Abstract
Human leukocyte antigen (HLA) genes are the most polymorphic in the human genome. They play a pivotal role in the immune response and have been implicated in numerous human pathologies, especially autoimmunity and infectious diseases. Despite their importance, however, they are rarely characterized comprehensively because of the prohibitive cost of standard technologies and the technical challenges of accurately discriminating between these highly related genes and their many allelles. Here we demonstrate a high-resolution, and cost-effective methodology to type HLA genes by sequencing, which combines the advantage of long-range amplification, the power of high-throughput sequencing platforms, and a unique genotyping algorithm. We calibrated our method for HLA-A, -B, -C, and -DRB1 genes with both reference cell lines and clinical samples and identified several previously undescribed alleles with mismatches, insertions, and deletions. We have further demonstrated the utility of this method in a clinical setting by typing five clinical samples in an Illumina MiSeq instrument with a 5-d turnaround. Overall, this technology has the capacity to deliver low-cost, high-throughput, and accurate HLA typing by multiplexing thousands of samples in a single sequencing run, which will enable comprehensive disease-association studies with large cohorts. Furthermore, this approach can also be extended to include other polymorphic genes.
Keywords: hematopoietic stem cell transplantation, sequence-based typing
Human leukocyte antigen (HLA) genes encode cell-surface proteins that bind and display fragments of antigens to T lymphocytes. This helps to initiate the adaptive immune response in higher vertebrates and thus is critical to the detection and identification of invading microorganisms (1). Six of the HLA genes (HLA-A, -B, -C, -DQA1, -DQB1, and -DRB1) are extremely polymorphic and constitute the most important set of markers for matching patients and donors for bone marrow transplantation (2, 3). Specific HLA alleles have been found to be associated with a number of autoimmune diseases, such as multiple sclerosis (4), narcolepsy (5), celiac disease (6), rheumatoid arthritis (7), and type I diabetes (3, 8). Alleles have also been noted to be protective in infectious diseases such as HIV (9, 10), and numerous animal studies have shown that these genes are often the major contributors to disease susceptibility or resistance (11–13).
HLA genes are among the most polymorphic in the human genome, and the changes in sequence affect the specificity of antigen presentation and histocompatibility in transplantation. A variety of methodologies have been developed for HLA typing at the protein and nucleic acid level. Whereas earlier HLA typing methods distinguished HLA antigens, modern methods such as sequence-based typing (SBT) determine the nucleotide sequences of HLA genes for higher resolution. However, due to cost and time constraints, HLA sequencing technologies have traditionally focused on the most polymorphic regions encoding the peptide-binding groove that binds to HLA antigens, i.e., exons 2 and 3 for class I genes and exon 2 for class II genes. The antigen-binding groove region of HLA molecules is the focus point of T-cell receptor and mediates transplant rejection and graft-versus-host diseases (GVHD). Regions other than the antigen-binding groove need to be typed because some of those polymorphic sites might affect or abrogate HLA protein expression such as the null allele HLA-A*02:53N with a single-base insertion in exon 4. Although the polymorphic regions of HLA genes predominantly cluster within these exons, an increasing number of alleles display polymorphisms in other exons and introns as well. Therefore, typing ambiguities can result from two or more alleles sharing identical sequences in the targeted exons, but differing in the exons that are not sequenced. Resolving these ambiguities is costly and labor intensive, which makes current SBT methods unsuitable for studies involving even a moderately large group of samples.
Here we demonstrate a unique method targeting a contiguous segment of each of four polymorphic HLA genes (HLA-A, -B, -C, and -DRB1), which define the minimal requirements for HLA matching for allogeneic hematopoietic stem cell transplantation (HSCT) (14). Each HLA gene is amplified from genomic DNA in a single long-range PCR spanning the majority of the coding regions and covering most known polymorphic sites. This approach has several advantages. First, more polymorphic sites are sequenced to provide genotyping information of higher definition and the physical linkage between exons can be determined to resolve combination ambiguity. Second, long-range PCR primers can be placed in less polymorphic regions, allowing for improved resolution of genetic differences. Third, exons of the same gene can be amplified in one fragment, thereby decreasing coverage variability. We calibrated this typing method on HLA-A, -B, -C, and -DRB1 genes using 40 reference cell-line samples in the sequence polymorphism reference panel provided by the International Histocompatibility Working Group (IHWG, www.ihwg.org) The overall concordance rate of 99% with previous results and verification of our HLA typing results in the three discordant alleles by an independent sequencing technology demonstrate that this low-cost, high-throughput HLA typing protocol provides a high level of reliability. In addition, we tested our method on 59 clinical samples and found three previously undescribed alleles (two short insertions and one single-base deletion), further illustrating the ability of this method to discover previously undescribed alleles.
