Abstract
Peptide hormones and neuropeptides have important roles in physiology and therefore the regulation of these bioactive peptides is of great interest. In some cases proteolysis controls the concentrations and signaling of bioactive peptides, and the peptidases that mediate this biochemistry have proven to be extremely successful drug targets. Due to the lack of any general method to identify these peptidases, however, the role of proteolysis in the regulation of most neuropeptides and peptide hormones is unknown. This limitation prompted us to develop an advanced peptidomics-based strategy to identify the peptidases responsible for the proteolysis of significant bioactive peptides. The application of this approach to calcitonin gene-related peptide (CGRP), a neuropeptide associated with blood pressure and migraine, revealed the endogenous CGRP cleavage sites. This information was then used to biochemically purify the peptidase capable of proteolysis of CGRP at those cleavage sites, which led to the identification of insulin-degrading enzyme (IDE) as a candidate CGRP-degrading enzyme. CGRP had not been identified as an IDE substrate before and we tested the physiological relevance of this interaction by quantitative measurements of CGRP using IDE null (IDE−/−) mice. In the absence of IDE, full-length CGRP levels are elevated in vivo, confirming IDE as an endogenous CGRP-degrading enzyme. By linking CGRP and IDE, this strategy uncovers a previously unknown pathway for CGRP regulation and characterizes an additional role for IDE. More generally, this work suggests that this may be an effective general strategy for characterizing these pathways and peptidases moving forward.
Bioactive peptides play a role in a broad range of physiology (1, 2), including pain sensation (3), blood pressure (4), and energy homeostasis (5, 6). The disruption of bioactive peptide signaling can lead to prevalent diseases such as diabetes (7). The regulation of bioactive peptides has therefore become an important area of research with implications for basic physiology as well as for medicine (3, 4, 8–11). Indeed, targeting the proteolytic pathways that regulate peptide levels has proven to be a successful strategy in the development of novel therapeutics (4, 8, 12). Dipeptidyl peptidase 4 (DPP4) inhibitors, for example, are a new class of antidiabetic drugs that prevent the degradation of glucagon-like peptide 1 (GLP-1), an insulinotropic peptide hormone, and thereby raise physiological insulin levels (8, 13, 14). The discovery of the peptidases that regulate other bioactive peptides will provide a deeper understanding of peptide regulation, identify new roles for enzymes, and may also reveal new opportunities in medicine.
The neuropeptide alpha calcitonin gene-related peptide (αCGRP, CGRP or CGRP1–37) was discovered as an alternatively spliced form of the calcitonin RNA (15, 16). Shortly after its discovery, pharmacological studies revealed CGRP to be a potent vasodilator (17), suggesting a role in blood pressure regulation. Genetic studies have shown that mice lacking a functional CGRP receptor have elevated blood pressure (18), supporting the role for this peptide in regulating blood pressure. CGRP has also been shown to actively participate in the onset of migraines. Analysis of plasma from migraine sufferers revealed elevated CGRP levels in comparison with control patients (19). In addition, current antimigraine therapeutics have been shown to lower CGRP levels (20). Together, these studies suggest that blockage of CGRP signaling could be of therapeutic benefit in treating migraine. The recent development of and clinical trials with CGRP-receptor antagonists support this hypothesis as patients taking CGRP-receptor antagonists have fewer migraines (21).
As a peptide with important biological and pathological roles, the underlying pathways that regulate CGRP levels and signaling are of great interest. Several aspects of CGRP regulation are understood—such as CGRP secretion following transient receptor potential cation channel subfamily V member 1 (TRPV1) activation (22)— but it is not known whether proteolysis controls physiological CGRP levels. There have been several unsuccessful attempts to identify peptidases responsible for CGRP proteolysis (23–26). In vitro experiments with recombinant enzymes, for example, have implicated several candidate peptidases (25, 26), but none of these enzymes have been shown to process CGRP in vivo. Immunoassays have also been used in an attempt to map the endogenous CGRP cleavage sites by identifying the endogenous CGRP fragments (23–25), but the inability of antibodies to distinguish proteolytic fragments (27, 28) limited this approach. The lack of a general approach to identify peptidases that regulate bioactive peptides and the desire to identify a CGRP-degrading enzyme inspired us to develop a peptidomics strategy to characterize a CGRP-degrading enzyme.
Results and Discussion
Peptidomics Analysis of Spinal Cords.
