Abstract
Many studies have evaluated the performance of risk assessment models for BRCA1/2 mutation carrier probabilities in different populations, but to our knowledge very few studies have been conducted in the German population so far. In the recent study, we validated the performance of three risk calculation models by names BRCAPRO, Myriad and BOADICEA in 183 German families who had undergone molecular testing of mutations in BRCA1 and BRCA2 with an indication based on clinical criteria regarding their family history of cancer. The sensitivity and specificity at the conventional threshold of 10% as well as for a threshold of 20% were evaluated. The ability to discriminate between carriers and non-carriers was judged by the area under the receiver operating characteristics curve. We further focused on the performance characteristic of these models in patients carrying large genomic rearrangements as a subtype of mutations which is currently gaining increasing importance. BRCAPRO and BOADICEA performed almost equally well in our patient population, but we found a lack of agreement to Myriad. The results obtained from this study were consistent with previously published results from other population and racial/ethnic groups. We suggest using model specific decision thresholds instead of the recommended universal value of 10%. We further suggest integrating the CaGene5 software package, which includes BRCAPRO and Myriad, in the genetic counselling of German families with suspected inherited breast and ovarian cancer because of the good performance of BRCAPRO and the substantial ease of use of this software.
Electronic supplementary material
The online version of this article (doi:10.1007/s10689-011-9498-y) contains supplementary material, which is available to authorized users.
Keywords: BOADICEA, BRCA1, BRCA2, BRCAPRO, LGRs, Myriad
Introduction
Breast cancer (BC) is the most frequently diagnosed type of cancer and the leading cause of cancer deaths of females in industrialized nations. Incidence rates are increasing in Germany, but sharply decreasing in other countries like the USA, UK, France and Australia since the beginning of the millennium [1]. The sharp reduction in BC incidence in these countries may be related to a national decline in the use of hormone replacement therapy (HRT) after publishing the results of the women’s health initiative (WHI) randomized trials of HRT use in 2002. Interestingly, the reduction in BC incidence was most pronounced in countries where peak prevalence of HRT use was quite high [2]. However, BC death rates have been decreasing in North America and several European countries over the past 25 years [3, 4]. Upto 5–10% of BC cases can be attributed to individuals with hereditary breast and ovarian cancer (HBOC). Unambiguously deleterious mutations in BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) contribute to BC and other associated tumors in large numbers of HBOC families [5]. The contribution of further tumor suppressor genes like p53 (MIM# 191140); PTEN (MIM# 601728), ATM (MIM# 208900) and RAD51C (MIM# 613399) to cancers in HBOC is very low [6, 7]. The prevalence of BRCA1 or BRCA2 germ line mutations varies widely among populations [8]. Identifying a person at high risk of carrying a BRCA1 or BRCA2 mutation on the basis of his familial pedigree poses a big challenge for the genetic counsellors. A common approach is the estimation of the carrier probability based on the familial history of a counselee, dealing especially with the number of family members affected by breast and/or ovarian cancer and the age of onset. Many European countries like UK, France, the Netherlands and Germany has established its own indication criteria and recommendations for genetic testing [9]. If families or single affected individuals fulfil these inclusion criteria for mutation screening, the molecular genetic analysis is offered. Several empiric and computer-based risk assessment models for BRCA1/2 mutation carrier probabilities have been developed to assist in pretest counseling, but independent validations of the performance of such models has produced variable results [10].
BRCA gene mutation probability thresholds used to establish the indication to perform DNA-analysis vary considerably among countries. In Germany the molecular genetic testing is recommended for families in which the mutation probability is 10% or larger [11]. The German consortium for hereditary breast and ovarian cancer (GC-HBOC) established clinical guidelines for genetic testing in suspected families based on their cancer history (Table 1). These guidelines obviously provide a rough estimation of the individual risk based on the results of empirical studies without any consideration of the degrees of relationship among affected family members, whereas mutation probability models are especially helpful to assess the individual carrier probability which underlies external variables such as pattern of inheritance, kind of tumors in family history, age-dependent incomplete penetrances, etc. [12]. However, mutation probability models have shown considerable discrepancies in performance across different racial/ethnic groups [13]. The aim of the present study is two-fold: (a) to compare performance and clinical applicability of three prediction models that performed best in former international studies in order to find the one that could best be integrated into the clinical course of our breast cancer consultancy, (b) To assess the performance characteristic of these models in patients carrying BRCA1- or BRCA2- large genomic rearrangements (LGRs), as a subtype of easily detectable Mutations.