Results
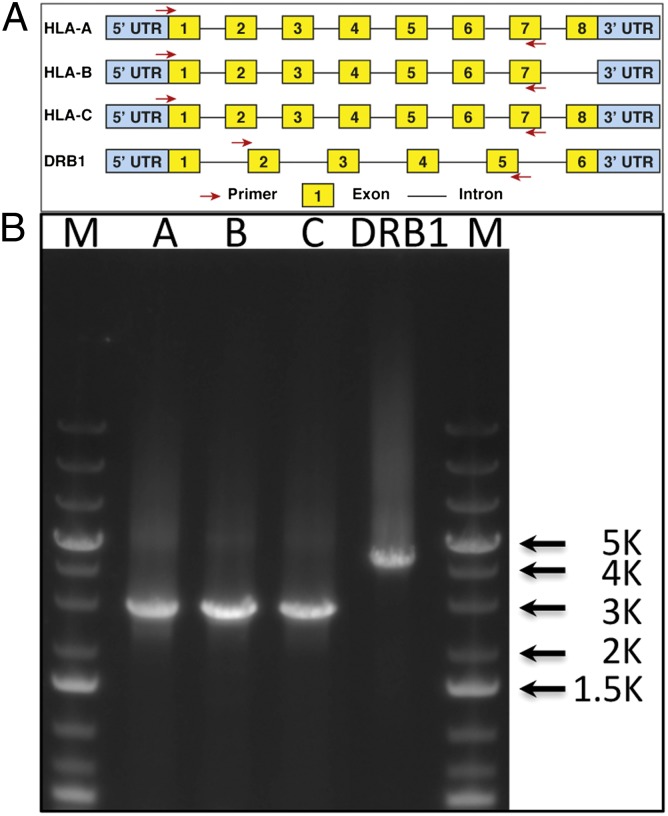
We designed PCR primers for each gene such that the most polymorphic exons and the intervening sequences could be amplified as a single product. For class I genes HLA-A, -B, and -C, primer sequences were selected to amplify the first seven exons. For HLA-DRB1, we designed primers to capture exons 2–5 and to avoid amplifying a large (approximately 8 kb) intron between exons 1 and 2 (Fig. 1). Equimolar amounts of the four HLA gene products were pooled to ensure equal representation of each gene and ligated together to minimize bias in the representation of the ends of the amplified fragments. These ligated products were then randomly sheared to an average fragment size of 300–350 bp and prepared for Illumina sequencing, after the addition of unique barcodes to identify the source of genomic DNA for each sample, using encoded sequencing adapters. Each sequencing adapter had a 7-base barcode between the sequencing primer and the start of the DNA fragment being ligated. The barcodes were designed such that at least 3 bases differed between any two barcodes. Samples sequenced in the same lane were pooled together in equimolar amounts. The sequences of 150 bases from both ends of each fragment for cell-line samples were determined using the Illumina GAIIx sequencing platform. For clinical samples, the sequences of 100 and 150 bases from both ends of each fragment were determined with the Illumina HiSeq2000 and MiSeq platforms, respectively.
Fig. 1.
Location of long-range PCR primers and PCR amplicons in HLA genes. (A) For class I HLA gene (HLA-A, -B, and -C), the forward primer is located in exon 1 near the first codon and the reverse primer is located in exon 7. For HLA-DRB1, the forward primer is located at the boundary between intron 1 and exon 2 and the reverse primer is located within exon 5. Note that the size of exons or introns in the drawing is not proportional to their actual size. (B) Agarose gel (0.8%) showing amplicons from long-range PCR. HLA-A, -B, and -C amplicons are 2.7 kb in length, and the -DRB1 amplicon is around 4.1 kb.
