Abstract
Bcl11b is a lineage-specific transcription factor expressed in various cell types and its expression is important for development of T cells, neurons and others. On the other hand, Bcl11b is a haploinsufficient tumor suppressor and loss of a Bcl11b allele provides susceptibility to mouse thymic lymphoma and human T-cell acute lymphoblastic leukemia. Although there are many transcription factors affecting both cell differentiation and cancer development, Bcl11b has several unique properties. This review describes phenotypes given by loss of Bcl11b and roles of Bcl11b in cell proliferation, differentiation and apoptosis, taking tissue development and lymphomagenesis into consideration.
Keywords: Bcl11b, T-cell development, haploinsufficient tumor suppressor, T-cell leukemia, thymic lymphoma
1. Introduction
Bcl11b (B-cell CLL/lymphoma 11b) belongs to Kruppel-like C2H2 type zinc finger transcription proteins, the largest family of transcription factors in eukaryotes.1) The gene encoding Bcl11b was first identified as a tumor suppressor gene by our study.2) In fact, mice lacking one Bcl11b allele are susceptible to thymic lymphomas. On the other hand, mice lacking both Bcl11b alleles, which die shortly after birth of unknown causes, exhibit many defects in different organs of newborn mice, including immune system, central nervous system (CNS), skin, teeth, and hair cells in cochlea.3–6) Therefore, Bcl11b plays critical roles in the development of those organs and possibly others.7) Recently, Liu et al. have reviewed roles for Bcl11b in T-cell development and maintenance of T-cell lineage commitment.1) Thus, this review provides a focus on the tumor suppressor role of Bcl11b rather than T-cell development.
Bcl11b is located on mouse chromosome 12 and on human chromosome 14. This gene is originally called Rit1 (radiation-induced tumor suppressor gene 1), because Bcl11b was isolated by scanning γ-ray induced mouse thymic lymphomas for losses of specific chromosomal DNA.8) More than 10 years ago, the scanning was performed for the 361 thymic lymphomas that were induced in mice crossed between BALB/c and MSM strains. The two strains belong to different mouse subspecies, Mus musculus domesticus and Mus musculus molossinus, respectively, and hence they carry many distinct alleles and DNA markers between the two. Genome-wide allelic loss or loss of heterozygosity (LOH) analysis using polymorphic DNA markers mapped several candidate tumor suppressor gene regions.8) Further analysis localized one of the regions on mouse chromosome 12 to a 2.9 cM interval between the D12Mit53 and D12Mit279 marker positions.9) Construction of a physical map consisting of 15 BAC clones in the vicinity contained informative boundaries of allelic losses, which allowed us to finally localize a 35 kb interval with a high frequency of allelic loss (62%). Sequence analysis of this interval led to the finding of Bcl11b gene, and mutation analysis identified this gene responsible for thymic lymphoma development.2)
Another candidate region was mapped on mouse chromosome 11, which harbored Ikaros gene. Mutation analysis of this gene in thymic lymphomas identified it as a tumor suppressor gene.10) Ikaros is the well-known gene that plays critical roles in the development of lymphoid tissues and lymphomas.11,12)
2. Bcl11b and Bcl11a
Bcl11a is another member of the Bcl11 family in the mouse and human genomes.13,14) Although Bcl11a and Bcl11b share some sequence homology, they are located on different chromosomes and have different exon-intron structures. Bcl11a and Bcl11b are also called Ctip1 and Ctip2, respectively,15) because they were independently isolated for their interaction with the chicken ovalbumin upstream promoter transcription factor (COUP-TF) of orphan nuclear receptors. COUP-TF family members play important roles in development,16) and they usually mediate transcriptional repression by recruiting nuclear receptor co-repressor (NCoR) and/or silencing mediator for retinoid and thyroid hormone receptor (SMRT) to the template.17) As a transcription factor, Bcl11a and Bcl11b are also associated with the nucleosome remodeling and histone deacetylase (NuRD) complex to repress target promoters.18,19) However, functional association between Bcl11a/Ctip1 or Bcl11b/Ctip2 and COUP-TF remains open.
Phylogenetic analysis of Bcl11-like genes suggests that a homolog of Bcl11b first appears in cartilaginous fishes.20–22) On the other hand, homologs of Bcl11a are already present in the genomes of amphioxus and sea lamprey. As previously pointed out,21,22) though not by Guo and Cooper et al.,20) the Bcl11 protein in sea lamprey, a jawless vertebrate, can be categorized into the Bcl11a cluster. This suggests that Bcl11b is segregated form the Bcl11a homolog at the vertebrate stage, and no Bcl11b homolog is present in invertebrates. This may be compatible with that Bcl11a is involved in the transcription of hemoglobin genes (see below) whereas Bcl11b regulates the development of T cells that are not present in sea lampreyor or in other invertebrates like sea urchin.
Bcl11a was originally called Evi9, named after a retroviral insertion site (Evi9) in myeloid leukemia tumors in BXH-2 recombinant inbred mice. Detailed analysis of the Evi9 site discovered this gene.23) Mice lacking Bcl11a exhibited neonatal lethality and impairments in B cell and lymphoid cell development.14) However, recent genetic studies of BCL11A in humans have shed new light on a complex regulatory process of fetal hemoglobin (HbF) expression. BCL11A is associated with persistent fetal hemoglobin in adult humans,24,25) which was provided by genome-wide association studies. This analysis identified BCL11A as a new HbF-associated gene on chromosome 2, by taking advantage of the natural variation in the level of HbF in various human populations. Subsequent studies established that BCL11A is a central mediator of γ-globin silencing and hemoglobin switching.26) An example of the finding in these studies is that down-regulation of BCL11A expression in adult human erythroid precursors led to robust induction of HbF,27) and mechanistically, BCL11A interacts with the Mi-2/NuRD chromatin remodeling complexes, as well as the erythroid transcription factors GATA1 and FOG1, in erythroid progenitors.27) Very recent studies revealed a network of transcription factors that the transcription factor KLF1 is a key activator of the BCL11A gene.28,29) Knockdown of KLF1 in human and mouse adult erythroid progenitors markedly reduced BCL11A levels and increased human gamma-globin/beta-globin expression ratios.
3. Bcl11b and trancription
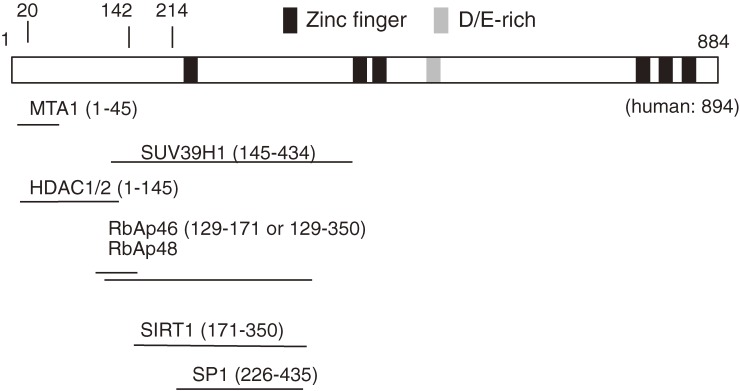
Bcl11b/Ctip2 was initially identified as a transcriptional repressor that either directly bound to a GC-rich consensus sequence of target genes and/or interacts with NuRD complex.18,19,30) On the other hand, Bcl11b was shown to activate the transcription of NF-κB target genes,31) suggesting that Bcl11b acts both as a transcriptional repressor and activator in a context dependent manner. Figure 1 displays the Bcl11b structure including DNA binding and protein-interacting regions. The Bcl11b gene consists of 4 exons and encodes two different isoforms, α-isoform consisting of 884 and β-isoform consisting of 812 lacking exon 3 in the mouse.2) The long exon 4 comprises all six zinc-finger domains, and the 2nd and 3rd domains are responsible for DNA binding. Recently, structural homology modeling has been performed as to canonical DNA binding of Bcl11b zinc fingers, which is based on the high-resolution crystal structure of the zinc finger domains of the transcription factor Egr1 in complex with DNA.32) The result reveals that mutations identified within the 2nd and 3rd domains disrupt the structure comprising conserved amino acids that are modeled to be required for the stability of the zinc finger domain or its binding to DNA. Apart from the DNA binding region, Bcl11b possesses domains responsible for interaction with proteins and protein complexes. Their catalogue has grown recently, including histone deacetylases (HDAC1 and HDAC2), and the ubiquitous transcription factor Sp1, and the Rb-associating proteins (RbAp46 and RbAp48), the member of surtuin family proteins (Sirt1), the heterochromatin protein 1 (HP1), and the histone methyltransferase SUV39H1.18,19,33–36)
Figure 1.