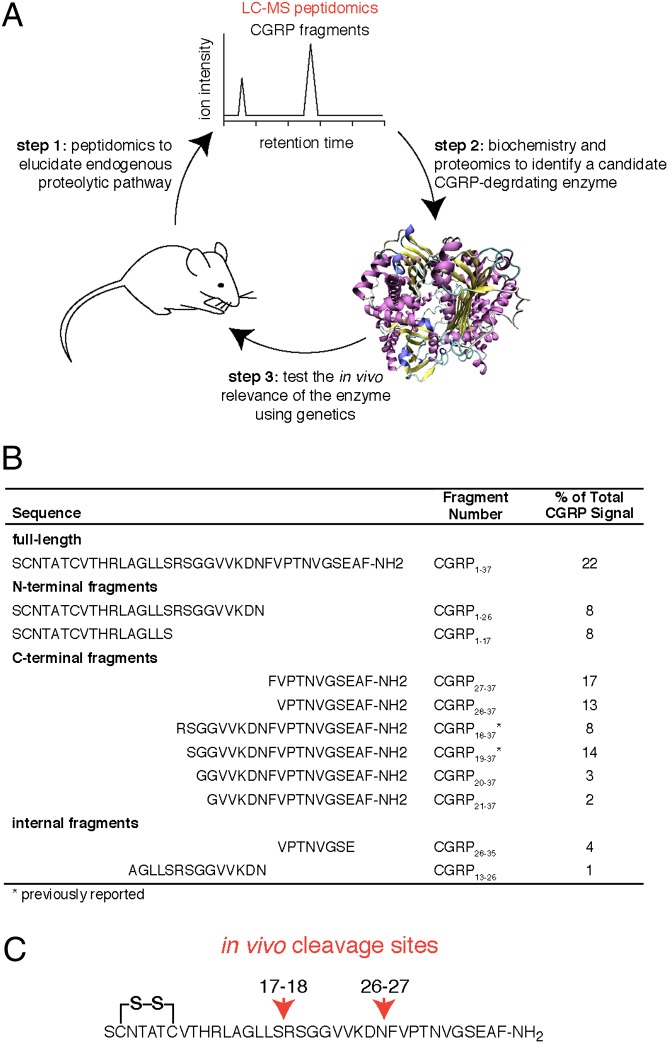
Our strategy can be divided into three key steps: (i) the characterization of CGRP fragments in tissues using peptidomics, (ii) the identification of the peptidase capable of producing those fragments from full-length CGRP (CGRP1–37), and (iii) the validation of the candidate CGRP-degrading enzyme through genetics in animal models (Fig. 1A). The distinguishing feature of our approach is the use of a liquid chromatography-tandem mass spectrometry (LC–MS/MS) peptidomics platform to detect CGRP fragments in tissues (29) and reveal the endogenous CGRP cleavage sites. Combining this peptidomics platform with biochemistry and genetics enables the discovery of the peptidases responsible for the proteolysis of important bioactive peptides.
Fig. 1.
Identifying endogenous CGRP cleavage sites. (A) Combination of peptidomics, biochemistry, and genetics provides a unique strategy to identify peptidases that process important bioactive peptides, such as CGRP. (B) Peptidomics analysis of mouse spinal cords reveals the largest number of CGRP fragments ever reported. These fragments were used to determine the endogenous CGRP cleavage sites. (Note: there is a disulfide between Cys2 and Cys7 for the full-length and N-terminal fragments). (C) Detection of the N-terminal CGRP fragments, CGRP1–17 and CGRP1–26, and the C-terminal fragments, CGRP18–37 and CGRP27–37, indicates that full-length CGRP1–37 is cleaved at the Ser17-Arg18 and Asn26-Phe27 sites.
Peptidomics analysis of mouse spinal cords detected a total of 10 CGRP fragments in addition to the full-length CGRP1–37 (Fig. 1B). Only two of these fragments, CGRP18–37 and CGRP19–37, had previously been reported (24), which illustrates the dramatic improvement in peptide coverage afforded by LC–MS/MS peptidomics. The two N-terminal fragments, CGRP1–17 and CGRP1–26, and two C-terminal fragments, CGRP18–37 and CGRP26–37, proved particularly useful in identifying the endogenous CGRP cleavage sites. These four peptides indicate that CGRP1–37 has two cleavage sites in vivo; Ser17-Arg18 and Asn26-Phe27 (Fig. 1C). These CGRP cleavage sites had not previously been reported. In addition, testing some of these CGRP fragments reveals a loss of activity against the CGRP receptor, indicating that this proteolytic pathway can inactivate CGRP signaling (Table S1).
Tissue Lysate Experiments.
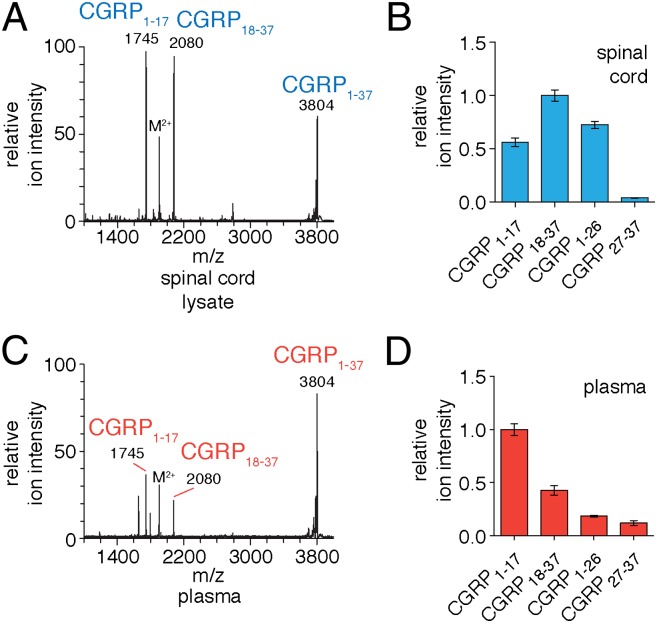
Further support for these CGRP cleavage sites was obtained through in vitro experiments with tissue lysates. Upon incubation with mouse spinal cord lysates (soluble fraction), CGRP1–37 is cleaved at Ser17-Arg18 and Asn26-Phe27 to produce the major endogenous CGRP fragments (i.e., CGRP1–17, CGRP18–37, CGRP1–26, and CGRP27–37) (Fig. S1 and Fig. 2). Further, we were interested in studying CGRP regulation in the blood where CGRP is also found in vivo. Examination of CGRP1–37 proteolysis in plasma demonstrated that the peptide is processed at Ser17-Arg18 and Asn26-Phe27, too (Fig. 2). The ratios of the CGRP fragments differ between the spinal cord lysates and plasma samples (Fig. 2 B and D). We believe these differences are likely due to the differing relative stabilities of these fragments in spinal cord lysates and plasma after they are produced, which leads to the different fragment ratios observed. The identification of identical cleavage sites in plasma and spinal cord lysates suggests that the same unidentified peptidase may be responsible for CGRP proteolysis in both settings. To confirm this hypothesis, we proceeded to identify the peptidase.
