Abstract
Resetting the peripheral clock and understanding the integration between the circadian rhythm and metabolic pathways are fundamental questions. To test whether insulin acts as a synchronizer for the hepatic clock by cell-autonomous mechanisms, the phase-resetting capabilities of insulin were investigated in cultured hepatic cells. We provide evidence that three-dimensional (3D) cell culture conditions that preserve the differentiated state of primary hepatocytes sustained the robustness of the molecular clock, while this robustness rapidly dampened under classical monolayer cell culture conditions. Herein, we established a 3D cell culture system coupled with a real-time luciferase reporter, and demonstrated that insulin directly regulates the phase entrainment of hepatocyte circadian oscillators. We found that insulin-deficient diabetic rats had a pronounced phase advance in their hepatic clock. Subsequently, a single administration of insulin induced phase-dependent bi-directional phase shifts in diabetic rat livers. Our results clearly demonstrate that insulin is a liver clock synchronizer.
Mammalian circadian rhythms are thought to be coordinated by a central pacemaker in the suprachiasmatic nucleus (SCN)1. Surprisingly, such molecular oscillators are operative not only in pacemaker cells such as SCN neurons but also in many peripheral tissues and even in cell lines2,3. Peripheral tissues contain self-sustained oscillators with a molecular composition similar to that of SCN neurons4,5,6,7. On the other hand, clock-resetting factors and properties that synchronize peripheral oscillators are not yet fully understood.
The glucocorticoid hormone is a potent phase-shifting cue in both tissue culture cells and peripheral tissues8. On the other hand, glucocorticoids are not required for the entrainment of liver circadian gene expression under steady-state conditions8, indicating the existence of other intrinsic timing cues for in vivo circadian oscillators.
The hepatic circadian clock is affected in insulin-resistant and diabetic animal models9,10,11,12. Changes to the circadian gene expression in the liver imply alterations to an input pathway. Oishi et al reports that the peak expression levels of Per2 were decreased in the liver by STZ, but Dbp was not affected by either the phase or the amplitude in any examined tissues of the diabetic mice13. Yanagihara et al reports that high fat diet had minimal effects on the expression of selected core circadian genes, and only Cyp7a1 circadian gene expression was attenuated by high fat diet14. Although these studies raise the possibility that either insulin or other insulin-controlled humoral factors act as a synchronizer for peripheral tissues, it remains to be clarified whether insulin plays a role in the phase entrainment of the peripheral circadian clock.
Here, we studied these issues by characterizing the circadian clock in the liver. In the peripheral tissues, the liver circadian rhythm plays important roles in metabolism and metabolism-related diseases such as hypercholesterolemia and diabetes15,16. We established a three-dimensional (3D) cell culture system of rat primary hepatocytes, and used it to investigate the ability of insulin to synchronize the hepatic circadian clock. This cell culture system outperformed the classical collagen-based method for monitoring the circadian clock in hepatocytes in vitro. We here conclude that insulin directly regulates the phase entrainment of hepatocyte circadian oscillators. We not only utilized primary and immortalized cell lines to describe this phenomenon but also supported it with in vivo data at the organ level. Our studies clearly demonstrate that insulin is a liver clock synchronizer.