For GAIIx sequence reads (counting each paired-end read as two independent reads), 91.8% of the sequence reads were parsed and separated according to their barcode tags. After stripping the barcode tags, 95.5% (∼54 million sequence reads) were aligned to genomic reference sequences from the International ImMunoGene Tics (IMGT)-HLA (http://www.ebi.ac.uk/imgt/hla/) database (15) with the National Center for Bioinformation Technology (NCBI) BLASTN program, resulting in an average of 10,600 reads per position (coverage), which was estimated on the basis of the number of reads mapped to genomic reference sequences without filtering. For clinical samples, 97.7% of the sequence reads from the HiSeq2000 instrument were parsed and separated according to their barcode tags. After stripping the barcode tags, 96.7% (around 152 million sequence reads) were aligned to genomic references, resulting in an estimated average of 10,000 reads per position.
Classical HLA Genotype Assignment.
Although genomic DNA was amplified and sequenced in our current approach, the standard genotype-calling algorithm relies mainly on the alignment to cDNA references from the IMGT-HLA database due to the lack of genomic reference sequences. Of 6,398 cDNA reference sequences for HLA-A, -B, -C, and -DRB1 genes in the IMGT-HLA database released on October 10, 2011, only 375 (5.8%) of them have genomic sequences. The IMGT-HLA database contains sequences of HLA genes, pseudogenes, and related genes, which allowed us to filter out sequences from pseudogenes or other nonclassical HLA genes, such as HLA-E, -F, -G, -H, -J, -K, -L, -V, -DRB2, -DRB3, -DRB4, DRB5, -DRB6, -DRB7, -DRB8, and -DRB9.
After mapping, the alignments were parsed in the following order: a best-match filter, a mismatch filter, a length filter, and a paired-end filter. The best-match filter only kept alignments with best bit scores. The mismatch filter eliminated alignments containing either mismatches or gaps. The length filter deleted alignments shorter than 50 bases in length if their corresponding exons were longer than 50 bases. It also removed any alignments shorter than their corresponding exons if those were less than 50 bases in length. Finally, the paired-end filter removed alignments in which references were mapped to only one end of a paired-end read, whereas at least one reference was mapped to both ends of the paired-end read.
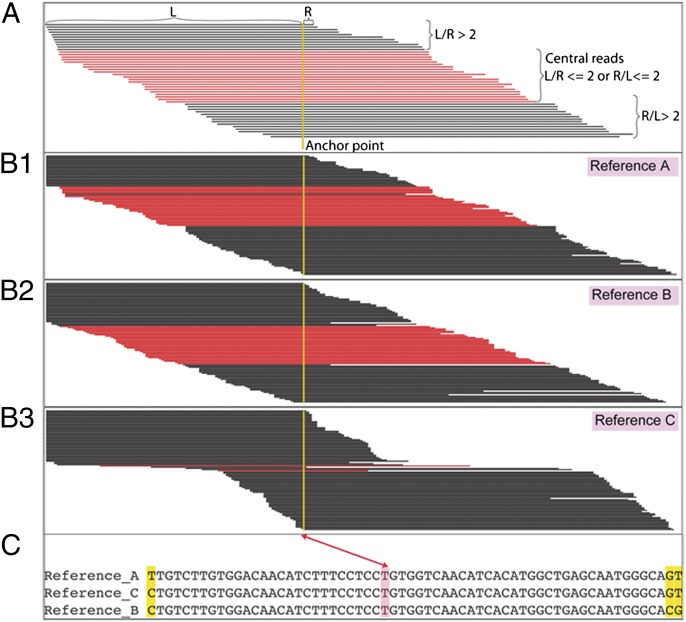
HLA genes share extensive similarities with each other, and many pairs of alleles differ by only a single nucleotide; it is this extreme allelic diversity that has made definitive SBT difficult and subject to misinterpretation. For instance, due to the short read lengths generated using the Illumina platform, it is possible for the same read to map to multiple references. In this study, sequencing was performed in the paired-end format so that the combined specificity of paired-end reads could be used to minimize misassignment to an incorrect reference. Also, because of sequence similarities among different alleles, combinations of different pairs of alleles could result in a similar pattern of observed nucleotide sequence, on the basis of the fortuitous mixture of sequences. We noted that when reads were mapped onto a correct reference sequence, they formed a continuous tiling pattern over the entire sequenced region (Fig. 2 B.1 and B.2). When reads were mapped onto an incorrect reference sequence, they formed a staggered tiling pattern at some positions of the sequenced region (Fig. 2 B.3.). To quantify this difference between the two alignment patterns, we counted the number of “central reads” for any given point. Central reads (Fig. 2A) were empirically defined as mapped reads for which the ratio between the length of the left arm and that of the right arm related to a particular point is between 0.5 and 2 (Fig. 2).