Fig. 2.
Biochemical assays with CGRP1–37 in spinal cord lysates and plasma. CGRP1–37 was incubated with spinal cord lysate and analyzed by (A) MALDI-TOF MS and (B) LC–MS/MS. MALDI-TOF MS analysis identified CGRP1–17 and CGRP18–37 as the major breakdown products of CGRP1–37, whereas the more sensitive LC–MS also identified CGRP1–26 and CGRP27–37. Incubation of CGRP1–37 with plasma and subsequent analysis by (C) MALDI-TOF MS and (D) LC–MS/MS identified the same fragments in plasma. (Error bars show SEM.)
Identification of Candidate Enzymes.
A search of the peptidase database (MEROPS) (30) identified a number of candidate peptidases that would likely cleave the Ser17-Arg18 and Asn26-Phe27 sites of CGRP (Table S2). This list included peptidases and proteases from every class (serine, cysteine, aspartyl, and metallo), as well as soluble and membrane-bound forms of the enzyme. According to this list, the metallopeptidases thimet oligopeptidase (THOP) and matrix metalloprotease 19 (MMP19) can cleave CGRP at both cleavage sites, suggesting that a single enzyme may regulate CGRP. Because the MEROPS database cannot account for uncharacterized peptidases or proteases, we opted to biochemically purify the CGRP-degrading enzyme from tissues to ensure the identification of the correct enzyme.
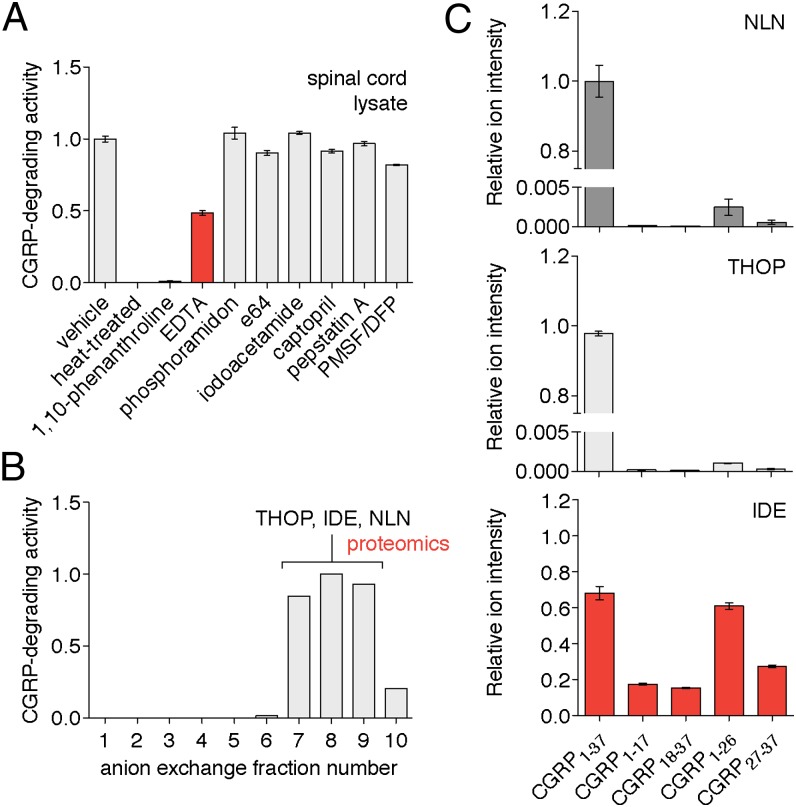
To assay CGRP-degrading activity, we used LC–MS/MS to quantitatively measure the production of CGRP1–17, CGRP18–37, or CGRP1–26 from full-length CGRP1–37. First, we used this assay to determine the peptidase class (i.e., serine, cysteine, aspartyl, or metallo) responsible for CGRP proteolysis. The use of different protease inhibitors in spinal cord lysates and plasma revealed that the CGRP-degrading enzyme is a metallopeptidase, with CGRP1–37 proteolysis inhibited by the presence of the metal chelators 1,10-phenanthroline or EDTA (Fig. 3 and Fig. S2). Metallopeptidase inhibitors also prevented the production of CGRP1–26 (Fig. S3), which indicates that metallopeptidases are responsible for processing CGRP1–37 at both cleavage sites.
Fig. 3.
Characterization of IDE as a candidate CGRP-degrading enzyme. CGRP-degrading activity was measured by quantitation of (A) CGRP1–17 or (B) CGRP18–37 produced from CGRP1–37 by LC–MS (i.e., area under the curve for these peaks in the LC–MS chromatogram). (A) Biochemical assays using spinal cord lysates treated with class selective protease inhibitors (aspartyl, cysteine, metallo, or serine) showed that the peptidase responsible for CGRP proteolysis is a metallopeptidase. Control reactions included a vehicle-treated sample that retained full activity and a heat-treated sample that lacked any activity. Metallopeptidase inhibitors 1,10-phenanthroline and EDTA were the only two compounds tested that inhibited the CGRP degradation. (B) Identification of candidate peptidases was carried out by anion-exchange chromatography of the spinal cord proteome followed by proteomics of the fractions with the highest levels of CGRP-degrading activity. Proteomics identified three metallopeptidases in these fractions; THOP, NLN, and IDE. (C) IDE was identified as the only peptidase with CGRP-degrading activity by incubating CGRP1–37 with recombinant murine THOP, NLN, and IDE followed by quantitation of CGRP1–37, CGRP1–17, CGRP18–37, CGRP1–26, and CGRP27–37. THOP and NLN barely showed any activity against CGRP (two orders of magnitude less than IDE). (Error bars show SEM.)