Structure of Bcl11b protein. The bar represents the β isoform of Bcl11b including Zinc-finger and D/E-rich domains on the bar. All these domains are within the long exon 4, and the exons 1–3 are 214 amino acids in length. The human BCL11B is longer than the mouse one and consists of 894 amino acids in the β isoform. The 2nd and 3rd Zinc-finger domains are for DNA binding and regions shown by lines are for various proteins binding to Bcl11b.
Several target genes of Bcl11b were discovered in cultured cells. Genes encoding cyclin-dependent kinase inhibitiors, p21/Cip2/Waf1 and p57/Kip2, are examples, and they are transcriptinally suppresssed by Bcl11b.19,37) In addition, we have identified HDM2(MDM2) as a new target, a ubiquitin ligase that downregulates a key tumor suppressor p53.38) Interestingly, Bcl11b inhibits HDM2 expression in a p53-dependent manner and modulates responses to radiation-induced DNA damages. A study of thymocytes in vivo using Chip-seq (Chromatin immunoprecipitation followed by DNA sequencing) method revealed several new target sequences and genes such as Th-Pok (Zbtb7b) and Runx3,39) expression of which is required for immature DP thymocytes (see Section 4.1 and Fig. 2 for abbreviation of thymocyte subsets) to further differentiate into mature thymocytes (CD4SP or CD8SP cells, respectively). Those genes are upregulated in the DP thymocytes lacking Bcl11b expression, suggesting suppressive role for Bcl11b in their transcription. Induction of Th-Pok expression occurs downstream of T-cell receptor signaling,40) whereas Runx3 contributes to Th-Pok repression in CD8SP committed cells.41) Hence, these suggest essential roles for Bcl11b in early silencing of Th-Pok and Runx3 genes. It may be also possible that Bcl11b cooperatively works with ThPok or Runx in the activation or suppression of some target genes. However, it is elusive of the mechanism and whether or not these repressions by Bcl11b are done through the association with NuRD or others.
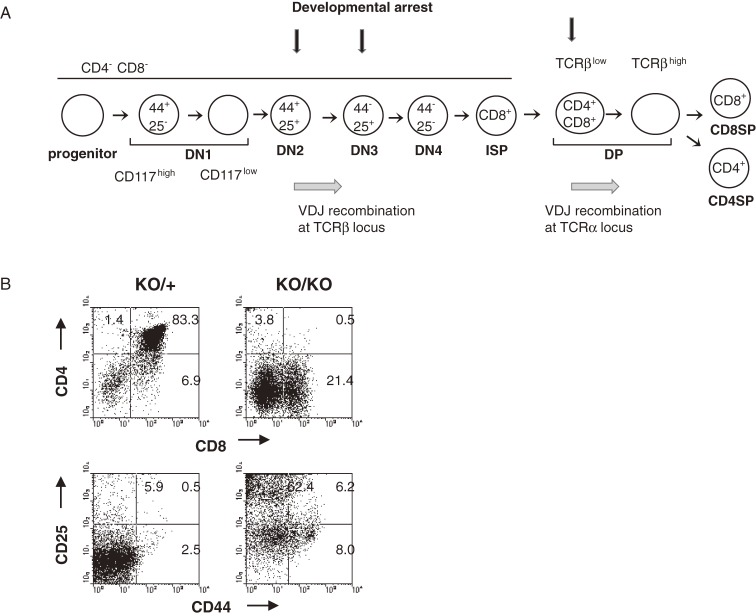
Figure 2.
(A) The diagram illustrates stages of T-cell development in thymus. Differentiation markers used here are CD117, CD44, CD25, CD4, CD8, and TCRβ on cell surface. See the text for details of development of αβ T cells. In brief, it proceeds in order of maturity, CD4−CD8− double negative (DN), CD4+CD8+ double positive (DP), and CD4+CD8− or CD4−CD8+ single positive cells (CD4SP or CD8SP cells, respectively). Vertical arrows indicate stages of developmental arrest given by loss of Bcl11b. (B) Flowcytometry of thymocytes in Bcl11bKO/+ and Bcl11bKO/KO mice using CD4, CD8, CD44, and CD25 markers. The vertical axis shows CD4 expression and the horizontal axis displays CD8 expression in total thymocytes (upper); the vertical axis shows CD25 expression and the horizontal axis displays CD44 expression in the CD4/CD8 double-negative quadrant (lower).
Recently, Chip-seq analysis for Bcl11b binding sequences was done in cells of striatal neurons. As a result, as many as 248 target genes were identified with the aid of gene expression profiling, which suggests the neurotrophic factor/neurotrophin signaling pathway as a primary target pathway for Bcl11b regulation.42)
4. Phenotypes of Bcl11b-deficient mice
Bcl11b is known to plays crucial roles in the development of several organs, including T cells, CNS, skin, and tooth. Bcl11b-deficient mice exhibit various developmental defects in these organs as follows.
4-1. T cells.
As described above, the defect in T-cell development given by Bcl11b deficiency is the phenotype firstly discovered. At present, defects have been identified at several distinct stages of development of thymocytes and T cells.3,22,43–47) Figure 2A illustrates T-cell development in thymus (see below for details) and indicates the stages of developmental arrest given by loss of Bcl11b. Bcl11b is a unique transcription factor that specifically functions for T-cell identity maintenance and another transcription factor of this type is Tcf1 (T-cell factor 1).22,46–48) The T cell specification pathway involves many different signalings, one of which is the signaling by the Notch1 receptors. The receptors are expressed by early progenitors, and activated to function upon interaction with cognate ligands (Delta-like proteins) expressed by thymic epithelial cells.49,50) The review by Liu et al.1) and others51,52) describe tissue-specific signals that direct developmental fates of thymocyte progenitors in the thymus. To avoid redundancy, I touch briefly on thymocyte development and roles for Bcl11b in controlling thymocyte differentiation and expansion.
T cells arise from hematopoietic progenitor cells that migrate from the bone marrow to the thymus, where they proliferate as thymocytes (Fig. 2A). Development of αβ T cells in the thymus proceeds through three major stages defined according to their expression pattern of CD4 and CD8 molecules on cell surface, i.e. in order of maturity, CD4−CD8− double negative (DN), CD4+CD8+ double positive (DP), and CD4+CD8− or CD4−CD8+ single positive cells (CD4SP or CD8SP cells, respectively). The CD4 and CD8 molecules are coreceptors of the T-cell receptor (TCR). Before DN thymocytes progress to the DP stage, they express CD8 but lack αβ TCR on cell surface. Those cells are highly proliferative and called immature CD8+ single positive (ISP) cells. Thymocytes at the DP stage express the αβ T-cell receptor (αβ TCR) complex after rearrangement at the TCRα locus, which allows engagement by intrathymic peptide/major histocompatibility complex (MHC) ligands. CD4 and CD8 molecules interact, respectively, with class II and I MHC molecules, thereby stabilizing or enhancing the interaction of TCRs with their MHC ligands. Co-expression of CD4 and CD8 at the DP stage allows thymocytes to receive optimal signals through either class I or II MHC-specific TCRs. Negative selection leads to death by apoptosis, while positive selection leads to thymocyte activation as evidenced by the upmodulation of activation markers, such as CD5 and CD69, and differentiation into SP T cells. Some thymocytes at the DP stage undergo signals to differentiate into natural killer T (NKT) cells. Gene expression analysis in thymocytes reveals that Bcl11b is upregulated at the transition from DN1 to DN2 and the expression is maintained in cells of αβ T cell lineages.
Immature DN thymocytes can be further divided into four subpopulations based on the surface expression of CD44 (or CD117/c-kit in case) and CD25, with the developmental progression being CD44+CD25− (DN1) to CD44+CD25+ (DN2) to CD44−CD25+ (DN3) and then to CD44−CD25− (DN4) cells (Fig. 2A). To make the developmental transition from DN3 to DP, proteins that are produced from productively rearranged genes at the TCRβ locus must be assembled into the preTCR complex and expressed on cell surface, which consists of a TCRβ-chain, the invariant pTα-chain, and CD3 components. At the DN4 stage, thymocytes re-enter into the cell cycle and rapidly proliferate.53,54) Since the preTCR confers survival signals for DN3/DN4 cells, only cells that have acquired a functional preTCR can transit from DN3 to DP, a process known as β-selection. Thymocyte progenitor cells at the DN1/DN2 stage retain the capability to generate cells of non-T-cell lineages such as myeloid and natural killer (NK) cells.1,51) However, DN3 thymocytes are committed T cells and have lost potentials to differentiate to other non-T-cell lineages.55,56)
Figure 2B shows flowcytomeric analysis of Bcl11bKO/KO thymocytes using CD4/CD8 and CD25/CD44 markers. The Bcl11bKO/KO thymus harbors DN and ISP cells but fail to produce DP thymocytes.3) Further analysis showed that thymocytes at the DN3 stage retain normal cellularity but not at the DN4 stage, and the DN4 thymocytes exhibit apoptosis, accompanying low expression of anti-apoptotic proteins, Bcl-xL and Bcl-2.57) As expected, Bcl11bKO/KO DN4 thymocytes lack preTCR on the surface, though containing some within the cell. These indicate that developmental progression is impaired before the DP stage and probably around the DN3 stage where cells start to express the preTCR complex on cell surface. Bcl11bKO/KO thymocytes exhibit less efficiency in DNA rearrangement of the TCRβ locus.57) Recombination between D and J segments normally occurs while subsequent recombination between V and DJ segments is reduced, which could contribute in part to the lack of expression of preTCR complex. The lack of preTCR expression in DN4 thymocytes may be the cause of the apoptosis, which has been considered as a mechanism to eliminate deleterious cells within the thymus.