Table 1.
Indication for molecular testing based on clinical features of the individual’s family history of cancer by the German consortium for hereditary breast and ovarian cancer (GC-HBOC)
| Number of affected relatives | Indication for performing DNA-analysis |
|---|---|
| 1 | |
| a | BC ≤ 35 years |
| b | Bilateral BC ≤ 50 years |
| c | BC and OC |
| d | BC/OC and male BC in the family |
| 2 | |
| a | Two cases of BC/OC, one case <51 years |
| b | Two cases OC |
| c | One case BC and one case OC |
| 3 | |
| a | Three cases of BC |
Materials and methods
Study population
Our study sample comprises all patients that attended interdisciplinary breast cancer consultancy in the Breast Cancer Center at the University Medical Center of Göttingen between 1999 and 2009. In total, we retrospectively analysed data of 183 unrelated families for which at least one affected member (the so called index-patient) was tested for mutations in the BRCA1 and BRCA2 genes. All these families met the inclusion criteria for genetic testing according to the clinical guidelines of GC-HBOC [14, 15].
The patient record is compiled by a genetic consultant and contains a complete personal medical history, the informed consent for molecular analysis, a three generation family pedigree as well as tumor and therapy related information. Diagnoses were confirmed using hospital records and pathology reports in most instances. Counselee’s first and second degree affected relatives were asked, when possible, to sign an informed consent to release their medical records. Both affected men and women were included in the study. Individuals with ductal carcinoma in situ (DCIS) were entered into the models as having had invasive BC [16]. Fallopian tube or peritoneal cancer were counted same as OC [17]. Individuals carrying an unambiguously deleterious mutation were exclusively considered to be BRCA1 or BRCA2 positive. Variants of unspecified significance (UVS) were assumed to be negative test results. The study was approved by the local ethical committee.
Mutation testing
Until 2005, molecular testing was accomplished using the denaturing high-performance liquid chromatography (DHPLC) method (group 1, 61 families). From 2005, direct DNA-sequencing was performed (group 2, 122 families). In the latter group, a subsequent multiplex ligation dependent probe amplification (MLPA) analysis for LGRs in case of negative sequencing results was carried out. The detected rearrangements were confirmed by using quantitative real-time polymerase chain reaction (QRT-PCR). All mutations and genetic variables were classified according to the breast cancer information core (BIC) database (http://research.nhgri.nih.gov/bic/) as well as to the human gene mutation database (HGMD, available at: http://www.biobaseinternational.com/Pages/index.php?id=hgmddatabase).
Mutation probability methods
The individual risk of carrying a BRCA1 or BRCA2 mutation was calculated for each patient by applying the three different models. The calculations for Myriad and BRCAPRO were carried out by using CaGene5 software package (http://www4.utsouthwestern.edu/breasthealth/cagene/). For BOADICEA, the corresponding software can be run online on the Cambridge University website (http://www.srl.cam.ac.uk/genepi/boadicea/boadicea_intro.html).
Statistical analysis
To evaluate the strength of the pairwise correlation between risks calculated by each model, we considered bias corrected Spearman’s rank correlation coefficients with exact 95% confidence intervals. To assess the agreement of classification, exceeding concordance by chance, we calculated kappa coefficients for decision thresholds of 10 and 20%. To evaluate the models’ ability to discriminate between mutation positive and negative individuals, we constructed sensitivity, specificity and ROC curves. Confidence intervals (CI) for sensitivity and specificity where derived by the method of Pearson and Clopper. ROC-curves were plotted and compared by means of AUC (area under the curve) according the method of DeLong [18], setting the level of significance to 5%. CI of thresholds given specific values of sensitivity or specificity cover all observed thresholds, where the value is included in the confidence interval of the considered measure. All calculations were performed using SAS 9.2 or standard programs for R.