To identify the peptidase, mouse spinal cord lysate was separated by anion exchange chromatography and fractions containing CGRP-degrading activity were analyzed by proteomics to identify any metallopeptidases (Fig. 3). The fractions with the highest CGRP-degrading activity contained the metallopeptidases neurolysin (NLN), thimet oligopeptidase (THOP) and insulin-degrading enzyme (IDE, insulysin). The addition of N-ethylmaleimide (NEM)—a cysteine-labeling reagent that inhibits IDE (31), THOP, and NLN (32)—to spinal cord lysates and plasma prevented CGRP proteolysis and supported a role for one or more of these enzymes in CGRP processing (Fig. S4). According to the MEROPS database, IDE should be able to cleave at the Ser17-Arg18 site, NLN at the Asn26-Phe27 site, and THOP at both sites (Table S2).
Identification of IDE as a CGRP-Degrading Enzyme Candidate.
To determine whether any of these peptidases is a CGRP-degrading enzyme—none of them have been implicated in CGRP proteolysis previously—we performed a series of biochemical assays with pure enzymes. When CGRP1–37 was incubated with each peptidase, only IDE cleaved CGRP1–37 at Ser17-Arg18 and Asn26-Phe27 efficiently, whereas NLN and THOP barely cleaved CGRP1–37 at all (Fig. 3C and Fig. S5). The discordance between these data and the predicted cleavage specificities is due to the fact that MEROPS only takes into account sequence. It is known that NLN and THOP prefer to cleave shorter peptides (<17 amino acids) (33). By contrast, IDE has been reported to cleave longer peptide substrates such as insulin (34, 35) and the beta amyloid peptide (Aβ) (34, 36) in vivo.
IDE Regulation of CGRP in TT Cells.
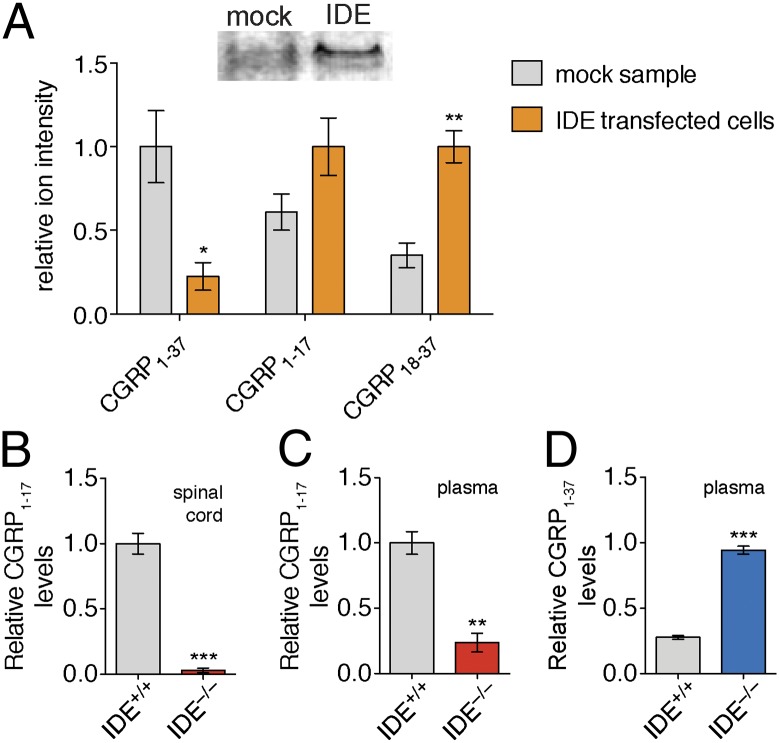
The hydrolysis of CGRP1–37 by pure IDE demonstrates that IDE is able to cleave CGRP1–37 in a physiologically relevant manner, but it was still necessary to ascertain whether IDE is responsible for the CGRP-degrading activity in complex proteomes (i.e., cell culture, spinal cord lysate, or plasma). Toward this goal, CGRP1–37 proteolysis was first analyzed in TT cells, which naturally produce CGRP (37). Overexpression of IDE in TT cells led to lower concentrations of CGRP1–37 and elevated levels of CGRP18–37 in conditioned media, demonstrating the IDE can mediate CGRP1–37 proteolysis in cell culture (Fig. 4A). Importantly, these experiments demonstrate that CGRP and IDE can interact in a biologically relevant environment. Although IDE is predominantly an intracellular enzyme, recent studies have demonstrated that it can be secreted from cells through a noncanonical exosome-mediated pathway (38, 39). Thus, even though IDE does not have a signal sequence indicating export from the cell, it is secreted and can interact with CGRP in the extracellular environment.
Fig. 4.