Results
Insulin induces acute and transient expression of clock genes in cultured H4IIE rat hepatoma cells
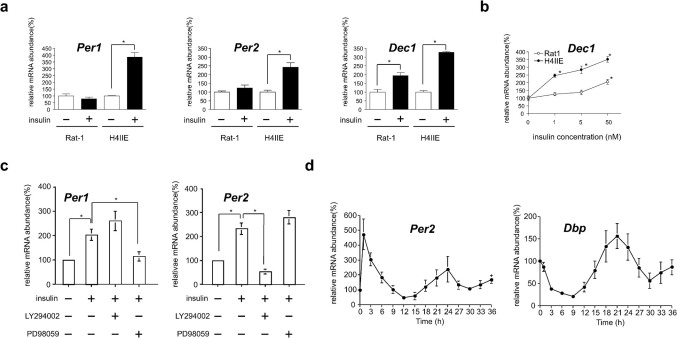
Cultured Rat-1 fibroblasts have frequently been used to study the peripheral clock-resetting mechanism2. Previously, insulin was found to induce Dec1 gene expression in the rat liver17. The present results confirm that insulin acutely induces the expression of Per1, Per2, and Dec1 genes in H4IIE cells (Fig. 1a). Insulin was also shown to increase the mPer1 and mPer2 promoter activity in H4IIE cells (data not shown). It was noteworthy that insulin could not induce the expression of Per1 and Per2 genes in Rat-1 fibroblasts (Fig. 1a). Balsalobre et al. reported that insulin could induce the expression of Per1 genes in Rat-1 fibroblasts18, but this induction was not observed 1 h after treatment with insulin. This difference can be attributed to the low insulin sensitivity of fibroblasts. To compare insulin sensitivity, Dec1 mRNA levels were investigated between Rat-1 fibroblasts and the H4IIE rat hepatoma cell line. A minimum concentration of 50 nM insulin was required to double Dec1 mRNA levels in Rat-1 fibroblasts (Fig. 1b). In contrast, a minimum concentration of only 1 nM insulin caused dose-dependent induction of Dec1 mRNA in H4IIE cells, by 3–4 folds (Fig. 1a, b). Therefore, although insulin induces the acute expression of Dec1 mRNA levels in both cell types, its potency and efficacy is minimal in Rat-1 fibroblasts, suggesting the existence of cell-specific entrainment factors. These results demonstrate that insulin acts as a resetting factor of circadian oscillation in insulin-sensitive cells such as hepatocytes.
Figure 1. Insulin acts as an acute entrainment signal in the cell-autonomous hepatic clock.
(a) Relative Per1, Per2, and Dec1 mRNA levels were determined by RT-PCR following addition of 50 nM insulin to the H4IIE and Rat-1 cell culture medium for 2 h. Each value was normalized to Gapdh. *p < 0.05 versus PBS-treated cells (Student's t-test). (b) Dose-dependent induction of Dec1 mRNA following 2-h insulin treatment of H4IIE and Rat-1 cells as determined by real-time RT-PCR. Each value was normalized to Gapdh. *p < 0.05 versus PBS-treated cells (Dunnett's test). (c) Relative Per1 and Per2 mRNA levels were determined by northern blotting in H4IIE cells for 30 min with inhibitors of proteins mediating the insulin signaling pathway (LY294002 and PD98059), followed by a 1-h treatment with 50 nM insulin. ApoE mRNA was used as an internal control. *p < 0.05 versus PBS- or insulin-treated cells (Student's t-test). (d) Temporal mRNA expression levels of Per2 and Dbp were determined by northern blotting following a shift of H4IIE cells to medium containing 50 nM insulin for 1 h at T 0, washing with PBS, and incubation in serum-free medium. Three time independent experiments were performed.
To determine how insulin-signaling pathways activate clock genes, H4IIE cells were stimulated with insulin in the presence or absence of the protein synthesis inhibitor cycloheximide. Inhibition of translation prevented stimulation of clock gene expression by insulin (data not shown), indicating that newly synthesized protein is required for insulin action. In contrast, we confirmed that dexamethasone was able to directly activate Per1 gene expression in H4IIE cells (data not shown), like in Rat-1 fibroblasts18. The molecular signaling underlying the induction of clock gene expression was also investigated. PD98059, an inhibitor of MAPK, inhibited Per1 mRNA accumulation, and LY294002, an inhibitor of PI3K, blocked accumulation of Per2 mRNA (Fig. 1c). These results indicate that MAPK and PI3K signaling is required for acute induction of Per1 and Per2 gene expression by insulin, respectively.