Results
DNA mutational analysis
Out of the 183 families that underwent BRCA1 and BRCA2 mutation testing, 48 families (26.2%) showed a deleterious BRCA-mutation. 35 families (19.1%) held a mutation in BRCA1 including 3 LGRs; and 13 families (7.1%) held a mutation in BRCA2 including 2 LGRs. The most frequently detected mutation was the c.5382insC in exon 20 of BRCA1 (8 families) followed by the mutation c.185delAG in exon 2 of BRCA1 which was found in 4 families. The whole mutations detected in our patient collective listed with family members affected by breast and/or ovarian cancer with age at diagnosis and personal risk figures calculated by BRCAPRO, BOADICEA and Myriad are given in Online Resource 1.
Correlation and agreement of calculated risk
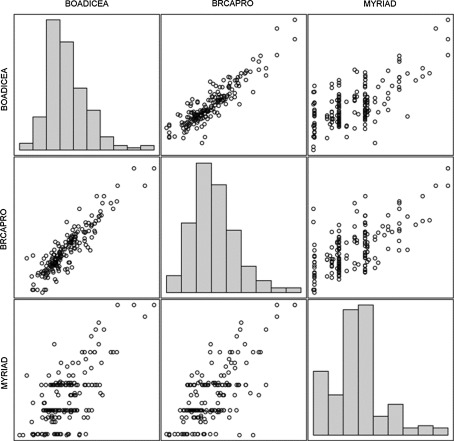
The risks calculated by BRCAPRO and BOADICEA were found to be strong correlated in mutation carriers (r = 0.93) and in non-carriers (r = 0.81), although a little less distinct in the latter. But risks calculated by Myriad show only a weak correlation to those of BOADICEA and BRCAPRO (Table 2). Typically the risks calculated by Myriad were lower than by those of BRCAPRO and BOADICEA, in mutation carrier (avg. risks: 28.65, 61.8 and 57.9%) as in non-carrier (avg. risks: 9.5, 17.3 and 17.8%). It can be seen from Fig. 1 that Myriad assigns the same risk to groups of individuals, where the other methods calculate wider ranges of risks.
Table 2.
Sensitivity and specificity of all three programs at a threshold of 10 and 20%
| Threshold | BRCAPRO | BOADICEA | Myriad |
|---|---|---|---|
| Mutation carrier, N = 48 | |||
| Sensitivity % (95% CI) | |||
| 10 | 85 (72–94) | 85 (72–94) | 88 (75–95) |
| 20 | 81 (67–91) | 81 (67–91) | 48 (33–63) |
| Average of calculated risk | 61.8 (52–72) | 57.9 (48–68) | 28.6 (22–35) |
| Correlation to | |||
| BRCAPRO | 1 | ||
| BOADICEA | 0.93 (0.88–0.96) | 1 | |
| Myriad | 0.56 (0.33–0.73) | 0.65 (0.45–0.79) | 1 |
| Inter-method agreement (kappa) at threshold 10% | |||
| BRCAPRO | 1 | ||
| BOADICEA | 83% (61–100%) | 1 | |
| Myriad | 56% (21–90%) | 56% (21–90%) | 1 |
| Mutation non-carrier, N = 135 | |||
| Specificity % (95% CI) | |||
| 10 | 62 (53–70) | 56 (47–64) | 67 (59–75) |
| 20 | 72 (63–79) | 70 (61–77) | 94 (89–97) |
| Average of calculated risk | 17.3 (14–21) | 17.8 (14–21) | 9.5 (8–11) |
| Correlation to | |||
| BRCAPRO | 1 | ||
| BOADICEA | 0.81 (0.74–0.86) | 1 | |
| Myriad | 0.42 (0.27–0.55) | 0.32 (0.16–0.46) | 1 |
| Inter-method agreement (kappa) at threshold 10% | |||
| BRCAPRO | 1 | ||
| BOADICEA | 74% (63–86%) | 1 | |
| Myriad | 34% (17–50%) | 26% (10–42%) | 1 |
Fig. 1.
Scatterplots: the risk stratifications of the three programs. Risk where transformed to logits for a more detailed presentation of small values
Regardless the high correlation of BRCAPRO with BOADICEA, the agreement of classification between these two at a decision threshold of 10% reaches 83% (95% CI: 61–100%) in mutation carrier but only 74% (95% CI: 62–86%) in non-carrier. In other words, there is still a noticeable amount of individuals classified differently by BRCAPRO and BOADICEA (2 of 48 mutation carriers and 19 of 135 non-carriers). The agreement of classification compared to MAYRIAD was moderate (about 50%) in mutation carrier and lower than 35% in non-carrier (Table 2).