IDE cleaves CGRP1–37 in complex proteomes. (A) TT cell line naturally produces and secretes CGRP. By Western blot (Inset, Top) TT cells have very little or no IDE. To study the impact of IDE on CGRP levels in TT cells, we transfected these cells with an empty vector (mock) or a vector containing the IDE gene (IDE transfected cells). CGRP1–37 levels were significantly lower in media from IDE transfected cells, whereas levels of the CGRP fragment, CGRP18–37, were elevated in media expressing IDE, indicating increased CGRP1–37 proteolysis. Incubation of CGRP1–37 with (B) spinal cord lysate and (C) plasma revealed higher CGRP1–17 in IDE+/+ samples, demonstrating that IDE is responsible for the majority of the CGRP-degrading activity in these lysates and plasma. (D) Incubation of CGRP1–37 with plasma revealed that CGRP1–37 is more stable in IDE−/− plasma, demonstrating that IDE processing can regulate CGRP1–37 levels in vitro. Relative quantification was accomplished by using the ion intensity of the different peptides in the LC–MS chromatogram. These ion intensities were then normalized to the largest peak to enable comparison of different peptides on the basis of relative changes in their abundance. (*P value < 0.05, **P value < 0.01, ***P value < 0.001; P values were derived from the two-tailed Student t test, n = 3. Error bars show SEM.)
To test that IDE is responsible for the majority of CGRP-degrading activity observed in tissue lysates, we performed ex vivo experiments using tissue lysates and plasma from IDE null (IDE−/−) mice (34). In the absence of IDE, the majority of CGRP-degrading activity was lost, confirming this enzyme as the major source of CGRP-degrading activity in both spinal cord lysates and plasma (Fig. 4). This result supports the in vitro assignment of IDE as the CGRP-degrading enzyme and validates the hypothesis that the same enzyme is responsible for the CGRP-degrading activity in spinal cord lysates and plasma. In addition, in these ex vivo experiments we find that CGRP levels are higher when incubated with plasma from IDE−/− mice. Of course, in vivo proteolysis will compete with other processes, such as renal clearance or tissue uptake, and therefore the net effect of IDE proteolysis on endogenous CGRP levels had to be assessed in animals.
IDE Regulation of CGRP Levels in Plasma.
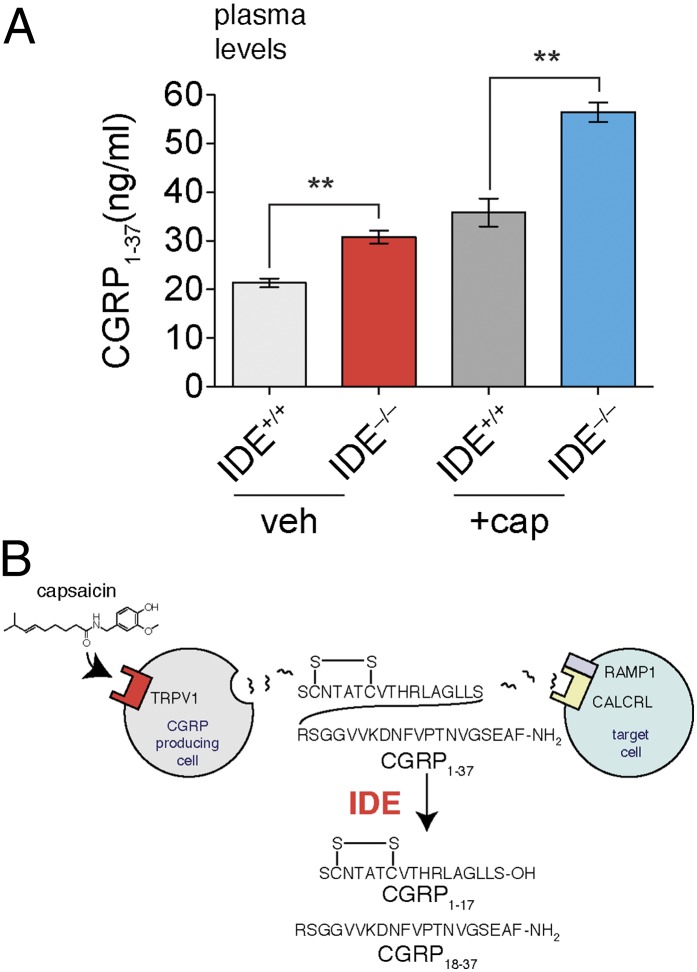
To determine whether IDE is a CGRP-degrading enzyme in vivo, we measured the absolute levels of CGRP1–37 in plasma from IDE+/+ and IDE−/− mice using an isotope-dilution mass spectrometry (IDMS) approach (40). We selected plasma for these measurements because IDE and CGRP1–37 are both present in blood and therefore they should be able to interact (22, 39). In addition, we compared CGRP1–37 levels in mice treated with capsaicin, a TRPV1 agonist that promotes CGRP1–37 secretion (22) (Fig. 4). The capsaicin experiments were included to establish whether IDE regulates CGRP1–37 under different conditions, such as the rapid increase in plasma CGRP1–37 levels associated with TRPV1 activation.
Comparison of plasma from IDE+/+ and IDE−/− mice revealed significantly higher CGRP1–37 levels in IDE−/− mice (40% greater than IDE+/+ mice) (Fig. 5). These data establish IDE as an endogenous regulator of CGRP and demonstrate a role for proteolysis in CGRP regulation. As expected, capsaicin increased the absolute amounts of CGRP1–37 in plasma regardless of genotype, but this increase did not impact the ability of IDE to regulate CGRP1–37. In the capsaicin-treated cohort, CGRP1–37 levels were 60% greater in plasma samples from IDE−/− mice than in IDE+/+ plasma samples (Fig. 4B). Interestingly, some phenotypes associated with capsaicin administration such as vasorelaxation (22) are mediated by CGRP. The fact that IDE−/− (red) samples had similar levels of CGRP1–37 to capsaicin-treated IDE+/+ samples (dark gray) (Fig. 5) suggests that IDE−/− mice may also have similar phenotypic responses as capsaicin-treated IDE+/+ mice in certain assays.