The treatment of cultured Rat-1 fibroblasts with high concentrations of serum induces the circadian expression whose transcription also oscillates in living animals2. We next examined whether insulin induces the circadian rhythm of gene expression in H4IIE cells. H4IIE cells were subjected to the same regimen of serum shock, and the levels of expression of several clock genes were monitored using northern blot analysis. Per1, Per2, and Dec1 expression were strongly induced by 2-h insulin treatment (Fig. 1a), after which Per2 expression was repressed. After 12 to 15 h, Per2 mRNA levels rose again and assumed circadian oscillations (Fig. 1d, supplementary Fig. 1). Dbp mRNA gradually diminished until minimal levels were reached 9 h after insulin treatment. Thereafter, the levels rose and also assumed circadian oscillation (Fig. 1d, supplementary Fig. 1). These results indicate that insulin can induce circadian rhythms of gene expression in H4IIE cells. To conclude that insulin acts as a synchronizer for the hepatic clock by cell-autonomous mechanisms, it is necessary to examine whether insulin resets hepatocyte circadian oscillators in a phase-dependent manner. Although circadian oscillations of Per2 and Dbp expression were observed in H4IIE cells, this robustness did not sustain itself over the course of several days in culture. Sustained and longer circadian oscillatory conditions as well as higher insulin sensitivity may help clarify the clock-resetting properties of insulin. We, therefore, employed primary cultured rat hepatocytes.
Primary hepatocytes sustain cell-autonomous circadian oscillations in an extracellular matrix-dependent manner
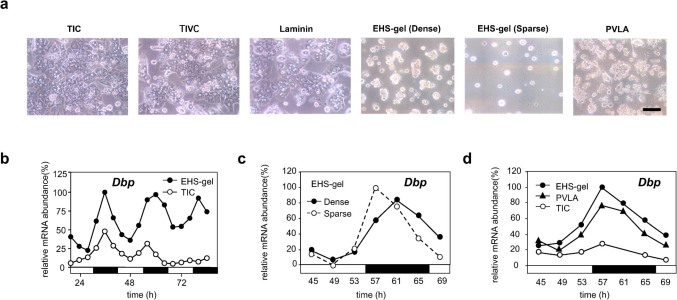
Once hepatocytes are disaggregated, monolayer hepatocytes rapidly lose their differentiation functions when cultured on type I collagen (TIC). In contrast, liver differentiation is well maintained by culturing on a basement membrane such as Engelbreth-Holm-Swarm sarcoma gel (EHS gel)19,20,21,22. Using a primary culture of rat hepatocytes, we were able to investigate interactions between the state of cellular differentiation and circadian oscillations. When freshly isolated rat hepatocytes were cultured on dishes coated with TIC, they spread and formed a confluent monolayer (Fig. 2a). In contrast, hepatocytes cultured on dishes coated with EHS gel retained a spherical cell shape (Fig. 2a), a finding consistent with previous observations20,21.
Figure 2. Spatial configuration of hepatocytes controls circadian time of the cells.
(a) Hepatocytes were cultured on dishes coated with TIC, TIVC, laminin, and PVLA at a high density (1 × 107 cells/dish). Hepatocytes were also cultured on dishes coated with EHS gel at both high and low densities (2.5 × 106 cells/dish). The morphological appearance of cultured hepatocytes at 48 h is shown. The scale bar indicates 100 μm. (b) Hepatocytes were plated on TIC- and EHS gel-coated dishes at a high density. Northern blotting was used to determine the Dbp mRNA level. Hepatocytes were collected at 4-h intervals. The open circles and filled circles represent TIC and EHS gel, respectively. The open and solid bars indicate light and dark conditions where the animals were kept before the preparation of hepatocytes. (c) Hepatocytes were cultured on an EHS gel-coated dish at high and low densities. Northern blotting was used to determine the Dbp mRNA level in rat primary hepatocytes cultured on EHS gel at high (solid lines) and low (dotted lines) densities. (d) Hepatocytes were cultured on dishes coated with TIC ( ), PVLA (
), PVLA ( ), and EHS gel (
), and EHS gel ( ) at a high density. Northern blotting was used to determine the Dbp mRNA level. Two time independent experiments were performed.
) at a high density. Northern blotting was used to determine the Dbp mRNA level. Two time independent experiments were performed.