Performance at pre-specified thresholds
Applying a decision threshold of 10% yielded a sensitivity of about 85% for all three methods, meaning that a genetic test was recommended to 41 or 42 of all 48 mutation carriers. On the other hand Myriad showed the largest specificity (67, 95% CI: 59–75%), meaning that genetic testing was recommended to 44 of all 135 non-mutation carriers. For BRCAPRO and BOADICEA (specificity of 62 and 56%) these were 51 and 60 non-mutation carriers respectively, hence an increase in 16 or 36% of unnecessary recommended genetic testing (Table 2).
Considering a decision threshold of 20%, the sensitivity of the methods decreases while the specificity increases as expected. For BRCAPRO and BOADICEA, we noticed small changes only (sensitivity: about −4-points, specifity: about +10-points). But for Myriad a remarkable reduction to 48% true positive rate together with a true negative rate of 94% were found.
In applying this threshold to BRCAPRO and BOADICEA, one would unnecessarily recommend genetic testing for 38, respectively 41, of all 135 non-carriers. Using Myriad, there were 8 non-carriers only. On the other hand, using Myriad, no genetic test would be recommended for 25 of all 48 mutation carriers, while using BRCAPRO or BOADICEA, there were only 9 mutation carriers.
Discrimination ability
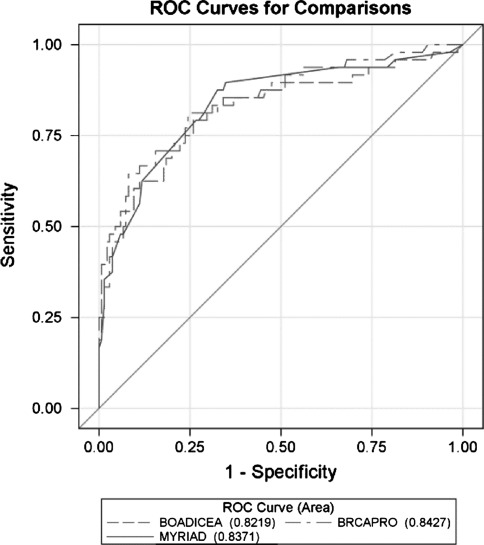
Comparing ROC-curves of the three methods does not reveal a significant difference in the overall ability to discriminate individuals at elevated and non-elevated risk (P = 0.5391) based on the sample at hand (Fig. 2). Hence we could not observe in general any advantage in sensitivity (true positive rate) given a false positive rate (specifity) for any of the methods. But the decision thresholds to achieve a certain couple of sensitivity/specificity vary noticeable, the lower the target false positive rate.
Fig. 2.
ROC curves: application of sensitivity against 1-specificity of BRCAPRO, BOADICEA and Myriad
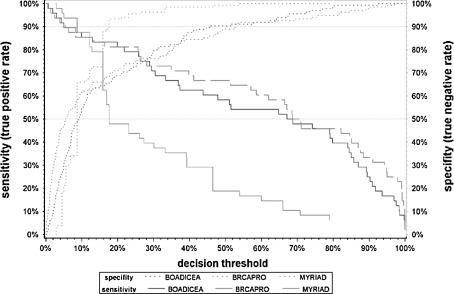
For a nominal specificity of 50%, a threshold of 9.2% needs to be applied using BOADICEA, 8.7% in using Myriad and 5.5% in using BRCAPRO, respectively. In doing so, a true positive rate of about 90% can be expected. But for a nominal specificity of 90% (genetic testing will be recommended to only 10% of non-mutation carriers), a threshold of 46% needs to be applied using BOADICEA, 18% in using Myriad and 53% in using BRCAPRO, respectively. The expected true positive will then range roughly between 50 and 60%. Vice versa, if one aims sensitivity of e.g. 80%, the decision threshold to apply for BOADICEA will be 22% and for BRCAPRO 26%. For Myriad the estimated threshold is significantly lower at 13% (95% CI: 8.7–16%).