Fig. 5.
IDE regulates CGRP1–37 in vivo. (A) IDE+/+ and IDE−/− mice were treated with vehicle or capsaicin, and absolute endogenous CGRP levels were measured by IDMS29 using a synthetic d18-CGRP1–37 internal standard. CGRP1–37 levels were significantly higher in the IDE−/− mice than in the IDE+/+ mice under both treatment conditions. (B) A unique model for CGRP regulation includes the IDE-mediated proteolysis pathway discovered here. (**P value < 0.01; P values were derived from the two-tailed Student t test, n = 3. Error bars show SEM.)
Concluding Remarks.
In conclusion, we have discovered an additional role for IDE as a CGRP-degrading enzyme and simultaneously elucidated a proteolytic pathway for CGRP regulation in vivo. CGRP is only the third substrate identified for IDE in vivo, along with insulin22,23 and Aβ22,24. The identification of insulin (34, 35) and Aβ (34, 36) as substrates for IDE helped explain the link between IDE and diabetes (41, 42) and Alzheimer’s disease (43–45), respectively. Moreover, the identification of Aβ as an IDE substrate has led to the development of small-molecule IDE activators in an attempt to develop novel treatments for Alzheimer’s disease (46). The identification of CGRP as an IDE substrate suggests a potential role for this enzyme in blood pressure as well as the etiology of migraine, two important processes regulated by CGRP. It will be interesting to test specific hypotheses regarding IDE and physiology as more tools [e.g., small-molecule IDE activators (46) and IDE inhibitors (47)] become available. More generally, this discovery indicates that our peptidomics-based strategy can reveal unknown pathways and peptidases that regulate significant bioactive peptides, such as CGRP. Given the success of peptidase inhibitors as drugs, the continued application of this approach may reveal new targets for the development of next-generation therapeutics.
Materials and Methods
LC–MS/MS Experiments to Detect CGRP Peptide Fragments.
Aliquots of peptide extracts from spinal cords were injected onto a Waters nanoAcquity HPLC configured with in-house packed 75-μm reverse-phase capillary trapping and analytical columns (New Objective) coupled to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific). The liquid chromatography gradient proceeded from 3 to 33% acetonitrile/water (0.1% formic acid) over 180 min. Mass spectra from 395 to 1,600 m/z were acquired in the Orbitrap with a resolution of 60,000, and the six most abundant ions were targeted for concurrent MS/MS in the linear ion trap with relative collision energy of 30%, 2.5-Da isolation width, and recurring ions dynamically excluded for 60 s. Sequencing was done using the SEQUEST algorithm with differential modifications including methionine oxidation, amidation, acetylation, phosphorylation, and disulfide. Peptides were accepted within 3 ppm of the expected mass if they also met a series of custom filters on ScoreFinal (Sf), –10 Log P and charge state that attained an average peptide false discovery rate (FDR) of <2% across datasets. Manual inspection of spectra, FDR calculation, and protein inference were performed in Proteomics Browser Suite 2.23 (Thermo Fisher Scientific).
MALDI-TOF MS and LC–MS/MS Analysis of in Vitro and in Vivo Peptides.
Quenched in vitro and in vivo peptides were desalted using a ZipTip C18 (Millipore; ZTC18S096) according to the manufacturer’s protocol before MALDI-TOF MS and LC–MS/MS analysis. MALDI-TOF MS was performed with α-cyano-4-hydroxycinnamic acid as the matrix. Reaction products were also monitored using LC–MS/MS to confirm the peptides observed in the MALDI spectra. For LC–MS analysis, MALDI samples were diluted 10-fold into 0.1% formic acid in water. Aliquots were injected onto an Eksigent nanoLC-2D HPLC configured with a prepacked IntegraFrit trapping column (ProteoPep II C18, 300 Å, 5 µm) and an in-house packed reverse-phase 75-μm picotip analytical column (New Objective) coupled to an LTQ mass spectrometer (ThermoFisher Scientific). The liquid chromatography gradient proceeded from 5 to 50% acetonitrile in water (0.1% formic acid) over 40 min. Mass spectra were acquired from 400 to 1,600 m/z followed by targeting the three most abundant ions for MS/MS. Sequencing was carried out using the SEQUEST algorithm with differential modifications including methionine oxidation, amidation, acetylation, phosphorylation, and disulfide.
Capsaicin Injection Experiments.
For capsaicin injection experiments, 5- to 7-mo-old male and female IDE−/− and IDE+/+ mice (n = 5; 3 females and 2 males) were fasted overnight. Injections were performed i.p. with a 10 μL/g injection of either vehicle or 0.1 mg/mL capsaicin in 5% Tween-80, 5% ethanol, and 90% saline (vol/vol) for a final dose of 1 mg/kg capsaicin. Mice were allowed to return to their cages for 5 min and then plasma was collected as described below in Quantitative CGRP Measurements from Plasma.
Quantitative CGRP Measurements from Plasma.
To prevent the breakdown of physiological peptides during standard blood processing, we used a modified version of the recently reported reduced temperatures, acidification, protease inhibition, isotopic exogenous controls, and dilution (RAPID) method (48). Mice were killed by CO2 inhalation, decapitated, and their trunk blood collected into an EDTA-coated tube (BD Biosciences). This tube was quickly inverted five times and aliquots of blood (200 μL) were rapidly transferred into ice-cold buffer (pH 3.6, 4 mL) containing 1 nM heavy-labeled CGRP1–37 (d18-CGRP1–37), 0.1 M ammonium acetate, 0.5 M NaCl, and enzyme inhibitors (1 μg/mL of LAF-237, e-64, antipain, leupeptin, chymostatin, 1,10-phenanthroline, N-ethylmaleimide) and then immediately centrifuged at 3,000 × g for 10 min at 4 °C. The supernatant was transferred into a new tube and frozen at −80 °C for 24–72 h.