We first investigated whether the interaction between the cells and their surrounding extracellular matrix influences circadian gene expression. Hepatocytes were cultured for 48 h on TIC- and EHS gel-coated dishes. Microarray analysis revealed that the expression of liver-specific genes (liver-enriched transcription factors; amino acid metabolism, lipid metabolism, and drug metabolism genes) was maintained at higher levels in the hepatocytes cultured on EHS gel than those cultured on TIC20 (Supplementary Fig. 2a, Supplementary Table 1). It is noteworthy that the expression levels of Dbp and Rev-erb alpha were significantly increased in hepatocytes cultured on EHS gel, whereas Dec1 and Dec2 were abnormally elevated in those cultured on TIC (Supplementary Fig. 2b, Supplementary Table 1). These observations imply that the hepatocyte differentiation function is related to circadian gene expression in hepatocytes. To confirm the mRNA levels observed by microarray, we monitored the cycles of high-amplitude expression of Dbp for 84 h after hepatocyte isolation. Northern blot analysis revealed that the rhythmic expression of Dbp mRNA rapidly declined by the fourth day of culture in the spread hepatocytes cultured on TIC, whereas the oscillations of Dbp expression persisted and remained robust during this period in hepatocytes cultured on the EHS gel (Fig. 2b, supplementary Fig. 3). To our knowledge, this study is the first to show that cell matrix contact is important for the maintenance of hepatic circadian gene expression.
EHS gel contains several extracellular matrix components such as laminin (60%), type IV collagen (TIVC) (30%), proteoglycan (3%), etc.19. We then assessed whether laminin and TIVC, which are the major components of the EHS gel, influence the circadian oscillation. The Dbp transcript cycle declined to low levels when hepatocytes were cultured on laminin and TIVC, and the Per1 and Per2 mRNA rhythms attenuated, leaving transcripts expressed at high levels after a 3-day culture period (Supplementary Fig. 4a). In contrast, the Dbp, Per1, and Per2 mRNA cycles persisted in the hepatocytes cultured on EHS gel (Supplementary Fig. 4a). These results indicate only a single extracellular matrix component of the EHS gel is not responsible for maintaining circadian oscillations.
Spatial configuration such as the 3D structure of the cell regulates its circadian time
Cell-to-cell interactions are one of the most remarkable features of epithelial cells. We next examined whether signaling between neighboring cells plays a role in circadian oscillations, because the hepatocytes cultured on EHS gel formed clusters. To decrease cell-to-cell interactions, we plated isolated hepatocytes at low density (Fig. 2a). Even in this sparse situation, the Dbp mRNA rhythm was maintained (Fig 2c, supplementary Fig. 5), suggesting that hepatocytes do not need to communicate directly to generate autonomous circadian oscillations. However, Dbp peak levels seemed to be slightly altered under sparse conditions (Fig 2c, supplementary Fig. 5). Cell-to-cell contact may, therefore, contribute to generating the correct phase.
The greatest difference between the hepatocytes cultured on the different matrices lies in morphological changes. Hepatocytes cultured on dishes coated with TIC, TIVC, and laminin were flattened and extended (Fig. 2a), whereas those cultured on EHS gel remained spherical in shape (Fig. 2a). Ingber and colleagues discovered a negative relationship between the differentiated state of hepatocytes and the adhesive island area: the rate of albumin secretion decreased with an increase in the adhesive island area23. We previously reported that cell shape regulates the differentiated state of hepatocytes, e.g., a spherical shape is required to maintain liver function24. To investigate the role of cell shape on hepatocyte rhythmicity, we attempted to maintain the morphology of hepatocytes by culturing them on a different matrix instead of EHS gel. When hepatocytes were cultured on a dished coated with a synthetic galactose-carrying polymer, poly-N-p-vinylvenzyl-d-lactonamide (PVLA)25, the cells formed an anchored spherical shape (Fig. 2a). Dbp mRNA rhythmicity was maintained over the 3-day culture period on PVLA, although the amplitude was slightly lower than that seen on EHS gel (Fig. 2d, supplementary Fig. 6). The Per1 mRNA patterns on the PVLA and EHS gel matrices were indistinguishable (Supplementary Fig. 4b). Although the present experiments cannot rule out a potential role of the actual materials composing the different ECMs, the liver clock is sustained when the 3D structure of the cell is maintained.