Loss of genes encoding a component of the preTCR complex in mice lacks the preTCR signaling and leads to the developmental arrest at DN3 stage As expected, introduction of the lost gene restores the ability to produce DP and SP cells.58–60) In those mice, interestingly, deletion of the apoptosis-promoting gene p53 abrogates the developmental arrest to produce DP cells.61–63) As for Bcl11bKO/KO mice, introduction of functional TCRβ gene did not restore the developmental arrest.64) This suggests that the preTCR complex formation alone cannot compensate the deficiency of Bcl11b. Furthermore, introduction of p53 deficiency into Bcl11bKO/KO mice did not affect the developmental arrest and failed to inhibit apoptosis.64) This suggests that p53-related apoptosis is not the reason for the failure of T-cell development upon loss of Bcl11b. These results suggest that Bcl11bKO/KO thymocytes have defects in not only the pre-TCR signaling but some other signaling required for survival and transition to DP stage of development. Thus, the exact cause of T-cell defects in Bcl11b-lacking mutant mice remains unresolved.
Bcl11b also plays a role in the differentiation from DP to SP cells. This was demonstrated by analysis of CD4-Cre;Bcl11bflox/flox mice, where loss of Bcl11b occurs in thymocytes after the DP stage.39,43) The mice exhibited the developmental arrest at DP stage and did not produce SP cells and NKT cells. This indicates that Bcl11b is required for DP cells to differentiate to SP and NKT cells. The DP thymocytes underwent rearrangement at the TCRα gene normally but they failed to display proximal TCR signaling that is required for initiation of positive selection. Thus, the DP cells lacking TCR signaling underwent apoptosis during the process of positive selection. Interestingly, susceptibility to the apoptosis in those DP cells was at least in part independent of the anti-apoptotic factor Bcl2, because the introduction of Bcl2 transgene to CD4-Cre;Bcl11bflox/flox mice did not fully prevent apoptosis of thymocytes.43) These results indicate that Bcl11b plays critical roles in the establishment of TCR signaling in DP cells that is required for producing precursor cells of CD4 and CD8 lineages and also thymic NKT precursors.43)
Recently, three independent reports showed that deletion of Bcl11b blocks the progression from DN2 cells to DN3 committed T cells, indicating a role for Bcl11b at an early stage in T-cell development and maintenance of T-cell lineage commitment.22,46,47) NK-like cells were generated in Bcl11bKO/KO mice, and they may be converted from cells of T-cell lineage. This suggests that Bcl11b plays a role on the maintenance of T cell lineage identity in T cell lineage committed thymocytes, and its absence leads to reprogram T cells into the NK cell lineage. Hence, Bcl11b may regulate the cell fate choice between cells of T cell lineage and NK cells. One of the papers by Ikawa et al. has succeeded in establishing a culture system that continuously cultures developmentally arrested and proliferating DN2 thymocytes in the presence of Delta-like 4 and the cytokine IL-7. Of importance, those DN2 thymocytes retain the potential to differentiate into multiple cell types, T cells, NK cells, dendritic cells, and macrophages. They discovered that the expression of Bcl11b in the proliferating DN2 cells leads to the relief of the differentiation arrest to differentiate into cells of T cell lineage. Also, they showed that simply reducing the concentration of IL-7 in the culture system stimulates robust T cell differentiation, suggesting that IL-7 controls the expression of Bcl11b and that Bcl11b is the critical T cell promoting transcription factor. Their study also includes the finding that identifies Bcl11b as a sensor that links cytokine signaling thresholds and T cell lineage commitment in early thymocyte progenitors.
4-2. Neuron.
As for CNS, expression of Bcl11b/Ctip2 was first detected in subcerebral projection neurons of the cerebral cortex, including developing corticospinal motor neurons (CSMN). Developmental analysis of Bcl11bKO/KO mice showed defects in axonal extension and pathfinding by the projection neurons, resulting in failure of the neurons to connect to the spinal cord.4) This indicates a critical role for Bcl11b in the development of corticospinal motor neurons. Interestingly, Bcl11bKO/+ heterozygous mice also show some subtle defects in CSMN fasciculation, suggesting haploinsufficiency of Bcl11b leading to phenotypic consequences. Further study showed expression of Bcl11b in GABAergic medium-sized spiny neurons (MSN) within the striatum that are derived from progenitors located in the germinal zone of the developing lateral ganglionic eminence.65) Loss of Bcl11b function results in the failure of differentiation of MSN, leading to disruption of the patch-matrix organization of MSN. This suggests roles for Bcl11b in the differentiation of MSN and establishment of cellular architecture of the striatum.65) Strial-enriched expression of Bcl11b was also demonstrated in adulthood,42) suggesting that Bcl11b plays important roles in the functioning and maintenance of mature medium spiny neurons.
Recent study has shown that Bcl11b is also expressed in the developing vomeronasal system in the accessory olfactory bulb of the mouse as well.66) The vomeronasal system detects pheromones to mediate social and reproductive behaviors in terrestrial vertebrates. In Bcl11bKO/KO mice, vomeronasal sensory neurons (VSNs) are generated during development in the correct number but selectively die due to apoptosis. As a consequence, the mice display various phenotypes such as disorganization of layer formation of the accessory olfactory bulb, impaired axonal projections of VSNs, and defective mature differentiation of VSNs. The VSNs can be classified into two major types of neurons having different receptors. Interestingly, loss of Bcl11b function results in an impaired balance of cells of the two VSN types, suggesting that Bcl11b regulates the cell fate choice between the two different VSN types of neuronal cells.
4-3. Skin.
Bcl11b/Ctip2 is highly expressed in some cell types in the developing epidermis whereas it is expressed at a lower level in the dermis. The expression can be also detected in adult skin but the level of expression is much lower. The development of the skin epidermis begins with the commitment of the primitive ectoderm to the keratinocyte cell fate. The subsequent processes of cellular proliferation, stratification and differentiation result in formation of the multilayered structure of epidermis. During embryonic development, keratinocytes of the innermost layer of the epidermis, the proliferative basal cell layer, undergo a program of the terminal differentiation, then exit the basal cell layer and migrate upward to the surface of the skin.67) Bcl11b/Ctip2KO/KO mice exhibit a hypoplastic epidermis with late differentiation events.5) The epidermis shows impairments in keratinocyte proliferation and the development of the epidermal permeability barrier. These indicate that Bcl11b plays critical roles during skin development. Bcl11bKO/KO mice exhibit an interesting feature, i.e., born with eyes open. The hypoplastic epidermis described above may account for this open eyelid.
4-4. Ameloblast in tooth.
Bcl11b/Ctip2 is expressed in the ectodermal components of the developing tooth, including enamel epithelial cells and cells of the ameloblast lineage. Bcl11b/Ctip2−/− mice exhibit multiple defects at the bell stage of embryonic tooth development.6) The early bell stage (embryonic day 15.5–16.5) is characterized by continued epithelial expansion and differentiation into the inner and outer enamel epithelium, stratum intermedium, and stellate reticulum.68) Mutant incisors and molars are reduced in size and exhibit hypoplasia of the stellate reticulum that probably harbors stem cells. A well known hallmark of mouse incisors, different from human incisors, is an asymmetric enamel formation, which results from differential distribution of ameloblasts around incisors during development. As a result, the lingual side of mouse incisors lacks ameloblasts and enamel. Interestingly, mutant incisors possess an ameloblast-like cell population at the lingual side. This suggests that Bcl11b functions as a critical regulator of epithelial cell fate and differentiation during tooth morphogenesis. Despite developmental roles delineated in incisors and molars, roles for Bcl11b in maintenance and homeostasis of tooth in adult remain open.