Fig. 2.
Mapping patterns of sequencing reads on correct and incorrect references. (A) Central reads of an anchor point are defined as mapped reads, where the ratio between the length of the left arm and that of the right arm related to a particular point is between 0.5 and 2 (highlighted in red). (B) Mapping pattern of sequencing reads onto correct references (A and B) and onto an incorrect reference (C). (C) Alignment of references A, B, and C around the anchor point shown in B. Anchor points are marked as two double-arrow line.
The genotype-calling algorithm is based on the assumption that more reads are mapped to correct reference(s) than to incorrect reference(s). We could, in a brute-force manner, enumerate all possible combinations of references and count the number of mapped reads for each combination. However, due to the large number of possible combinations, this approach is very inefficient. Therefore, we applied a heuristic approach to eliminate those implausible references first. We computed the minimum coverage of overall reads (MCOR) and the minimum coverage of central reads (MCCR) for each reference. We ignored the MCCR values for 30 bases near intron/exon boundaries, which were always zero, on the basis of the definition of central reads and the cutoff length (Fig. 2). We eliminated the references with an MCOR less than 20 and an MCCR less than 10, as they were unlikely to be correct. From the remaining references, we enumerated all possible combinations of either one reference (homozygous allele) or two references (heterozygous alleles) of the same locus, and counted the number of distinct reads that mapped to each combination. To compensate for a single reference (homozygous allele), the number of distinct reads was multiplied with an empirical value of 1.05 to avoid miscalls due to spurious alignments. The member(s) in the combination with maximum number of distinct reads were assigned as the genotype of that particular sample.
The aforementioned procedure only used the sequence information in the aligned region to do genotype calling. Such a process necessarily introduces bias in the interpretation, because it relies on existing reference data. However, unmapped nucleotides outside aligned regions could also have important sequence information for previously undescribed alleles. To ensure that they were taken into consideration, we implemented a program named EZ_assembler, which carries out de novo assembly of mapped reads including their unmapped regions. Briefly, we partitioned the mapped reads, including unmapped regions, into tiled 40-base fragments with a 1-base offset. We built a directed, weighted graph where each distinct fragment was represented as a node and two consecutive fragments of the same read were connected, and an edge between two nodes was weighted with the frequency of reads from the two connected nodes. A contig was constructed on the path with the maximum sum of weights. By comparing a contig with its corresponding reference sequence, we were able to identify differences between a contig built from reads and its closest reference. We applied the de novo assembly procedure for each candidate allele to verify the accuracy of the HLA typing and to detect novel alleles.
Genotyping Four Highly Polymorphic HLA Genes in 40 Cell Lines.
A total of 40 cell-line–derived DNA samples of known HLA type were obtained from IHWG and sequenced at four loci (HLA-A, -B, -C, and -DRB1). We compared our predictions with the genotypes reported in the public database for those cell lines. Out of 229 alleles from the 40 cell lines typed for HLA-A, -B, -C, and -DRB1 loci, the concordance of our approach with previously determined HLA types was 99% (226/229, see Dataset S1). To further test the accuracy of our approach, we evaluated these discordant alleles by using an independent long-range PCR amplification, and sequenced the PCR products using Sanger sequencing. The HLA-DRB1 locus in the cell-line FH11 (IHW09385) was previously reported as 01:01/11:01:02, which we found to be 01:01/11:01:01. One nucleotide, 12 bases upstream from the end of exon 2, differentiated HLA-DRB1*11:01:01 from HLA-DRB1*11:01:02. Sanger sequencing verified that the HLA-DRB1 locus of the cell-line FH11 is 01:01/11:01:01 (Fig. S1). The reference alleles listed for the HLA-B locus of the cell-line FH34 (IHW09415) are 15/15:21 and, on the basis of our sequencing data, we are able to extend the resolution to 15:35/15:21. Our data showed that Illumina sequencing reads were aligned to both HLA-B*15:21/15:35 references continuously. HLA-B*15:21 and HLA-B*15:35 were different in three positions in exon 2, and seven positions in exon 3. The Sanger sequencing chromatogram indicated the presence of a mixture in the corresponding positions at exon 2, matching the expected combination of HLA-B*15:21/15:35 (Fig. S2). The HLA-B locus of the cell-line ISH3 (IHW09369) was reported as homozygous for 15:26N in the IHWG cell-line database. Our Illumina sequencing reads mapped to exons 2–5, but not exon 1 of the HLA-B*15:26N reference. Instead, the reads mapped to exons 1, 3, 4, and 5, but not exon 2 of the HLA-B*15:01:01:01 reference. There is no reference sequence available where the Illumina reads could tile continuously across the reference sequence. The Sanger sequencing data confirmed that ISH3 HLA-B allele had the exon 1 sequence as that of 15:01:01:01 and the sequence of exons 2–5 of 15:26N (Fig. S3). This finding suggests that either there is an error in the exon 1 region of B*15:26N reference sequence or that it represents yet another previously undescribed B*15 null allele.