Cell morphology may reflect the organization of the intracellular cytoskeletal network, which consists of 3 kinds of cytoskeleton filaments: actin filaments, microtubules, and intermediate filaments. We confirmed that the peak Dbp mRNA level was diminished by treatment with microtubule-interfering agents such as colchicine, vinblastine sulfate, and nocodazole, whereas it was maintained during treatment with the microtubule-stabilizer paclitaxel (Supplementary Fig. 7). This implies that the hepatic circadian rhythm is dependent on microtubule organization but not on microtubule dynamics. An actin filament-inhibitor (cytochalasin b) and an intermediate filament-inhibitor (IDPN) had little effect on the peak Dbp mRNA level compared to the microtubule-interfering agents (Supplementary Fig. 7). Our results demonstrate that the network of cytoplasmic microtubules, but not those of actin and intermediate filaments, significantly influences the cell shape-related regulation of the circadian rhythm.
Insulin is a phase entrainment factor in hepatocytes under ex vivo culture conditions in the Per2-dLuc reporter transgenic rat
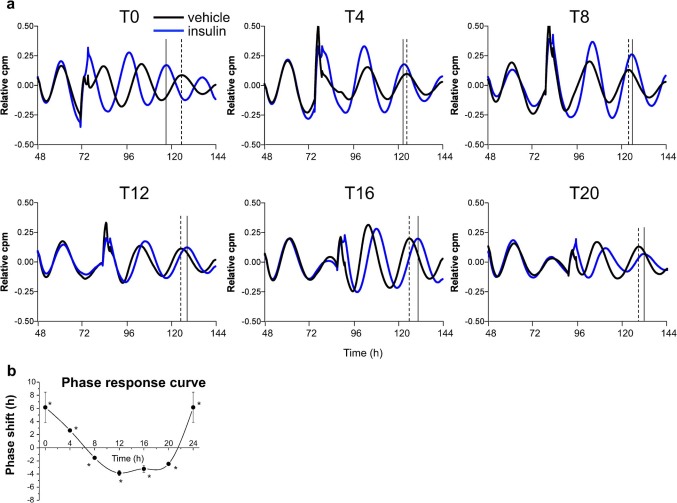
We established herein that spherical hepatocytes exhibited remarkably stable oscillations under more differentiated conditions in primary hepatocytes. By using this culture system, we then established an ex vivo luminescence reporter system in the hepatocytes prepared from a Per2-dLuc transgenic rat26. Isolated rat parenchymal hepatocytes were plated on an EHS gel-coated dish. Hepatocytes can be cultured in a serum- and hormone-free medium, which is beneficial when studying the relationship between hormones and phase entrainment. The hepatocytes were found to exhibit remarkably stable oscillations in Per2 expression over 7 days. Insulin was added to the hepatocytes at 6 different time points: T0, T4, T8, T12, T16, and T20, for 2-h period in each case, with circadian T0 defined as the time at which minimal bioluminescence was observed (Fig. 3a). Insulin treatment caused a phase advance at T0 and T4, but a phase delay at T8, T12, T16, and T20 (Fig. 3a). An apparent phase-shift response curve for hepatocytes could be obtained from these data (Fig. 3b). In particular, treatment with insulin during the subjective “night,” when the serum insulin levels of the rodents peak, produced a pronounced phase delay. To validate this Per2 phase response effect of insulin, hepatocytes were treated with dexamethasone, which has been reported to phase shift Dbp rhythms in the liver8. This produced a phase advance in the T0 treatment, but a phase delay in the T12 treatment (data not shown). These results indicate that insulin acts as a synchronizer for the hepatic clock by cell-autonomous mechanisms.
Figure 3. Insulin directly regulates the phase entrainment of hepatocyte circadian oscillators throughout the 24-h day.
(a) Hepatocytes derived from Per2-dLuc transgenic rats were isolated and plated onto an EHS gel-coated dish in serum- and hormone-free medium. At the indicated circadian time, hepatocytes were treated for 2 h with 50 nM insulin (blue curves), or with vehicle (black curves), and bioluminescence levels were measured. Black and dotted lines indicate peaks after insulin and vehicle treatment, respectively. (b) The phase response curve associated with insulin treatment at each circadian time point (T0, T4, T8, T12, T16, and T20). *p < 0 .05 versus vehicle -treated cells (Student's t-test). Three time independent experiments were performed.