5. Bcl11b tumor suppressor gene
Tumor suppressor genes act as inhibitory signals for uncontrolled cell growth and some play a role in DNA repair or cell survival.69) p53 is one of the most important tumor suppressors, often called the guardian of the genome because of its central role in maintaining the integrity of the cell’s DNA by controlling cell cycle inhibition, repair and apoptosis.70) APC, another important tumor suppressor, is a negative regulator on the Wnt/β-catenin signaling pathway.71) The signaling in intestines is required for differentiation of enterocytes and secretory cells and also for maintaining stem cells and progenitors within the intestinal crypt.71–74) APC inactivating mutations or activating β-catenin mutations impair the balance between cell proliferation, differentiation and apoptosis that affects the net number of cells in the tissue. As a consequence, it leads to formation of benign polyps or adenoma cells.
Bcl11b is a tumor suppressor and loss of a Bcl11b allele contributes to thymic lymphoma development. On the other hand, Bcl11b is a lineage-specific transcription factor probably responsible for turning on cell type-specific genes. Regulated expression of Bcl11b is important for differentiation of T cells and other types of cells, as described above. There are many other transcription factors affecting both cell differentiation and cancer development, and a well known precedent is β-catenin. Bcl11b and β-catenin have similar properties in cell proliferation, differentiation and apoptosis, and hence deregulation of these properties might be the contribution of loss of a Bcl11b allele to lymphomagenesis. In the following sections I will describe haploinsufficiency of Bcl11b for tumor suppression and how Bcl11b is involved in apoptosis, proliferation and differentiation.
Although Bcl11b was identified as a tumor suppressor gene in the mouse model, genetic changes were also observed in human malignancy.32,75,76) Mutations or deletion of BCL11B gene were found in approximately 10%–16% of human T-cell acute lymphoblastic leukemia (T-ALL).32,76,77) This indicates the involvement of Bcl11b in human malignancy as well.
6. Haploinsufficiency of Bcl11b for tumor suppression
As for tumor suppressor genes, loss of two alleles normally contributes to tumorigenesis, and this is known as the Knudson’ two-hit theory. However, the Bcl11b tumor suppressor gene is exceptional, belonging to a class of haploinsufficient tumor suppressor genes.78,79)
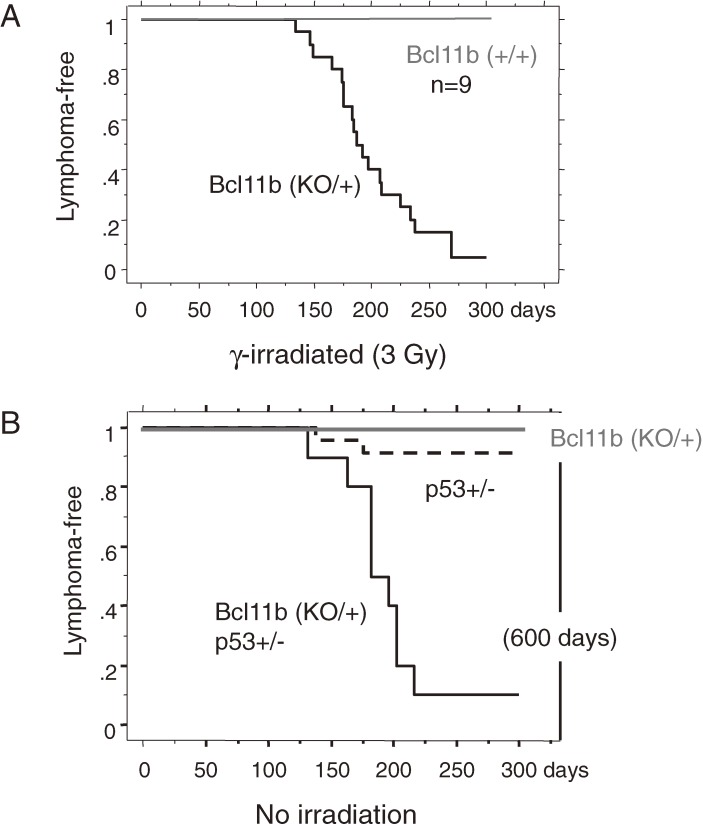
Figure 3A shows cumulative incidences of thymic lymphomas in Bcl11b+/+ mice and Bcl11bKO/+ mice after γ-irradiation.2) The thymic lymphoma incidence was much higher in Bcl11bKO/+ mice than wild-type mice. Figure 3B shows cumulative incidences of spontaneously developed thymic lymphomas in Bcl11bKO/+ mice, p53KO/+ mice and Bcl11bKO/+p53KO/+ doubly heterozygous mice. The incidences in Bcl11bKO/+ mice and p53KO/+ mice were low until one year of the age whereas the incidence in Bcl11bKO/+p53KO/+ mice was very high. These results suggest that loss of one Bcl11b allele does not affect lymphomagenesis in basal conditions but contributes to lymphomagenesis in radiation-induced injury conditions or in the p53KO/+ heterozygous genetic background. One characteristic of the tumor suppressor gene Bcl11b is that its suppressive capacity is haploinsufficient, one wild-type allele being insufficient for tumor suppression. This is based on the finding that most of the thymic lymphomas developed in Bcl11bKO/+ mice retained the wild-type allele although the thymic lymphomas developed in wild-type mice showed loss of one Bcl11b allele at a high frequency.2,80) The retention of the wild-type allele was also observed in spontaneously developed thymic lymphomas in Bcl11bKO/+p53KO/+ mice.81) Of importance, the retention of one BCL11B allele was also observed in human T-ALLs having mutations on the BCL11B gene.32,76) Because loss of one allele is an easier event and much more frequent than loss of two alleles, the haploinsufficiency of Bcl11b/BCL11B tumor suppressor is important from the point of etiological view.
Figure 3.
The cumulative frequency distributions of thymic lymphomas (Kaplan-Mier analysis). (A) Analyzed are the thymic lymphomas that were induced by 3Gy γ-irradiation in Bcl11bKO/+ mice and Bcl11b+/+ mice. The curve of Bcl11bKO/+ mice is shown in black and that of Bcl11b+/+ mice in gray. (B) Thymic lymphomas spontaneously developed in Bcl11bKO/+p53+/− mice at a high frequency in black, but vary rare in Bcl11b+/+p53+/− mice in black dashed line or Bcl11b+/−p53+/+ mice in gray.
7. Bcl11b and apoptosis
Keeping one Bcl11b wild-type allele in most thymic lymphomas may be related to the apoptosis that is often seen in cells with loss of two Bcl11b alleles. As described above apoptosis occurs in not all but certain types of thymocytes in Bcl11b-knockout mice. An anti-apoptotic property with Bcl11b has been demonstrated using Bcl11b knock-down (KD) Jurkat cells, a T-cell culture line. When Bcl11b expression was heavily reduced using siRNA method, Bcl11b-KD cells underwent apoptosis.82,83) On the other hand, we obtained KD cell lines retaining Bcl11b expression at certain levels that were viable and able to proliferate in the serum concentration of 5%.82) When the serum concentration was increased from 5% to 10% serum, the Bcl11b-KD cell lines showed growth inhibition due to cell death, accompanying decreased expressions of the CDK inhibitor p27 and the anti-apoptotic protein Bcl-xL. This suggests that the level of apoptosis is related to the cell proliferation rate, which can be controlled by serum concentration in the culture medium.
Further analysis showed that apoptosis occurred in S phase of the cell cycle and impaired activation of the cell-cycle checkpoint kinase Chk1. Activated Chk1 through phosphorylation, which is induced by DNA replication stress and subsequent formation of single-stranded DNA, leads to the arrest of cell cycle progression to allow DNA repair. Failure of the cell cycle arrest may be a cause of apoptosis in Bcl11b-KD cells by inducing proapoptotic signals. Consistently, radiation with UV, an agent producing single-stranded DNA, enhanced apoptosis more in those Bcl11b-KD Jurkat cell lines than in control Jurkat cells. Therefore, the apoptosis may be a reflection of deregulated cell cycle progression leading to DNA replication stress and of an accumulation of DNA damages during the S phase. These suggest that Bcl11b plays a role in the recovery for DNA replication stress. The anti-apoptotic property of Bcl11b may contradict the tumor suppressor function because apoptosis has been considered as a mechanism to eliminate deleterious cells with damaged DNA. However, a different interpretation is also possible. Hyperplastic or dysplastic cells in premalignant lesions often exhibit apoptotic phenotype together with high mitotic index.84,85) Therefore, the apoptosis may be a phenotype of precancerous cells, and the cells susceptible to apoptosis can be progressed to a rapidly progressive tumor when they acquire the ability to escape apoptosis.