Genotyping Four Highly Polymorphic HLA Genes in 59 Clinical Samples.
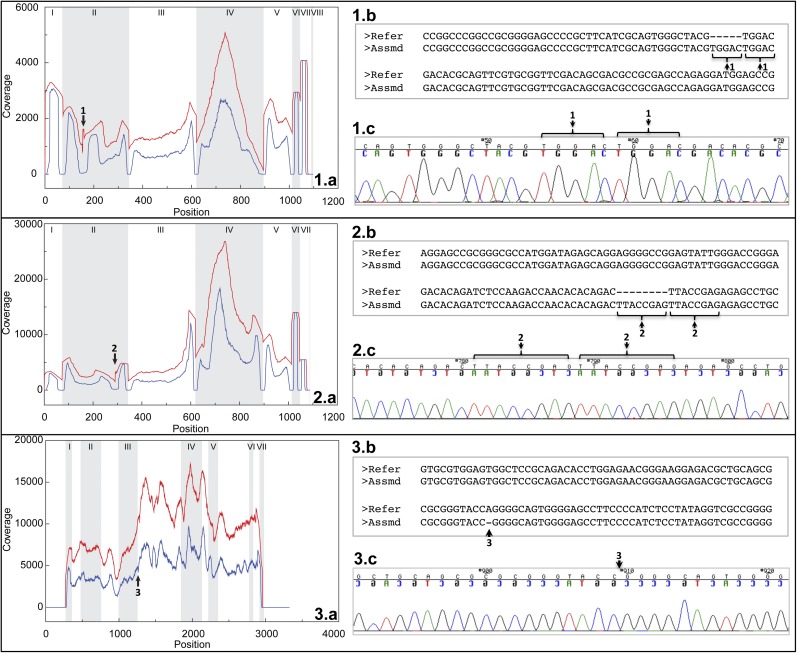
To test increased throughput using our approach, we pooled 59 clinical samples and typed HLA-A, -B, -C, and -DRB1 in a single HiSeq2000 lane. Of these, 47 samples (samples 1–47, Dataset S2) from an HLA disease association study were typed both by our methodology and an oligonucleotide hybridization assay. Even though the resolution of the probe-based assay was lower, the pairwise comparisons of possible genotypes showed overlap in at least one possible genotype for all loci in all samples. There were no allele dropouts in testing by either methodology. Twelve additional samples included specimens of HSCT patients or donors that presented less common or unique allele types (samples 48–59, Dataset S2). In this group, two samples with insertions of 5 and 8 exonic nucleotide insertions were concordantly typed by both classic Sanger sequencing and by the methodology described in the present study (Fig. 3 1.a–c and 2.a–c). The occurrence of these insertions shows a change in the reading frame with the occurrence of premature termination codons; therefore, the corresponding mature HLA proteins of these alleles are not expressed on the cell surface (null). In conventional sequencing, both heterozygous alleles are coamplified and sequenced. However, when one of the alleles contains an insertion or deletion, it results in an off-phase heterozygous sequence and the readout is cumbersome and laborious; in contrast, the readout obtained by the unique methodology was straightforward. The precise identification of the type of insertion/deletion in these unique alleles is of crucial importance in clinical histocompatibility practice. The allele containing the insertion or deletion may not be expressed because the reading frame may include changes in the amino acid sequence, resulting in the occurrence of premature termination codons or it may have altered expression if the mutations are close to mRNA splicing sites (Fig. 3.3). If a mutation of this nature is overlooked, the evaluation of the HLA typing match between a patient and an unrelated donor could easily be incorrect.