We found that a small change in the decision threshold has much more effect on the discrimination characteristics for Myriad than for BRACAPRO or BOADICEA (Table 3; Fig. 3). To achieve the same values in sensitivity or specificity, the thresholds for Myriad need to be smaller than for the other two methods, with the exception of requested high sensitivity/low specificity.
Table 3.
model specific decision thresholds to achieve given sensitivity or specificity for both mutation and non-mutation carriers
| Nominal specificity % | Threshold % (95% CI) | Sensitivity % | ||||
|---|---|---|---|---|---|---|
| BOADICEA | BRCAPRO | Myriad | BOADICEA | BRCAPRO | Myriad | |
| 50 | 9.2 (7.1–11) | 5.5 (3.7–8.8) | 8.6 (6.8–8.8) | 87.5 | 90.0 | 90.0 |
| 60 | 11.6 (9.4–19) | 9.4 (6.0–17) | 8.7 (8.0–11) | 85.4 | 85.4 | 90.0 |
| 70 | 22 (13–30) | 19 (11–33) | 12.2 (8.7–16) | 81.3 | 81.3 | 81.3 |
| 80 | 30 (25–39) | 35 (22–47) | 15.9 (15–16) | 70.8 | 70.8 | 62.5 |
| 90 | 46 (38–67) | 53 (39–76) | 18 (16–21) | 60.4 | 64.6 | 47.9 |
| Nominal sensitivity % | Threshold % (95% CI) | Specificity % | ||||
|---|---|---|---|---|---|---|
| BOADICEA | BRCAPRO | Myriad | BOADICEA | BRCAPRO | Myriad | |
| 50 | 67 (37–85) | 69 (51–88) | 18 (16–33) | 94.1 | 92.6 | 93.3 |
| 60 | 47.9 (29–77) | 62.0 (28–82) | 16.9 (16–21) | 90.4 | 91.9 | 88.9 |
| 70 | 30.4 (13–51) | 39.8 (13–67) | 15.9 (12–17) | 80.0 | 84.4 | 88.1 |
| 80 | 22 (5.0–32) | 26 (5.4–40) | 13 (8.7–16) | 70.4 | 75.6 | 72.6 |
| 90 | 5.0 (0.6–22) | 5.4 (0.7–25) | 8.7 (2.9–12) | 30.4 | 48.9 | 65.2 |
Fig. 3.
Sensitivity and specificity of BRCAPRO, BOADICEA and Myriad
Large genomic rearrangements (LGRs)
We separately evaluated the risk values of the 5 probands carrying a LGR. The three probands that carry a large deletion or duplication in BRCA1 exhibited a high (≥70%) risk profile in BRCAPRO (71, 97 and 100%, respectively). Likewise BOADICEA calculated high risk values (84.5, 97 and 99.97%), whereas Myriad assessed rather moderate (≥30 to <70%) risk figures for these individuals (39.2, 39.2 and 79%).
For the carrier of the exon 15–16, duplication in BRCA2 both BRCAPRO and BOADICEA calculated moderate risk values (34.2 and 25.77%, respectively) while Myriad assessed a false negative risk <10% (8.7%). For the last proband who carries the BRCA2 exon 22–27 deletion, none of the models computed an over 10% elevated risk (BRCAPRO: 1.5%, BOADICEA: 1.66% and Myriad: 4.5%).
Discussion
We conducted the current study to evaluate the performance and elucidate the clinical practicability of the above-described assessment models for mutation carrier probabilities in 183 German families previously tested for mutations in BRCA1 and BRCA2 gene. Many studies have been carried out to evaluate the performance of such models in different populations [19–23], but very few studies dealt with their reliability and applicability in the German population so far [24, 25].
We obtained a mutation detection rate of 26%, which is comparable to the mutation detection rate of GC-HBOC of 27% [26]. The most frequently detected mutation was the BRCA1 c.5382insC mutation known to be frequent in German BC patients [27] and displays the second most described mutation in the BIC database. Furthermore, the mutation c.185delAG in exon 2 of BRCA1 was found 4 times in our patient population. It is described as an Ashkenasim Jewish founder mutation [28], but was also found frequently in other racial/ethnic groups [29, 30]. It is also the most listed mutation of BRCA1 in the BIC database. Unfortunately we cannot reconstruct the proportion of consulters who are from Ashkenasim Jewish descent.