8. Premalignancy and cell proliferation
Premalignant conditions are recognizable lesions that are strongly associated with the development of malignant neoplasia. They differ from normal conditions and hence may possess properties unique to them. Normal cells possess checkpoint function that can perceive and arrest aberrant cell cycle triggered by cancer-promoting stimuli. The checkpoint functions as an inducible barrier against clonal cell expansion and genomic instability leading to tumor development. Accordingly, premalignancy might be related to impairment or inability of the checkpoint barrier function. Indeed, premalignant legions in human tissues exhibit signatures of persistent functioning of checkpoint, for instance, elevated protein expressions relevant for DNA damage responses.84,85)
γ-irradiation to Bcl11bKO/+ mice confers the thymus atrophic, the cell number being reduced approximately one tenth, and the atrophic thymus will develop thymic lymphomas. This implies that the atrophic thymus is in a premalignant condition and comprises a lesion harboring premalignant cells. Supporting evidence is that the mice that had received thymocytes from the atrophic thymus developed thymic lymphomas at a high frequency.86,87) Other studies also show the presence of premalignant cells in atrophic thymus.88,89) In general, a small thymus with increased apoptosis and an expanded proportion of immature DN thymocytes characterizes the premalignant cells. For instance, transgenic mice expressing the oncogene Lmo2 develop T-cell leukemia after a long latency period, keeping atrophic thymus before the development.90,91) Of note, the induction of atrophic thymus is caused by γ-irradiation but not a direct consequence of irradiation, because the thymus is recovered from damages within one week after γ-irradiation and atrophy of the thymus begins approximately three weeks after.92) To characterize γ-ray induced atrophic thymus is important to elucidate how loss of Bcl11b contributes to premalignancy and tumor development at initial stages.
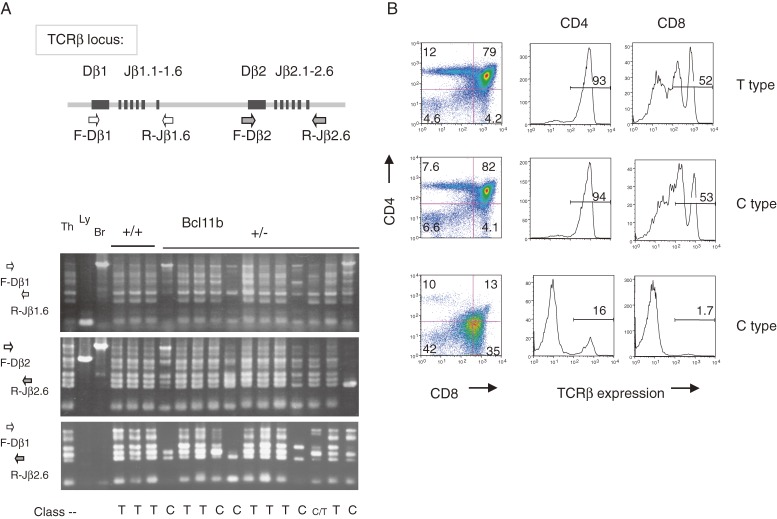
Cell proliferation of clonal origin is a hallmark of premalignant cells. Clonal proliferation of certain thymocytes was observed in about a half of atrophic thymuses in Bcl11bKO/+ mice at as early as 60 days after irradiation.92) Clonality was determined by assaying specific V(D)J rearrangements with three primer sets designed for the TCRβ locus. Recombination leading to the V(D)J rearrangements occurs in thymocytes at DN3 stage before β-selection. Figure 4A shows positions of primers and band patterns of PCR products in gel electrophoresis. Normal thymocytes within a thymus exhibit a DJ rearrangement pattern of all six distinct bands reflecting polyclonal origin of thymocytes. However, atrophic thymuses showed a few prominent band patterns of rearrangement (Fig. 4A), indicating clonal expansion of a few parental thymocytes having passed β-selection. The thymus is here designated as C-type thymus (C stands for clonal expansion) and the other thymuses are called T-type thymus (T stands for normal thymus).
Figure 4.
Clonal proliferation of thymocytes in atrophic thymuses after γ-irradiation of Bcl11bKO/+ mice. (A) The upper diagram shows part of the TCRβ locus and the relative location of PCR primers. The lower panel shows gel electrophoresis of PCR products with three different sets of primers, F-Dβ1 and R-Jβ1.6 (top), F-Dβ2 and R-Jβ2.6 (middle), and F-Dβ1 and R-Jβ2.6 (bottom). Thymuses are classified into two group, T type thymus (similar to normal thymus) and C type thymus (showing clonal expansion) depending on band patterns of PCR products. The T type thymus shows all six different bands as normal thymus does, whereas the C type thymus shows skewed band patterns, a few bands being more prominent than the other bands. The latter pattern indicates the presence of clonally proliferating thymocytes. (B) Flowcytometry of CD4, CD8 and TCRβ expression on thymocytes, showing differences in differentiation level. The vertical axis shows CD4 expression and the horizontal axis displays CD8 expression (left); the vertical axis shows cell number and the horizontal axis displays TCRβ expression of thymocytes in the CD4 quadrant (middle) and in the CD8 quadrant (right). Some C type thymuses showed differentiation arrest and lower expression of TCRβ on surface.
The C-type thymus or clonal expansion was also detected in atrophic thymuses that were γ-ray induced in Bcl11b+/+ wild-type mice.93) In this case, irradiation is required 4 times of 2.5 Gy at one-week interval for efficient lymphoma development, because Bcl11b+/+ mice are much less susceptible to thymic lymphomas than Bcl11bKO/+ mice. Approximately a half of those C type thymuses showed allelic loss at Bcl11b locus, suggesting that the allelic loss contributes to lymphomagenesis and possibly to clonal expansion of thymocytes. However, those atrophic thymuses did not exhibit the activation of DNA damage checkpoints such as γH2AX, Chk1, Chk2 or p53,93) which is a hallmark of human precancerous cells.84,85) This was an unexpected result to us. Further study is necessary to elucidate relationship among the atrophic thymus, premalignancy and the activation of checkpoint function. Collectively, these results suggest that loss of one Bcl11b allele in Bcl11bKO/+ mice contributes to clonal cell proliferation at an early stage of thymic lymphoma development.
9. Bcl11b and cell differentiation
Figure 4B shows flowcytometry of T-type and C-type thymuses. Despite clonal cell expansion, a half of the C-type Bcl11bKO/+ thymuses comprised thymocyte subtypes in the same proportion as normal thymus, mostly consisting of CD4+CD8+ DP cells. This indicates their retention of the capability to differentiate from DN3 cells to DP cells and further to SP cells. Accordingly, the C-type thymocytes have properties to undergo many rounds of cell division cycle within the thymus and to capable to differentiate. These capabilities must have been acquired at a developmental stage after β-selection. It is because, if not, the clonally proliferating thymocytes would have shown normal, but not skewed, distribution of the V(D)J recombination patterns.
The other half of the C-type Bcl11bKO/+ thymuses consisted mostly of immature thymocytes with differentiation arrest at DN or/and ISP stages (the third low in Fig. 4B). This suggests that this class of C-type thymocytes lacks the capability to differentiate to DP cells. The differentiation arrest, a hallmark of cancer, was found only in this subgroup of the Bcl11bKO/+ C-type thymocytes, but not seen in the Bcl11b+/+ C-type thymocytes.91) Of note, nevertheless approximately a half of the Bcl11b+/+ C-type thymocytes lost one allele of Bcl11b. This suggests that acquired loss of a Bcl11b allele did not affect the differentiation arrest in those thymocytes. Though it is not clear at which developmental stage the acquisition of loss occurred, it is likely that the Bcl11b allelic loss took place at the stage after V(D)J recombination because of limited V(D)J recombination patterns detected. Li et al.46) demonstrated that the expression of Bcl11b begins at the early DN1 cell stage in thymus. The DN1 cell population is multipotent and has the potential to differentiate into macrophage, NK cells, T cells and others. Actual T cell precursors within the DN1 population are CD117high, whereas CD117low DN1 cells are precursors for NK and γδT cells or others.94) Bcl11b expression was observed in CD117low DN1 progenitor cells but not in CD117high DN1 cells.46) Hence, Bcl11b heterozygosity can affect lymphomagenesis at CD117high DN2 stage after CD117high DN1 stage. Though impaired differentiation is a hallmark of cancer, it would not alone be sufficient for DN or ISP thymocytes to acquire malignancy. The number of thymocytes in C-type thymuses is low (one tenth in average relative to normal thymus) and hence the C-type thymocytes are obviously not fully malignant. To establish full malignancy, some of the C-type thymocytes must acquire an additional genetic or epigenetic change(s). Together, these findings suggest that the differentiation arrest is related to Bcl11b+/− genotype and may be ascribed to Bcl11b loss in immature thymocytes after early CD117low DN1 stage and possibly before the ISP or DP stage.