Fig. 3.
Identification and verification of three unique alleles with insertions and deletions. (1.a) Coverage of overall reads (red) and central reads (blue) mapped onto HLA-A*02:01:01:01 cDNA reference in one clinical sample. (1.b) Partial alignment between a contig derived from reads mapped onto HLA-A*02:01:01:01 reference and HLA-A*02:01:01:01 reference. (1.c) Chromatogram of Sanger sequence on a clone derived from HLA-A PCR product from the same sample. Black arrow 1 highlights a 5-base “TGGAC” insertion in coverage plot (1.a), alignment (1.b), and chromatogram (1.c). (2.a) Coverage of overall reads (red) and central reads (blue) mapped onto HLA-B*40:02:01 cDNA reference in one clinical sample. (2.b) Partial alignment between a contig derived from reads mapped onto HLA-B*40:02:01 reference and HLA-B*40:02:01 reference. (2.c) Chromatogram of Sanger sequence on a clone derived from HLA-B PCR product from the same sample. Black arrow 2 highlights an 8-base “TTACCGAG” insertion in coverage plot (2.a), alignment (2.b) and chromatogram (2.c). (3.a) Coverage of overall reads (red) and central reads (blue) mapped onto HLA-B*51:01:01 genomic reference in one clinical sample. (3.b) Partial alignment between a contig derived from reads mapped onto HLA-B*51:01:01 reference and HLA-B*51:01:01 reference. (3.c) Chromatogram of Sanger sequence on a clone derived from HLA-B PCR product from the same sample. Black arrow 3 highlights a single-base “A” deletion in coverage plot (3.a), alignment (3.b), and chromatogram (3.c). In the coverage plots, exon regions are indicated with Roman numerals.
In the present study, we identified the alleles B*40:01:02, A*23:17, and C*07:01:02, which are thought to be rare (Dataset S2). However, from the data presented here, it is likely that some of them may be the predominant allele of their group (B*40:01:02) or more common than previously thought.
Discussion
Recently, several laboratories (16–20) have developed high-throughput HLA genotyping methodologies using massively parallel sequencing strategies such as Roche/454 sequencing (21). In all these high-throughput HLA-genotyping studies, with the exception of the study by Lind et al. (19), a few polymorphic exons were amplified separately and sequenced in a multiplexed manner. In our approach, a large genomic region of each gene including introns and the most polymorphic exons was amplified in a single PCR and sequenced with a large excess of independent paired-end reads. There are two major ambiguities/uncertainties that arise from conventional SBT methods for HLA genotyping: uncertainties that are commonly seen in typing protocols where alleles vary outside the targeted regions, and combination ambiguities that are frequently encountered where different allele combinations yield the same sequence pattern (22). As more exons of a gene were sequenced, our method (Fig. S4), which sequenced exons 1–7 for HLA class I genes and exons 2–5 for HLA-DRB1, substantially enhanced the allele resolution and dramatically improved the combination resolution in comparison with the conventional SBT method, which sequences exons 2 and 3 for HLA class I genes and exon 2 alone for HLA-DRB1. In addition, the extensive sequence coverage allowed us to largely overcome genotype calling artifacts. The paired-end sequencing strategy extends the read length effectively to 400–500 bases, which matches that of the Roche/454 platform, while allowing much higher throughput. The paired-end reads facilitated the determination of linkage phase across 400 bases in each DNA fragment, and together with polymorphic sites in intron regions, provided us with important phasing information that was useful to resolve combination ambiguities.