To delineate the mechanism for how insulin entrains the hepatic clock, we used PD98059, an inhibitor of MAPK, and LY294002, an inhibitor of PI3K, in our ex vivo culture system. While insulin caused phase shift of the hepatic clock, both PD98059 and LY294002 blocked this phase shift (Supplementary Fig. 8). These results indicated that both MAPK and PI3K pathways are important mediators of the phase entrainment of the hepatic clock by insulin.
Phase advance of the hepatic circadian gene expression in STZ-induced diabetic rats
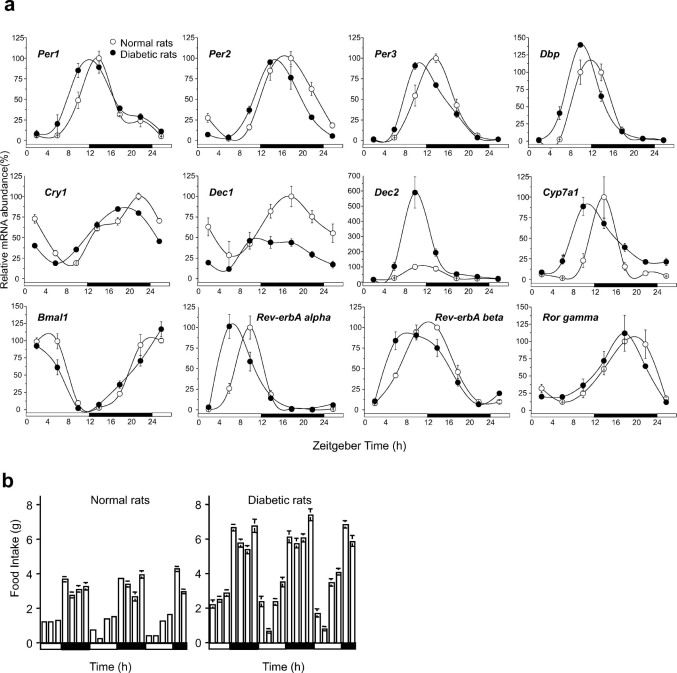
To verify the role of insulin in phase entrainment in vivo, the effects of streptozotocin (STZ)-induced diabetes mellitus on daily circadian gene expression in the liver were investigated. The success of diabetes induction was assessed by measuring serum glucose levels, which were 3-fold higher in STZ-injected rats compared to control rats (Supplementary Fig. 9). Real-time RT-PCR was used to observe similar changes in the mRNA levels of several clock-regulated genes, which showed a pronounced phase advance in STZ-induced diabetic rat livers compared to control livers (Fig. 4a, Supplementary Table 2). Interestingly, Dec1 mRNA levels dramatically declined, and Dec2 mRNA rhythmicity was maintained but was significantly elevated in STZ rat livers (Fig. 4a). These findings reveal that STZ-induced diabetes resulted in changes to the expression of circadian clock genes in a gene-specific manner. Kuriyama et al. reported that the rhythmic expression of Per2 was dampened in the STZ mouse liver12. In the hearts isolated from STZ-induced rats, the phases of circadian rhythms were altered, compared to those observed for control hearts27. Herichova et al reported that a phase advance in the rhythm of Dbp expression in both, the liver and the heart, although it did not observe a significant daily rhythm in the expression of Per2 in the liver of STZ-treated animals9. To our knowledge, the profound phase advance seen in a majority of these clock genes has never been observed before in STZ rat livers. A question raised by the results presented here is why previous studies did not demonstrate that the majority of clock genes exhibited a profound phase advance in STZ rat livers compared to control livers. One possible explanation would be that sampling in the previous studies was conducted to examine a longer period (6–7-h intervals) than those chosen in the present study. With the present method, it is possible to detect subtle effects such as an approximate 3-h phase advance in STZ-induced diabetic rat livers.
Figure 4. STZ-induced diabetic rats have a pronounced phase advance in their hepatic clock.
(a) Hepatic circadian gene expression profiles in STZ-induced diabetic rats. Livers were harvested every 4 h from control and STZ-induced rats. Transcripts of clock-related genes in the liver were analyzed using real-time PCR after normalization to β-actin expression levels. Values and bars represent mean ± SEM of 3–4 control (open circles) or STZ (filled circles) rats. Open and solid horizontal bars indicate light and dark periods, respectively. (b) Food intake of control (left panel) and STZ (right panel) rats was measured at 3-h intervals.