At the recommended universal decision threshold of 10%, all three methods are comparable in terms of sensitivity (about 85%). All models demonstrated a strong ability to discriminate between carriers and non-carriers with an area under the ROC (AUC) between 0.77 and 0.80 (Fig. 2). Our results are consistent with previously published results from other population and racial/ethnic groups which showed reasonably similar performance and AUCs of all the models [12–16].
The exclusion of a high proportion of non-mutation carriers from expensive molecular analysis is thought to be an advantage of the risk stratifying programs over clinical guidelines. At the decision threshold of 10%, genetic tests would not be recommended to between 55 and 67% of all non-mutation carriers. The models would help in such a way as to save the limited healthcare resources [31]. On the other hand, a primary disadvantage is their consistent tendency to underpredict the number of mutations in families with low a priori risk [21].
The specificity of the models could be increased by choosing a higher threshold level which would admittedly go along with a loss of sensitivity and simultaneously the occurrence of more false negative test results (mutation carriers with a risk <10%). We found that the methods behave quite differently in such a change of the cut-off point. At a universal decision threshold of 20%, the empirical model Myriad showed a sensitivity of 95%, while only every second mutation carrier would be detected. The two mathematical models (BRCAPRO and BOADICEA) showed only an inert change in their ability to discriminate.
The median mutation probabilities in our patient population differ between the two mathematical models and the empirical model (Table 2). The high correlation between BRCAPRO and BOADICEA and the weak correlation to Myriad support this picture (Fig. 1). In contrast to the other two programs, Myriad stratifies the consulter’s risk on the basis of a strongly condensed family history and allocates it into several risks categories. Thus for several individuals, the same risk value is predicted by this program. This circumstance becomes visually clear in Fig. 1 as the risk values were assessed by Myriad are situated in groups.
The imperfect agreement within calculated risks of BC indicates that the use of an universal decision threshold for any model, as the recommended 10% threshold of GC-HBOC, can lead to quite different conclusions. The calculated risks should not be considered as solely based on the family history but also on the mathematical rules and the model used. Our data show that an appropriate decision threshold should be derived from diagnostic accuracy measures rather than defined directly by any BC risk. From a clinical point of view, a high sensitivity, i.e. detecting a higher number of mutation carriers, is more important than a high specificity which would economically reduce expenses for molecular analysis. For instance, if one seeks a sensitivity of 80%, a threshold of ~25% for BRCAPRO and BOADICEA would be equivalent to a threshold of 13% for Myriad. This is accompanied by a specificity of ~75%. In contrast to the recommended threshold of 10%, less mutation non-carriers will be sent to genetic testing by almost comparable sensitivity.
The contribution of LGRs to the whole BRCA1/2 mutations varies widely among populations from almost no presence in the African till 27% in the Dutch population. A small to no contribution of BRCA2 LGRs in HBOC have been reported in the majority of publications [32]. LGRs in BRCA1 accounted for 9.6% of all BRCA1-mutations in the study of Engert et al. on 1,506 German families with suspected HBOC [33]. In the course of this study, a subgroup of 412 high-risk individuals has been screened for LGRs in BRCA2 with negative results. Veschi et al. have reported that LGRs in BRCA1 are associated with high BRCAPRO—a priori risks [34]. To verify this hypothesis, we separately evaluated the risk values of the 5 probands carrying a LGR in our patients’ collective. Similar high risk figures for the three BRCA1-LGRs-carriers have been assessed by BRCAPRO and BOADICEA in our study. Myriad has delivered moderately elevated risk figures for these probands. Interestingly, two LGRs in the BRCA2 have been found in our collective (Online Resource 1). For the previously described duplication of exon 15–16, the three models assessed a moderate to low risk. For the other patient who carries the novel deletion of exon 22–27, all programs calculated false negative risk values. Indeed, she had a negative family history concerning BRCA-associated tumors. The indication for DNA analysis was the occurrence of BC at an age younger than 35 years (indication 1a, Table 1). DNA samples from parents were not available for testing on this deletion. Indeed, due to a small number of detected LGRs in our study, a possible correlation between this type of BRCA mutation and high or moderate a- priori risk estimated by BRCAPRO and BOADICEA should be interpreted with discretion. However, screening for LGR in both BRCA1 and BRCA2 should be considered to complete the genetic analysis in HBOC patients with no apparent BRCA1/2 point mutations.