10. Cancer stem cells and C-type thymocytes
Recently, McCormack et al. have shown an interesting finding of premalignant thymocytes in atrophic thymus, using the Lmo2-transgenic mouse model.90) The LMO2 oncogene causes a subset of human T cell lymphoblastic leukemias. They used a combination of in vivo cell fate mapping, in which the Lmo2 gene was constitutively expressed in the thymus but not in the bone marrow (BM), and transplantation of thymocytes from young Lmo2-transgenic mice. As a result, they found self-renewing thymocytes in atrophic thymus of Lmo2-transgenic mice that were committed T cells at the DN3 stage. Of interest, these self-renewing DN3 cells possessed many features of cancer stem cells, including the ability to serially transplant, the ability to generate mature T cells, and the expression of several genes typical of hematopoietic stem cells including stem cell marker. Thymic atrophy in Lmo2-transgenic mice is probably caused by loss of the entry of progenitor cells from BM due to the development of self-renewing cells within the thymus. They also showed that the Lmo2-induced premalignant thymocytes can survive after a high-dose irradiation, consistent with cancer stem cell hypothesis. The importance of cancer stem cells in leukemia therapy has been pointed out in relapsed acute lymphoblastic leukemia in humans.91,95)
As described above, some of the C-type thymocytes in atrophic thymus have two properties, to undergo many rounds of cell cycle within the thymus and to capable to differentiate into DP and SP cells. In the respect of self-renewal and differentiation, they are similar to the Lmo2-induced premalignant thymocytes. Analyses of Bcl11bKO/KO mice22,46) showed that the arrested Bcl11b-KO DN2 cells started to self-renew. This may implicate Bcl11b deficiency in the generation of cancer stem cells or premalignancy. Interestingly, a subtype of human leukemias, CML (chronic myeloid leukemia), possesses self-renewal and cell lineage capabilities, and thereby the cells are assumed to be leukemia-initiating or cancer stem cells.95,96) Some of the C-type thymocytes have the phenotypes of CML. Therefore, those C-type thymocytes might be lymphoma stem cells. If so, they will be a model for therapy.
Acknowledgments
The author thanks Dr. Hiroshi Hiai for critical reading this manuscript and helpful comments. The author is also grateful for people in his laboratory who have contributed to works described here.
Competing financial interests
The authors declare no completing interests.
Profile
Ryo Kominami was born in 1947 and graduated from Okayama University School of Medicine at 1971. He started his research career at the Department of Biochemistry under the direction of Professor Masami Muramatsu at Tokushima University School of Medicine in 1972 and he received his PhD in 1976. After two years of study as a postdoctoral fellow at Frederic Cancer Research Institute near Washington DC in U.S.A., and then returned to Cancer Institute in Tokyo in 1979 and served as Researcher. He became Assistant Professor at the University of Tokyo in 1982 and was promoted to Associate Professor in 1983. He studied repetitive sequences, including minisatellites that were useful for individual identification. He is Professor of Biochemistry at Niigata University School of Medicine since 1987 and serving as a Vice Dean of the Graduate School of Medical and Dental Sciences at Niigata University. He isolated the Bcl11b tumor suppressor gene by analyzing radiation-induced mouse thymic lymphomas and found a regulatory role for Bcl11b in T-cell development. He was awarded the Japan Radiation Research Society Award in 2007.
References
- 1).Liu P., Li P., Burke S. (2010) Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol. Rev. 238, 138–149 [DOI] [PubMed] [Google Scholar]
- 2).Wakabayashi Y., Inoue J., Takahashi Y., Matsuki A., Kosugi-Okano H., Shinbo T., Mishima Y., Niwa O., Kominami R. (2003) Homozygous deletions and point mutations of the Rit1/Bcl11b gene in γ-ray induced mouse thymic lymphomas. Biochem. Biophys. Res. Commun. 301, 598–603 [DOI] [PubMed] [Google Scholar]
- 3).Wakabayashi Y., Watanabe H., Inoue J., Takeda N., Sakata J., Mishima Y., Hitomi J., Yamamoto T., Utsuyama M., Niwa O., Aizawa S., Kominami R. (2003) Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat. Immunol. 4, 533–539 [DOI] [PubMed] [Google Scholar]
- 4).Arlotta P., Molyneaux B.J., Chen J., Inoue J., Kominami R., Macklis J.D. (2005) Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 [DOI] [PubMed] [Google Scholar]
- 5).Golonzhka O., Liang X., Messaddeq N., Bornet J.M., Campbell A.L., Mertzger D., Chambon P., Ganguli-Indra G., Leid M., Indra A. (2009) Dual role of COUP-TF-interacting protein 2 in epidermal homeostasis and permeability barrier formation. J. Invest. Dermatol. 129, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Golonzhka O., Metzger D., Bornert J.M., Bay B.K., Gross M.K., Kioussi C., Leid M. (2009) Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 4278–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Okumura H., Miyasaka Y., Morita Y., Nomura T., Mishima Y., Takahashi S., Kominami R. (2011) Bcl11b heterozygosity leads to age-related hearing loss and degeneration of outer hair cells of the mouse cochlea. Exp. Anim. 60, 355–361 [DOI] [PubMed] [Google Scholar]
- 8).Matsumoto Y., Kosugi S., Shinbo T., Chou D., Ohashi M., Wakabayashi Y., Sakai K., Okumoto M., Mori N., Aizawa S., Niwa O., Kominami R. (1998) Allelic loss analysis of γ-ray-induced mouse thymic lymphomas: two candidate tumor suppressor gene loci on chromosomes 12 and 16. Oncogene 16, 2747–2754 [DOI] [PubMed] [Google Scholar]
- 9).Shinbo T., Matsuki A., Matsumoto Y., Kosugi S., Takahashi Y., Niwa O., Kominami R. (1999) Allelic loss mapping and physical delineation of a region harboring a putative thymic lymphoma suppressor gene on mouse chromosome 12. Oncogene 18, 4131–4136 [DOI] [PubMed] [Google Scholar]
- 10).Okano H., Saito Y., Miyazawa T., Shinbo T., Chou D., Kosugi S., Takahashi Y., Odani S., Niwa O., Kominami R. (1999) Homozygous deletions and point mutations of the Ikaros gene in γ-ray-induced mouse thymic lymphomas. Oncogene 18, 6677–6683 [DOI] [PubMed] [Google Scholar]
- 11).Georgopoulos K., Moore D.D., Derfler B. (1992) Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258, 808–812 [DOI] [PubMed] [Google Scholar]
- 12).Winandy S., Wu P., Georgopoulos K. (1995) A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83, 289–299 [DOI] [PubMed] [Google Scholar]
- 13).Satterwhite E., Sonoki T., Willis T.G., Harder L., Nowak R., Arriola E.L., Liu H., Price H.P., Gesk S., Steinemann D., Schlegelberger B., Oscier D.G., Siebert R., Tucker P.W., Dyer M.J.S. (2001) The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood 98, 3413–3420 [DOI] [PubMed] [Google Scholar]
- 14).Liu P., Keller J.R., Ortiz M., Tessarollo L., Rachel R.A., Nakamura T., Jenkins N.A., Copeland N.G. (2003) Bcl11a is essential for normal lymphoid development. Nat. Immunol. 4, 525–532 [DOI] [PubMed] [Google Scholar]
- 15).Avram D., Fields A., Pretty On Top K., Nevrivy D.J., Ishmael J.E., Leid M. (2000) Isolation of a novel family of C2H2 zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275, 10315–10322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Wang L.H., Tsai S.Y., Cook R.G., Beattie W.G., Tsai M.J., O’Malley B.W. (1989) COUP transcription factor is a member of the steroid receptor superfamily. Nature 340, 163–166 [DOI] [PubMed] [Google Scholar]
- 17).Shibata H., Nawaz Z., Tsai S.Y., O’Malley B.W., Tsai M.J. (1997) Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT). Mol. Endocrinol. 11, 714–724 [DOI] [PubMed] [Google Scholar]
- 18).Cismasiu V.B., Adamo K., Gecewicz J., Duque J., Lin Q., Avram D. (2005) BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24, 6753–6764 [DOI] [PubMed] [Google Scholar]
- 19).Topark-Ngarm A., Golonzhka O., Peterson V.J., Barrett B.Jr., Martinez B., Crofoot K., Filtz T.M., Leid M. (2006) CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J. Biol. Chem. 281, 32272–32283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Guo P., Hirano M., Herrin B.R., Li J., Yu C., Sadlonova A., Cooper M.D. (2009) Dual nature of the adaptive immune system in lampreys. Nature 459, 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Bajoghli B., Guo P., Aghaallaei N., Hirano M., Strohmeier C., McCurley N., Bockman D.E., Schorpp M., Cooper M.D., Boehm T. (2009) Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 138, 186–197 [DOI] [PubMed] [Google Scholar]