The circadian fluctuations of serum corticosterone levels are greatly elevated by STZ under ad libitum feeding13. Moreover, continuous administration of the synthetic glucocorticoid into mice constitutively increases hepatic Per1 mRNA levels, which attenuates the oscillation of the expression of other clock genes28. However, the present study confirmed that Per1 gene expression was phase advanced, rather than increased, in STZ rat livers (Fig. 4a), suggesting that the phase advance in STZ-induced rat livers is not due to abnormal corticosterone levels. Previous work has shown that Dec2 mRNA levels are elevated in Dec1 knockout mice, suggesting that the disruption in Dec1 expression may be compensated for by its paralog, i.e., Dec229. Consistent with this hypothesis, the authors found that disruption of a regular feeding rhythm induced Dec1 mRNA expression and caused a dramatic decline in Dec2 mRNA rhythmicity16. Taken together, these observations suggest an interrelationship between serum insulin concentrations and Dec1 mRNA levels. Moreover, a recent study showed that Dec1 deficiency advanced the phase of the E-box-containing clock genes30. Therefore, the disruption of Dec1 expression may, in part, contribute to the phase advance of clock genes in STZ-induced diabetic rat livers.
Circadian rhythm of food consumption is maintained in STZ-induced rats
Contribution of the central clock to desynchronization in STZ rat livers was investigated. Previously, the melatonin rhythm, which reflects the phase of the master clock, was shown to be unaffected by STZ application9, and the rhythmicity of locomotor activity was also maintained in STZ-induced mice11. In addition, the amplitude of Per2 expression in the SCN was unaffected by STZ administration12. Furthermore, we and other investigators have shown that a disruption of the regular feeding rhythm can cause a slight phase advance in the circadian rhythmicity of the liver16,31. In the present study, the food intake of STZ-induced and control rats was measured every 3 h, and it was found that STZ-induced rats maintained a circadian feeding rhythm, although the total daily food consumption of the STZ-induced rats was higher than that of control rats (Fig. 4b). These results demonstrate that the liver clock is desynchronized with the central nervous system clock in STZ-induced diabetic rats, although central nervous system-related circadian activities are quite normal.
Insulin acts as an in vivo phase entrainment factor for the liver circadian clock
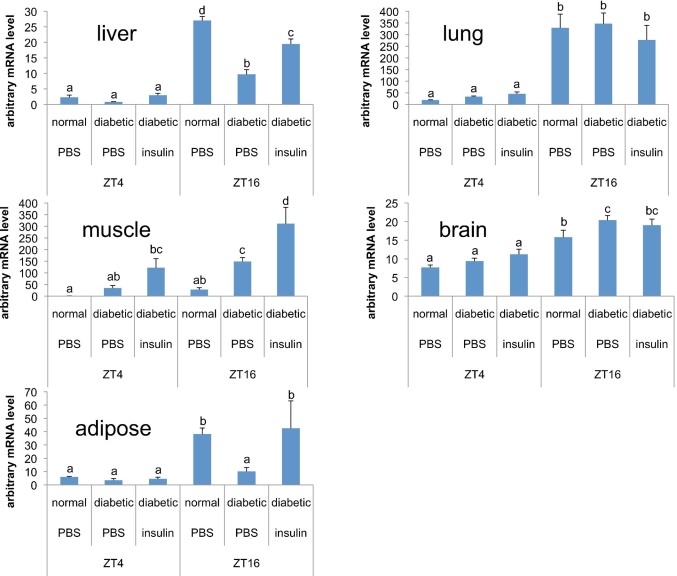
Effects of STZ on daily circadian gene expression in other tissues were investigated. We confirmed that Per2 mRNA peak and trough levels were maintained in the lung and brain compared to insulin sensitive organ such as the liver, muscle and adipose tissues (Fig. 5). Our studies indicate that diabetes leads to tissue-specific abnormalities in the circadian rhythm.