As there are no strong discrepancies in the performance characteristic of these models, the ease of use gains a strong impact for basing a decision as to which model could be implied in the genetic counseling of German families with suspected HBOC. We suggest integrating the CaGene5 software package because of the good performance of BRCAPRO and the substantially ease of use of CaGene5 if compared with BOADICEA. The Myriad model is included within this package so that there is a double coverage in the case of uncertainty. Finally, to draw more precise conclusions about LGRS, further studies with larger number of BRCA 1\2 LGRs-carriers are needed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Since 2011 the Breast Cancer Center at the Medical University Göttingen is a member of the GC-HBOC. We would like to thank Prof. R. Schmutzler from GC-HBOC for her great support. We would also like to thank Prof. W. Engel and A. Lou for their careful review of the manuscript.
Conflict of interest
The authors wish further to acknowledge that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The authors S. M. Schneegans and A. Rosenberger contributed equally to this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Zbuk K, Anand SS. Declining incidence of breast cancer after decreased use of hormone-replacement therapy: magnitude and time lags in different countries. J Epidemiol Community Health. 2012;66(1):1–7. doi: 10.1136/jech.2008.083774. [DOI] [PubMed] [Google Scholar]
- 3.Autier P, Boniol M, La Vecchia C, et al. Disparities in breast cancer mortality trends between 30 European countries: retrospective trend analysis of WHO mortality database. BMJ. 2010;341:c3620. doi: 10.1136/bmj.c3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Center MM, DeSantis C, EM Ward. Global patterns of cancer incidence, mortality rates, trends. Cancer Epidemiol Biomarkers Prev. 2010;19(8):1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Ford D, Easton DF, Stratton M, et al. Genetic heterogeneity, penetrance analysis of the BRCA1, BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am J Hum Genet. 1998;62(3):676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll JC, Cremin C, Allanson J, et al. Hereditary breast and ovarian cancers. Can Fam Physician. 2008;54(12):1691–1692. [PMC free article] [PubMed] [Google Scholar]
- 7.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast, ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 8.Stegel V, Krajc M, Zgajnar J, et al. The occurrence of germline BRCA1, BRCA2 sequence alterations in Slovenian population. BMC Med Genet. 2011;12:9. doi: 10.1186/1471-2350-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadzicki D, Evans DG, Harris H, et al. Genetic testing for familial/hereditary breast cancer—comparison of guidelines and recommendations from the UK, France, The Netherlands and Germany. J Community Genet. 2011;2:53–69. doi: 10.1007/s12687-011-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102(10):680–691. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 11.Kuschel B, Hauenstein E, Kiechle M, Meindl A. Hereditary breast and ovarian cancer—current clinical guidelines in Germany. Breast Care. 2006;1:8–14. doi: 10.1159/000091305. [DOI] [Google Scholar]
- 12.Katki H. Incorporating medical interventions into carrier probability estimation for genetic counseling. BMC Med Genet. 2007;8:13. doi: 10.1186/1471-2350-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurian AW, Gong GD, John EM, et al. Performance of prediction models for BRCA mutation carriage in three racial/ethnic groups: findings from the Northern California breast cancer family registry. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1084–1091. doi: 10.1158/1055-9965.EPI-08-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmutzler R, Beckmann M, Kiechle M. Prevention: hereditary breast and ovarian cancer. Dtsch Arztebl. 2002;99:A1372–A1378. [Google Scholar]
- 15.Kreienberg R, Kopp I, Albert U et al. (2008) Interdisziplinare S3-leitlinie fur die diagnostik, therapie und nachsorge des mammakarzinoms. Munich, W. Zuckschwerdt Verlag GmbH
- 16.Bayraktar S, Elsayegh N, Gutierrez Barrera AM et al. (2011) Predictive factors for BRCA1/BRCA2 mutations in women with ductal carcinoma in situ. Cancer [DOI] [PMC free article] [PubMed]