A Sep-Pak C18 cartridge 6 mL (Waters; WAT043395) was preconditioned by washing three times with acetonitrile (18 mL total) and four times with 0.1% TFA in water (24 mL total). Frozen supernatants from the RAPID plasma collection were thawed and 2 mL of this supernatant was applied to the Sep-Pak C18 cartridge. The loading step was repeated three times to ensure that all of the material bound to the column. The Sep-Pak was then washed with 0.1% TFA in water (24 mL total) and eluted with a 70:30 mixture of acetonitrile/water containing 0.1% TFA (2 mL total) into LoBind Eppendorf tubes. The sample was then dried using a speed vacuum concentrator at medium temperature. After concentration, the sample was dissolved in water with 0.1% formic acid (100 μL), centrifuged at 13,000 × g (10 min, 4 °C), and a part of the supernatant (20 μL) was further desalted using a ZipTip C18 (Millipore; ZTC18S096) according to the manufacturer’s instructions.
For the LC–MS/MS analysis, the samples were injected at an isocratic flow rate of 2 μL/min for 10 min onto a trapping column. The sample was then analyzed using a flow rate of 300 nL/min and a gradient of 5–65% B in 40 min (mobile phase A: 0.1% formic acid in water, mobile phase B: 0.1% formic acid in acetonitrile). The peptides were detected in the positive mode and CGRP levels were quantified by comparing the peak area for the +5 ion for CGRP1–37 and d18-CGRP1–37.
Reagents, Mouse Maintenance, Enzyme Assays, Biochemistry, and Cell Culture.
See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Cammi Valdez for assistance with maintenance of the mouse colony. This work was supported by a Forris Jewitt Moore Fellowship sponsored by Amherst College (to A.M.L.), National Institutes of Health (NIH) Training Grant GM007598 (to A.M.L and W.M.N.), a Searle Scholar Award (to A.S.), a Burroughs Wellcome Fund Career Award in the Biomedical Sciences (to A.S.), NIH Grant 1DP2OD002374 (to A.S.), and the Sloan Foundation (A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203195109/-/DCSupplemental.
References
- 1.Coll AP, Farooqi IS, O’Rahilly S. The hormonal control of food intake. Cell. 2007;129:251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hökfelt T, Bartfai T, Bloom F. Neuropeptides: Opportunities for drug discovery. Lancet Neurol. 2003;2:463–472. doi: 10.1016/s1474-4422(03)00482-4. [DOI] [PubMed] [Google Scholar]
- 3.De Felipe C, et al. Altered nociception, analgesia and aggression in mice lacking the receptor for substance P. Nature. 1998;392:394–397. doi: 10.1038/32904. [DOI] [PubMed] [Google Scholar]
- 4.Patchett AA, et al. A new class of angiotensin-converting enzyme inhibitors. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 5.Woods SC, Seeley RJ, Porte D, Jr, Schwartz MW. Signals that regulate food intake and energy homeostasis. Science. 1998;280:1378–1383. doi: 10.1126/science.280.5368.1378. [DOI] [PubMed] [Google Scholar]
- 6.Ueno H, Yamaguchi H, Kangawa K, Nakazato M. Ghrelin: A gastric peptide that regulates food intake and energy homeostasis. Regul Pept. 2005;126:11–19. doi: 10.1016/j.regpep.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 8.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–568. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 9.Che FY, et al. Identification of peptides from brain and pituitary of Cpe(fat)/Cpe(fat) mice. Proc Natl Acad Sci USA. 2001;98:9971–9976. doi: 10.1073/pnas.161542198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM. Proprotein convertases: Lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 11.Zhou A, Webb G, Zhu X, Steiner DF. Proteolytic processing in the secretory pathway. J Biol Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 12.Gradman AH, et al. Aliskiren, a novel orally effective renin inhibitor, provides dose-dependent antihypertensive efficacy and placebo-like tolerability in hypertensive patients. Circulation. 2005;111:1012–1018. doi: 10.1161/01.CIR.0000156466.02908.ED. [DOI] [PubMed] [Google Scholar]
- 13.Marguet D, et al. Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proc Natl Acad Sci USA. 2000;97:6874–6879. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lankas GR, et al. Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: Potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes. 2005;54:2988–2994. doi: 10.2337/diabetes.54.10.2988. [DOI] [PubMed] [Google Scholar]
- 15.Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 16.Mason RT, et al. Release of the predicted calcitonin gene-related peptide from cultured rat trigeminal ganglion cells. Nature. 1984;308:653–655. doi: 10.1038/308653a0. [DOI] [PubMed] [Google Scholar]
- 17.Brain S, Williams T, Tippins J, Morris H. Calcitonin gene-related peptide is a potent vasodilator. Nature. 1985;313:54–56. doi: 10.1038/313054a0. [DOI] [PubMed] [Google Scholar]
- 18.Tsujikawa K, et al. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci USA. 2007;104:16702–16707. doi: 10.1073/pnas.0705974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashina M, Bendtsen L, Jensen R, Schifter S, Olesen J. Evidence for increased plasma levels of calcitonin gene-related peptide in migraine outside of attacks. Pain. 2000;86:133–138. doi: 10.1016/s0304-3959(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 20.Tepper SJ, Stillman MJ. Clinical and preclinical rationale for CGRP-receptor antagonists in the treatment of migraine. Headache. 2008;48:1259–1268. doi: 10.1111/j.1526-4610.2008.01214.x. [DOI] [PubMed] [Google Scholar]