- 22).Ikawa T., Hirose S., Masuda K., Kakugawa K., Satoh R., Shibano-Satoh A., Kominami R., Katsura Y., Kawamoto H. (2010) An essential developmental checkpoint for production of the T cell lineage. Science 329, 93–96 [DOI] [PubMed] [Google Scholar]
- 23).Nakamura T., Yamazaki Y., Saiki Y., Moriyama M., Largaespada D.A., Jenkins N.A., Copeland N.G. (2000) Evi9 encodes a novel zinc finger protein that physically interacts with BCL6, a known human B-cell proto-oncogene product. Mol. Cell. Biol. 20, 3178–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Menzel S., Garner C., Gut I., Matsuda F., Yamaguchi M., Heath S., Foglio M., Zelenika D., Boland A., Rooks H., Best S., Spector T.D., Farrall M., Lathrop M., Thein S.L. (2007) A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat. Genet. 39, 1197–1199 [DOI] [PubMed] [Google Scholar]
- 25).Uda M., Galanello R., Sanna S., Lettre G., Sankaran V.G., Chen W., Usala G., Busonero F., Maschio A., Albai G., Piras M.G., Sestu N., Lai S., Dei M., Mulas A., Crisponi L., Naitza S., Asunis I., Deiana M., Nagaraja R., Perseu L., Satta S., Cipollina M.D., Sollaino C., Moi P., Hirschhorn J.N., Orkin S.H., Abecasis G.R., Schlessinger D., Cao A. (2008) Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc. Natl. Acad. Sci. U.S.A. 105, 1620–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–1842 [DOI] [PubMed] [Google Scholar]
- 27).Sankaran V.G., Xu J., Ragoczy T., Ippolito G.C., Walkley C.R., Maika S.D., Fujiwara Y., Ito M., Groudine M., Bender M.A., Tucker P.W., Orkin S.H. (2009) Developmental and species-divergent globin switching are driven by BCL11A. Nature 460, 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Borg J., Papadopoulos P., Georgitsi M., Gutiérrez L., Grech G., Fanis P., Phylactides M., Verkerk A.J., van der Spek P.J., Scerri C.A., Cassar W., Galdies R., van Ijcken W., Özgür Z., Gillemans N., Hou J., Bugeja M., Grosveld F.G., von Lindern M., Felice A.E., Patrinos G.P., Philipsen S. (2010) Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 42, 801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Zhou D., Liu K., Sun C.W., Pawlik K.M., Townes T.M. (2010) KLF1 regulates BCL11A expression and γ- to β-globin gene switching. Nat. Genet. 42, 742–744 [DOI] [PubMed] [Google Scholar]
- 30).Avram D., Fields A., Senawong T., Topark-Ngarm A., Leid M. (2002) COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 368, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Cismasiu V.B., Duque J., Paskaleva E., Califano D., Ghanta S., Young H.A., Avram D. (2009) BCL11B enhances TCR/CD28-triggered NF-κB activation through up-regulation of Cot kinase gene expression in T-lymphocytes. Biochem. J. 417, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Gutierrez A., Kentsis A., Sanda T., Holmfeldt L., Chen S.C., Zhang J., Protopopov A., Chin L., Dahlberg S.E., Neuberg D.S., Silverman L.B., Winter S.S., Hunger S.P., Sallan S.E., Zha S., Alt F.W., Downing J.R., Mullighan C.G., Look A.T. (2011) The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 118, 4169–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Marban C., Redel L., Suzanne S., Van Lint C., Lecestre D., Chasserot-Golaz S., Leid M., Aunis D., Schaeffer E., Rohr O. (2007) Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Senawong T., Peterson V.J., Avram D., Shepherd D.M., Frye R.A., Minucci S., Leid M. (2003) Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 278, 43041–43050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Senawong T., Peterson V.J., Leid M. (2005) BCL11A-dependent recruitment of SIRT1 to a promoter template in mammalian cells results in histone deacetylation and transcriptional repression. Arch. Biochem. Biophys. 434, 316–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Cismasiu V.B., Ghanta S., Duque J., Albu D.I., Chen H.M., Kasturi R., Avram D. (2006) BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood 108, 2695–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Cherrier T., Suzanne S., Redel L., Calao M., Marban C., Samah B., Mukerjee R., Schwartz C., Gras G., Sawaya B.E., Zeichner S.L., Aunis D., Van Lint C., Rohr O. (2009) p21WAF1 gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene 28, 3380–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Obata M.Kominami, R. and Mishima, Y. (2012) BCL11B tumor suppressor inhibits HDM2 expression in a p53-dependent manner. Cell. Signal. 24, 1047–1052 [DOI] [PubMed] [Google Scholar]
- 39).Kastner P., Chan S., Vogel W.K., Zhang L.J., Topark-Ngarm A., Golonzhka O., Jost B., Le Gras S., Gross M.K., Leid M. (2010) Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur. J. Immunol. 40, 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J. (2005) The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 [DOI] [PubMed] [Google Scholar]
- 41).Setoguchi R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., Taniuchi I. (2008) Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 [DOI] [PubMed] [Google Scholar]
- 42).Tang B., Di Lena P., Schaffer L., Head S.R., Baldi P., Thomas E.A. (2011) Genome-wide identification of Bcl11b gene targets reveals role in brain-derived neurotrophic factor signaling. PLoS One 6 (9), e23691, doi:10.1371/journal.pone.0023691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Albu D.I., VanValkenburgh J., Morin N., Califano D., Jenkins N.A., Copeland N.G., Liu P., Avram D. (2007) BCL11B is required for positive selection and survival of double-positive thymocytes. J. Exp. Med. 204, 3003–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Zhang S., Rozell M., Verma R.K., Albu D.I., Califano D., VanValkenburgh J., Merchant A., Rangel-Moreno J., Randall T.D., Jenkins N.A., Copeland N.G., Liu P., Avram D. (2010) Antigen-specific clonal expansion and cytolytic effector function of CD8+T lymphocytes depend on the transcription factor Bcl11b. J. Exp. Med. 207, 1687–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Albu D.I., VanValkenburgh J., Morin N., Califano D., Jenkins N.A., Copeland N.G., Liu P., Avram D. (2011) Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 6211–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Li P., Burke S., Wang J., Chen X., Ortiz M., Lee S.C., Lu D., Campos L., Goulding D., Ng B.L., Dougan G., Huntly B., Gottgens B., Jenkins N.A., Copeland N.G., Colucci F., Liu P. (2010) Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Li L., Leid M., Rothenberg E.V. (2010) An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Weber B.N., Chi A.W., Chavez A., Yashiro-Ohtani Y., Yang Q., Shestova O., Bhandoola A. (2011) A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Maillard I., Fang T., Pear W.S. (2005) Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23, 945–974 [DOI] [PubMed] [Google Scholar]
- 50).Taghon T.N., David E.S., Zūñiga-Pflücker J.C., Rothenberg E.V. (2005) Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19, 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Rothenberg E.V., Moore J.E., Yui M.A. (2008) Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Rothenberg E.V., Zhang J., Li L. (2010) Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238, 150–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Gounari F., Aifantis I., Khazaie K., Hoeflinger S., Harada N., Taketo M.M., von Boehmer H. (2001) Somatic activation of β-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat. Immunol. 2, 863–869 [DOI] [PubMed] [Google Scholar]
- 54).Xue L., Nolla H., Suzuki A., Mak T.W., Winto A. (2008) Normal development is an integral part of tumorigenesis in T cell-specific PTEN-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 105, 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Wada H., Masuda K., Satoh R., Kakugawa K., Ikawa T., Katsura Y., Kawamoto H. (2008) Adult T-cell progenitors retain myeloid potential. Nature 452, 768–772 [DOI] [PubMed] [Google Scholar]
- 56).Bell J.J., Bhandoola A. (2008) The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 452, 764–767 [DOI] [PubMed] [Google Scholar]
