda A, Pérez-Ortín JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Graves JA, Henry SA. Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics. 2000;154:1485–1495. doi: 10.1093/genetics/154.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods. 2009;48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto I, Wu WH, Na JG, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 29.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun ZW, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho EJ, Buratowski S. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J Biol Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Fairley JA, Roberts SG. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu WH, Pinto I, Chen BS, Hampsey M. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medler S, et al. Evidence for a complex of TFIIB with poly(A) polymerase and cleavage factor I subunits required for gene looping. J Biol Chem. 2011;286:33709–33718. doi: 10.1074/jbc.M110.193870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 38.Jansa P, Burek C, Sander EE, Grummt I. The transcript release factor PTRF augments ribosomal gene transcription by facilitating reinitiation of RNA polymerase I. Nucleic Acids Res. 2001;29:423–429. doi: 10.1093/nar/29.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 41.Spitalny P, Thomm M. A polymerase III-like reinitiation mechanism is operating in regulation of histone expression in archaea. Mol Microbiol. 2008;67:958–970. doi: 10.1111/j.1365-2958.2007.06084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Lykke-Andersen S, Mapendano CK, Jensen TH. An ending is a new beginning: Transcription termination supports re-initiation. Cell Cycle. 2011;10:863–865. doi: 10.4161/cc.10.6.14931. [DOI] [PubMed] [Google Scholar]
- 44.Cheng TH, Chang CR, Joy P, Yablok S, Gartenberg MR. Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res. 2000;28:E108. doi: 10.1093/nar/28.24.e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 46.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 47.Hirayoshi K, Lis JT. Nuclear run-on assays: Assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 1999;304:351–362. doi: 10.1016/s0076-6879(99)04021-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.