- 21.Olesen J, et al. BIBN 4096 BS Clinical Proof of Concept Study Group Calcitonin gene-related peptide receptor antagonist BIBN 4096 BS for the acute treatment of migraine. N Engl J Med. 2004;350:1104–1110. doi: 10.1056/NEJMoa030505. [DOI] [PubMed] [Google Scholar]
- 22.Yang D, et al. Activation of TRPV1 by dietary capsaicin improves endothelium-dependent vasorelaxation and prevents hypertension. Cell Metab. 2010;12:130–141. doi: 10.1016/j.cmet.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama M, et al. Catabolism of calcitonin gene-related peptide and substance P by neutral endopeptidase. Peptides. 1991;12:563–567. doi: 10.1016/0196-9781(91)90102-u. [DOI] [PubMed] [Google Scholar]
- 24.Le Grevès P, Andersson K, Silberring J. Isolation and identification of CGRP C-terminal fragments in the rat spinal cord. Neuropeptides. 1997;31:19–23. doi: 10.1016/s0143-4179(97)90014-7. [DOI] [PubMed] [Google Scholar]
- 25.Le Grevès P, Nyberg F, Hökfelt T, Terenius L. Calcitonin gene-related peptide is metabolized by an endopeptidase hydrolyzing substance P. Regul Pept. 1989;25:277–286. doi: 10.1016/0167-0115(89)90176-6. [DOI] [PubMed] [Google Scholar]
- 26.Tam EK, Caughey GH. Degradation of airway neuropeptides by human lung tryptase. Am J Respir Cell Mol Biol. 1990;3:27–32. doi: 10.1165/ajrcmb/3.1.27. [DOI] [PubMed] [Google Scholar]
- 27.Clynen E, Baggerman G, Husson S, Landuyt B, Schoofs L. Peptidomics in drug research. Expert Opinion on Drug Discovery. 2008;3:425–440. doi: 10.1517/17460441.3.4.425. [DOI] [PubMed] [Google Scholar]
- 28.Jankowski V, et al. Mass-spectrometric identification of a novel angiotensin peptide in human plasma. Arterioscler Thromb Vasc Biol. 2007;27:297–302. doi: 10.1161/01.ATV.0000253889.09765.5f. [DOI] [PubMed] [Google Scholar]
- 29.Tinoco AD, Tagore DM, Saghatelian A. Expanding the dipeptidyl peptidase 4-regulated peptidome via an optimized peptidomics platform. J Am Chem Soc. 2010;132:3819–3830. doi: 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rawlings ND, Barrett AJ, Bateman A. MEROPS: The peptidase database. Nucleic Acids Res. 2010;38(Database issue):D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duckworth WC, Heinemann MA, Kitabchi AE. Purification of insulin-specific protease by affinity chromatography. Proc Natl Acad Sci USA. 1972;69:3698–3702. doi: 10.1073/pnas.69.12.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray K, Hines CS, Rodgers DW. Mapping sequence differences between thimet oligopeptidase and neurolysin implicates key residues in substrate recognition. Protein Sci. 2002;11:2237–2246. doi: 10.1110/ps.0216302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira V, et al. Substrate specificity characterization of recombinant metallo oligo-peptidases thimet oligopeptidase and neurolysin. Biochemistry. 2001;40:4417–4425. doi: 10.1021/bi002715k. [DOI] [PubMed] [Google Scholar]
- 34.Farris W, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller BC, et al. Amyloid-beta peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc Natl Acad Sci USA. 2003;100:6221–6226. doi: 10.1073/pnas.1031520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi H, et al. Peptidomic identification and biological validation of neuroendocrine regulatory peptide-1 and -2. J Biol Chem. 2007;282:26354–26360. doi: 10.1074/jbc.M701665200. [DOI] [PubMed] [Google Scholar]
- 38.Glebov K, Walter J. Statins in Unconventional Secretion of Insulin-Degrading Enzyme and Degradation of the Amyloid-β Peptide. Neurodegener Dis. 2012;10:309–312. doi: 10.1159/000332595. [DOI] [PubMed] [Google Scholar]
- 39.Tamboli IY, et al. Statins promote the degradation of extracellular amyloid beta-peptide by microglia via stimulation of exosome-associated insulin-degrading enzyme (IDE) secretion. J Biol Chem. 2010;285:37405–37414. doi: 10.1074/jbc.M110.149468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keshishian H, et al. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 42.Zeggini E, et al. Wellcome Trust Case Control Consortium (WTCCC) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bertram L, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 44.Farris W, et al. Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol. 2004;164:1425–1434. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blomqvist ME, et al. Sequence variants of IDE are associated with the extent of beta-amyloid deposition in the Alzheimer’s disease brain. Neurobiol Aging. 2005;26:795–802. doi: 10.1016/j.neurobiolaging.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Cabrol C, et al. Small-molecule activators of insulin-degrading enzyme discovered through high-throughput compound screening. PLoS ONE. 2009;4:e5274. doi: 10.1371/journal.pone.0005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leissring MA, et al. Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin. PLoS ONE. 2010;5:e10504. doi: 10.1371/journal.pone.0010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stengel A, et al. The RAPID method for blood processing yields new insight in plasma concentrations and molecular forms of circulating gut peptides. Endocrinology. 2009;150:5113–5118. doi: 10.1210/en.2009-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.