- 57).Inoue J., Kanefuji T., Okazuka K., Watanabe H., Mishima Y., Kominami R. (2006) Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβT cells in Bcl11bKO/KO mice. J. Immunol. 176, 5871–5879 [DOI] [PubMed] [Google Scholar]
- 58).Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., Alt F.W. (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 [DOI] [PubMed] [Google Scholar]
- 59).Mombaerts P., Clarke A.R., Rudnicki M.A., Iacomini J., Itohara S., Lafaille J.J., Wang L., Ichikawa Y., Jaenisch R.M., Hooper L., Tonegawa S. (1992) Mutations in T-cell antigen receptor genes α and block thymocyte development at different stages. Nature 360, 225–231 [DOI] [PubMed] [Google Scholar]
- 60).Shinkai Y., Koyasu S., Nakayama K., Murphy K.M., Loh D.Y., Reinherz E.L., Alt F.W. (1993) Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science 259, 822–825 [DOI] [PubMed] [Google Scholar]
- 61).Bogue M.A., Zhu C., Aguilar-Cordova E., Donehower L.A., Roth D.B. (1996) p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 10, 553–565 [DOI] [PubMed] [Google Scholar]
- 62).Jiang D., Lenardo M.J., Zuniga-Pflucker J.C. (1996) p53 prevents maturation to the CD4+CD8+ stage of thymocyte differentiation in the absence of T cell receptor rearrangement. J. Exp. Med. 183, 1923–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Haks M.C., Krimpenfort P.J., van den Brakel H., Kruisbeek A.M. (1999) Pre-TCR signaling and inactivation of p53 induces crucial cell survival pathways in pre-T cells. Immunity 11, 91–101 [DOI] [PubMed] [Google Scholar]
- 64).Okazuka K., Wakabayashi Y., Kashihara M., Inoue J., Sato T., Yokoyama M., Aizawa S., Aizawa Y., Mishima Y., Kominami R. (2005) p53 prevents maturation of T cell development to the immature CD4–CD8+ stage in Bcl11b−/− mice. Biochem. Biophys. Res. Commun. 328, 545–549 [DOI] [PubMed] [Google Scholar]
- 65).Arlotta P., Molyneaux B.J., Jabaudon D., Yoshida Y., Macklis J.D. (2008) Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J. Neurosci. 28(3), 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Enomoto T., Ohmoto M., Iwata T., Uno A., Saitou M., Yamaguchi T., Kominami R., Matsumoto I., Hirota J. (2011) Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. J. Neurosci. 31 (28), 10159–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Mack J.A., Anand S., Maytin E.V. (2005) Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res. C Embryo Today 75, 314–329 [DOI] [PubMed] [Google Scholar]
- 68).Tummers M., Thesleff I. (2003) Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 69).Kinzler K.W., Vogelstein B. (1996) Lessons form hereditary colorectal cancer. Cell 87, 159–170 [DOI] [PubMed] [Google Scholar]
- 70).Vousden K.H., Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 71).Clevers H. (2006) Wnt/β-catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 72).Polakis P. (2000) Wnt signaling and cancer. Genes Dev. 14, 1837–1851 [PubMed] [Google Scholar]
- 73).Fevr T., Robine S., Louvard D., Huelsken J. (2007) Wnt/β-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 27, 7551–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74).Li L., Clevers H. (2010) Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Przybylski G.K., Dik W.A., Wanzeck J., Grabarczyk P., Majunke S., Martin-Subero J.I., Siebert R., Dölken G., Ludwig W.D., Verhaaf B., van Dongen J.J., Schmidt C.A., Langerak A.W. (2005) Disruption of the BCL11B gene through inv(14)(q11.2q32.31) results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T-ALL. Leukemia 19, 201–208 [DOI] [PubMed] [Google Scholar]
- 76).De Keersmaecker K., Real P.J., Gatta G.D., Palomero T., Sulis M.L., Tosello V., Van Vlierberghe P., Barnes K., Castillo M., Sole X., Hadler M., Lenz J., Aplan P.D., Kelliher M., Kee B.L., Pandolfi P.P., Kappes D., Gounari F., Petrie H., Van der Meulen J., Speleman F., Paietta E., Racevskis J., Wiernik P.H., Rowe J.M., Soulier J., Avran D., Cavé H., Dastugue N., Raimondi S., Meijerink J.P., Cordon-Cardo C., Califano A., Ferrando A.A. (2010) The TLX1 oncogene drives aneuploidy in T cell transformation. Nat. Med. 16, 1321–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Paganin M., Ferrando A. (2011) Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 25, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., Jacks T. (2009) Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23, 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Alimonti A., Carracedo A., Clohessy J.G., Trotman L.C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W.J., Brogi E., Richardson A.L., Zhang J., Pandolfi P.P. (2010) Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80).Ohi H., Mishima Y., Kamimura K., Maruyama M., Sasai K., Kominami R. (2007) Multi-step lymphomagenesis deduced from DNA changes in thymic lymphomas and atrophic thymuses at various times after γ-irradiation. Oncogene 26, 5280–5289 [DOI] [PubMed] [Google Scholar]
- 81).Kamimura K., Mishima Y., Obata M., Endo T., Aoyagi Y., Kominami R. (2007) Lack of Bcl11b tumor suppressor results in vulnerability to DNA replication stress and damages. Oncogene 26, 5840–5850 [DOI] [PubMed] [Google Scholar]
- 82).Kamimura K., Ohi H., Kubota T., Okazuka K., Yoshikai Y., Wakabayashi Y., Aoyagi Y., Mishima Y., Kominami R. (2007) Haploinsufficiency of Bcl11b for suppression of lymphomagenesis and thymocyte development. Biochem. Biophys. Res. Commun. 355, 538–542 [DOI] [PubMed] [Google Scholar]
- 83).Grabarczyk P., Przybylski G.K., Depke M., Völker U., Bahr J., Assmus K., Bröker B.M., Walther R., Schmidt C.A. (2007) Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene 26, 3797–3810 [DOI] [PubMed] [Google Scholar]
- 84).Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 85).Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr., Kastrinakis N.G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T.D. (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 [DOI] [PubMed] [Google Scholar]
- 86).Sado T., Kamisaku H., Kubo E. (1991) Bone marrow-thymus interactions during thymic lymphomagenesis induced by fractionated radiation exposure in B10 mice: Analysis using bone marrow transplantation between Thy 1 congenic mice. J. Radiat. Res. 32, 168–180 [DOI] [PubMed] [Google Scholar]
- 87).Kominami R., Niwa O. (2006) Radiation carcinogenesis in mouse thymic lymphomas. Cancer Sci. 97, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Larson R.C., Lavenir I., Larson T.A., Baer R., Warren A.J., Wadman I., Nottage K., Rabbitts T.H. (1996) Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 15, 1021–1027 [PMC free article] [PubMed] [Google Scholar]
- 89).Chervinsky D.S., Lam D.H., Melman M.P., Gross K.W., Aplan P.D. (2001) scid Thymocytes with TCRβ gene rearrangements are targets for the oncogenic effect of SCL and LMO1 transgenes. Cancer Res. 61, 6382–6387 [PubMed] [Google Scholar]
- 90).McCormack M.P., Young L.F., Vasudevan S., de Graaf C.A., Codrington R., Rabbitts T.H., Jane S.M., Curtis D.J. (2010) The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879–883 [DOI] [PubMed] [Google Scholar]
- 91).Curtis D.J., McCormack M.P. (2010) The molecular basis of Lmo2-induced T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 16, 5618–5623 [DOI] [PubMed] [Google Scholar]
- 92).Go R., Hirose S., Morita S., Yamamoto T., Katsuragi Y., Mishima Y., Kominami R. (2010) Bcl11b heterozygosity promotes clonal expansion and differentiation arrest of thymocytes in γ-irradiated mice. Cancer Sci. 101, 1347–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Yamamoto T., Morita S., Go E., Obata M., Katsuragi Y., Fujita Y., Maeda Y., Yokoyama M., Aoyagi Y., Ichikawa H., Mishima Y., Kominami R. (2010) Clonally expanding thymocytes having lineage capability in γ-ray induced mouse atrophic thymus. Int. J. Radiation Oncology Biol. 77, 235–243 [DOI] [PubMed] [Google Scholar]
- 94).Lai J.C., Wlodarska M., Liu D.J., Abraham N., Johnson P. (2010) CD45 regulates migration, proliferation, and progression of double negative 1 thymocytes. J. Immunol. 185, 2059–2070 [DOI] [PubMed] [Google Scholar]
- 95).Calabretta B., Perrotti D. (2004) The biology of CML blast crisis. Blood 103, 4010–4022 [DOI] [PubMed] [Google Scholar]
- 96).Clarke M.F., Dick J.E., Dirks P.B., Eaves C.J., Jamieson C.H., Jones D.L., Visvader J., Weissman I.L., Wahl G.M. (2006) Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 66, 9339–9344 [DOI] [PubMed] [Google Scholar]