We validated this long-range PCR amplification and next-generation sequencing approach by retyping the 40 different IHWG reference cell lines. The accuracy of this approach was demonstrated with a high degree (overall 99%) of concordance between our results and those reported in the reference databases. The Sanger sequencing data confirmed our genotype-calling results in the discordant alleles in all cell lines. Although the number of new alleles in public databases has increased dramatically in the past few years, the list is far from being exhaustive as many ethnic groups have yet to be sequenced in depth. In particular, populations from areas with high pathogen diversity are expected with increased HLA diversity in relation to their average genomic diversity (23). Therefore, the ability of a HLA genotyping method to discover previously undescribed alleles is significant. Our approach demonstrates the ability to identify previously undescribed alleles that have insertions, deletions, and substitutions. In particular, our strategy of using PCR primers outside polymorphic regions for long-range PCR increases the chance of capturing previously undescribed alleles.
Finally, we were interested in optimizing our approach to accommodate more samples in a single instrument run. Of all alleles from 59 clinical samples typed in a single HiSeq2000 lane, 99.3% of alleles meet the minimum coverage of 100, and the majority of them are beyond 900 (Fig. S5). The ratios of minimum coverage of heterozygous alleles of a gene in the same sample were under four in all but two samples, indicating that heterozygous alleles of the same gene were amplified with similar efficiencies and coverage variation are largely due to pooling unevenness. Our simulation experiment showed that a minimum coverage of 20 could provide reliable information for genotype calling. With an optimized protocol to improve the pooling evenness, we project that for HLA typing of four genes, we can pool about 180 samples in one lane of Illumina HiSeq2000 or 2,700 samples in one HiSeq2000 instrument run (15 lanes), respectively.
In conclusion, we demonstrate here a successful approach for determining accurate HLA genotypes in a high-throughput manner for large numbers of clinical samples simultaneously. Having such a high throughput effectively lowers the cost per sample. Indeed, in the setting of testing many subjects simultaneously, the cost for high-resolution typing by this methodology is significantly lower than classical Sanger sequencing and it in the same range or lower than the cost of probe-based assays, which have a much lower typing resolution. Therefore, the combination of high-resolution, high-throughput, and low cost will enable comprehensive disease-association studies with large cohorts. The HLA typing approach described here may also be useful in obtaining high-resolution HLA results of donors and cord blood units recruited or collected by registries of potential volunteer donors for bone marrow transplantation and cord blood banks. Successful outcomes of allogeneic hematopoietic stem cell transplantation correlate well with close HLA matching between the patient and the selected donor unit (14, 24). Also, in many diseases early treatment including hematopoietic stem cell transplantation soon after diagnosis, correlates with superior outcomes (25). Listing donors and units with the corresponding high resolution HLA type can dramatically accelerate the identification of optimally compatible donors. On the other hand, we have also demonstrated that the same approach can be adapted to accommodate the need for quick turnaround for urgent samples. With the Illumina Miseq, we can type a few samples within 5 d. As improved sequencing technologies are developed, we can adapt the typing method to suit any sequencing platform, as the alignment algorithms and HLA genotype calling are independent of the sequencing method.
The present study shows that the current knowledge of sequence variation in the HLA system can rapidly be expanded by the application of the latest nucleotide sequencing technologies. In the present study we were able to analyze comprehensively segments of the HLA genes that have not been tested routinely. The testing of these areas will allow us to gain insight into the fine details of the possible evolutionary pathways of the HLA variation. Furthermore, these methodologies may allow us to refine the mapping of susceptibility factors, and potentially of immunity-enabling features. In this regard, it may be possible to extend this approach to all HLA genes to discern patient-specific factors that may influence future vaccination strategies. Similarly, we may be able to obtain more precise evaluation of the HLA match grade between patients and unrelated donors in solid organ and hematopoietic stem cell transplantation.
Materials and Methods
HLA typing reference cell lines were obtained from the IHWG (IHWG, www.ihwg.org) at the Fred Hutchinson Cancer Research Center, Seattle. The sequence polymorphism reference panel was used for validating the Illumina HLA typing technology. The 47 clinical samples (samples 1–47, Dataset S2) were drawn from the molecular genetics of schizophrenia I linkage sample (26), which is part of the National Institute of Mental Health Center for Genetic Studies repository program (http://nimhgenetics.org). The other 12 clinical samples (samples 48–59, Dataset S2) were from specimens of HSCT patients or donors that presented less common or novel allele types. Each clinical specimen was collected after subjects signed a written informed consent.
PCR Primer Design.