- 17.Cremin C, Wong N, Buzaglo K, Paradis AJ, Foulkes W. Nonovarian pelvic cancers in BRCA1/2 mutation carriers and the BRCAPRO statistical model. J Clin Oncol. 2002;20(18):3936. doi: 10.1200/JCO.2002.02.130. [DOI] [PubMed] [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 19.Antoniou AC, Hardy R, Walker L, et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: validation of BOADICEA, BRCAPRO, IBIS, Myriad, the Manchester scoring system using data from UK genetics clinics. J Med Genet. 2008;45(7):425–431. doi: 10.1136/jmg.2007.056556. [DOI] [PubMed] [Google Scholar]
- 20.Kurian AW, Gong GD, Chun NM, et al. Performance of BRCA1/2 mutation prediction models in Asian Americans. J Clin Oncol. 2008;26(29):4752–4758. doi: 10.1200/JCO.2008.16.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindor NM, Lindor RA, Apicella C, et al. Predicting BRCA1 and BRCA2 gene mutation carriers: comparison of LAMBDA, BRCAPRO, Myriad II, and modified Couch models. Fam Cancer. 2007;6(4):473–482. doi: 10.1007/s10689-007-9150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HH, Williams R, Leary J, Ringland C, Kirk J, Ward R. Evaluation of models to predict BRCA germline mutations. Br J Cancer. 2006;95(7):914–920. doi: 10.1038/sj.bjc.6603358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panchal SM, Ennis M, Canon S, Bordeleau LJ. Selecting a BRCA risk assessment model for use in a familial cancer clinic. BMC Med Genet. 2008;9:116. doi: 10.1186/1471-2350-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasching PA, Bani MR, Nestle-Kramling C, et al. Evaluation of mathematical models for breast cancer risk assessment in routine clinical use. Eur J Cancer Prev. 2007;16(3):216–224. doi: 10.1097/CEJ.0b013e32801023b3. [DOI] [PubMed] [Google Scholar]
- 25.Beckmann MW, Bani MR, Fasching PA, Strick R, Lux MP. Risk and risk assessment for breast cancer: molecular and clinical aspects. Maturitas. 2007;57(1):56–60. doi: 10.1016/j.maturitas.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Kast K, Schmutzler RK, Distler W et al. (2009) Prevalence rates of pathogenic mutations in the BRCA1 and BRCA2 genes in families with different disease histories: results from the German consortium for hereditary breast and ovarian cancer. Cancer Res 69(24):731S-S
- 27.Backe J, Hofferbert S, Skawran B, et al. Frequency of BRCA1 mutation 5382insC in German breast cancer patients. Gynecol Oncol. 1999;72(3):402–406. doi: 10.1006/gyno.1998.5270. [DOI] [PubMed] [Google Scholar]
- 28.Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1, BRCA2. Nat Genet. 1996;14(2):185–187. doi: 10.1038/ng1096-185. [DOI] [PubMed] [Google Scholar]
- 29.Vaidyanathan K, Lakhotia S, Ravishankar HM, Tabassum U, Mukherjee G, Somasundaram K. BRCA1, BRCA2 germline mutation analysis among Indian women from south India: identification of four novel mutations, high-frequency occurrence of 185delAG mutation. J Biosci. 2009;34(3):415–422. doi: 10.1007/s12038-009-0048-9. [DOI] [PubMed] [Google Scholar]
- 30.Mullineaux LG, Castellano TM, Shaw J, et al. Identification of germline 185delAG BRCA1 mutations in non-Jewish Americans of Spanish ancestry from the San Luis Valley, Colorado. Cancer. 2003;98(3):597–602. doi: 10.1002/cncr.11533. [DOI] [PubMed] [Google Scholar]
- 31.Zanna I, Rizzolo P, Sera F, et al. The BRCAPRO 5.0 model is a useful tool in genetic counseling, clinical management of male breast cancer cases. Eur J Hum Genet. 2010;18(7):856–858. doi: 10.1038/ejhg.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1, BRCA2 genes: review of the literature, report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 33.Engert S, Wappenschmidt B, Betz B, et al. MLPAscreening in the BRCA1 gene from 1, 506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, occurrence in single early-onset cases. Hum Mutat. 2008;29(7):948–958. doi: 10.1002/humu.20723. [DOI] [PubMed] [Google Scholar]
- 34.Veschi S, Aceto G, Scioletti AP, et al. High prevalence of BRCA1 deletions in BRCAPRO-positive patients with high carrier probability. Ann Oncol. 2007;18(6):vi86–vi92. doi: 10.1093/annonc/mdm233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.