To design gene-specific primers, we have analyzed all available sequences and chosen primers that would ensure the amplification of all known alleles for each gene. We have avoided regions of high variability, and where necessary, have designed multiple primers to ensure amplification of all alleles. For class I HLA gene (HLA-A, -B, and -C), the forward primer was located in exon 1 near the first codon, and the reverse primer was located in exon 7. Only a limited number of genomic sequences were available for HLA-DRB1 genes. Therefore, the PCR primer for HLA-DRB1 genes were placed in less divergent exons. Taking into consideration the size of the PCR amplicons and completeness of genes, the forward primer for HLA-DRB1 was placed at the boundary between intron 1 and exon 2, and the reverse primer within exon 5. To ensure the robustness of the PCR, the first exon of DRB1 was not included to avoid amplifying intron 1, which is about 8 kb in length.
Supplementary Material
Acknowledgments
Funding from National Institutes of Health (NIH) Grants U19AI090019, P01HG000205, GM62119 and Defense Threat Reduction Agency Grant HDTRA1-11-1-0058 (to R.W.D. and M.M.D.) and from U19AI090019 and the Howard Hughes Medical Institute (to M.M.D.) supported this work. L.F.S. is supported by an NIH K08 Award (K08 AR059760-01).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRA051897).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206614109/-/DCSupplemental.
References
- 1.Siegrist CA. [Molecular basis for detection of infectious agents] Schweiz Med Wochenschr. 1996;126:246–254. [PubMed] [Google Scholar]
- 2.Marks C. Immunobiological determinants in organ transplantation. Ann R Coll Surg Engl. 1983;65:139–144. [PMC free article] [PubMed] [Google Scholar]
- 3.Davies JL, et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 4.Oksenberg JR, Barcellos LF. Multiple sclerosis genetics: Leaving no stone unturned. Genes Immun. 2005;6:375–387. doi: 10.1038/sj.gene.6364237. [DOI] [PubMed] [Google Scholar]
- 5.Mignot E, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–699. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sollid LM, et al. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 8.Hanis CL, et al. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet. 1996;13:161–166. doi: 10.1038/ng0696-161. [DOI] [PubMed] [Google Scholar]
- 9.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 10.Carrington M, et al. HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 11.O’Neill TP. HLA-B27 transgenic rats: Animal model of human HLA-B27-associated disorders. Toxicol Pathol. 1997;25:407–408. doi: 10.1177/019262339702500411. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, et al. Characterization of HLA DR3/DQ2 transgenic mice: A potential humanized animal model for autoimmune disease studies. Eur J Immunol. 2003;33:172–182. doi: 10.1002/immu.200390020. [DOI] [PubMed] [Google Scholar]
- 13.Nabozny GH, et al. HLA-DQ8 transgenic mice are highly susceptible to collagen-induced arthritis: a novel model for human polyarthritis. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SJ, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 15.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2009;37(Database issue):D1013–1017. doi: 10.1093/nar/gkn662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holcomb CL, et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens. 2011;77:206–217. doi: 10.1111/j.1399-0039.2010.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley G, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel C, et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum Immunol. 2009;70:960–964. doi: 10.1016/j.humimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Lind C, et al. Next-generation sequencing: The solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol. 2010;71:1033–1042. doi: 10.1016/j.humimm.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Erlich RL, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens HA. HLA and other gene associations with dengue disease severity. Curr Top Microbiol Immunol. 2010;338:99–114. doi: 10.1007/978-3-642-02215-9_8. [DOI] [PubMed] [Google Scholar]
- 23.Prugnolle F, et al. Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 2005;15(11):1022–1027. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 24.Flomenberg N, et al. Impact of HLA class I and class II high-resolution matching on outcomes of unrelated donor bone marrow transplantation: HLA-C mismatching is associated with a strong adverse effect on transplantation outcome. Blood. 2004;104(7):1923–1930. doi: 10.1182/blood-2004-03-0803. [DOI] [PubMed] [Google Scholar]
- 25.Guinan EC. Diagnosis and management of aplastic anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:76–81. doi: 10.1182/asheducation-2011.1.76. [DOI] [PubMed] [Google Scholar]
- 26.Suarez BK, et al. Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: Suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. American Journal of Human Genetics. 2006;78(2):315–333. doi: 10.1086/500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.