Figure 5. Diabetic mellitus is a disorder with disintegrated body clocks occurring through the lack of insulin sensitivity.
The liver, muscle, adipose tissue, brain, and lung were harvested at ZT4: 16 each from control and STZ-induced rats. STZ-induced rats were injected with insulin (50 U·mg−1·kg−1) and killed 2 h after the injection for measurement of mRNA levels. Per2 mRNA levels were determined by RT-PCR. Values and bars represent mean ± SEM of 6 control or STZ-induced rats. The statistical significance of differences of differences among values was analyzed ANOVA and then by Duncan's multiple range test. Values with different letters indicate a statistically significant difference (p < 0 .05).
We also investigated whether insulin could act as a synchronizer capable of inducing the core clock gene in its sensitive organs. Insulin rapidly induced Per2 gene expression in the liver, muscle, and adipose tissues from STZ-induced rats, whereas this induction was not observed it in the lung and brain (Fig. 5). PER2 also were used indicators for hepatic clock through cell-autonomous mechanisms because they are introduced as clock-resetting gene in brain32. Induction of this gene shall turn on the signals for hepatic circadian functions.
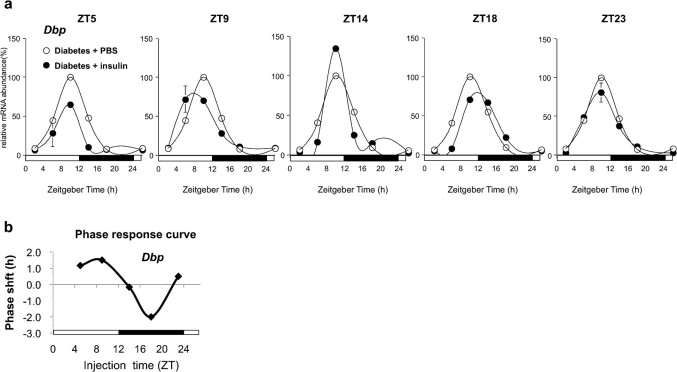
STZ-induced rats were injected intraperitoneally with insulin to determine whether insulin could produce a phase shift in the liver circadian rhythms. Insulin, or a vehicle, was injected at zeitgeber times (ZT) 5, ZT9, ZT14, ZT18, and ZT23, and the livers were harvested at 4-h intervals. Insulin administration at ZT18 delayed the phase shift of Dbp and Per1 in the liver of STZ-diabetic rats (Fig. 6, Supplementary Fig. 10). Importantly, the administration of insulin at ZT9 to STZ-diabetic rats further advanced the phase of Dbp gene expression (Fig. 6a). A phase response curve was established for the liver clock by injecting insulin into STZ-induced rats at various times of the day (Dbp, Fig. 6b; Per1, Supplementary Fig. 10). These findings clearly demonstrate that insulin acts as an in vivo phase entrainment factor for the rat liver circadian clock.
Figure 6. A single administration of insulin induces phase-dependent bi-directional phase shifts in diabetic rat livers.
(a) Temporal Dbp mRNA expression levels were determined by northern blot analysis of the livers from control or STZ-induced rats harvested over a 24-h period. Rats were maintained under light-dark conditions and injected intraperitoneally at the indicated times with insulin (50 U·mg−1·kg−1) dissolved in PBS or in PBS alone. Values represent mean ± SEM of 2–3 PBS- (open circles) or insulin-administered (filled circles) rats. (b) The phase response curve of circadian liver gene expression was obtained by recording the circadian Dbp mRNA after injecting insulin into STZ-induced rats at the indicated times.
Discussion
As in vivo analysis does not distinguish whether the phase shift is dependent on direct action of insulin or its indirect action, further investigation was required to determine whether insulin plays a direct role in the phase entrainment of cultured cells. We first report that maintaining the spherical shape of the hepatocyte by coating the plates with a particular extracellular matrix (EHS gel) is critical for maintaining the robustness of the molecular clock over the course of several days in culture. Herein, we established this cell culture system, coupled with real-time luciferase reporter, which is commonly used to monitor the molecular clock in cultured cells. The results from this cell culture system better support insulin's role in peripheral clock setting, and provide a potentially useful procedure for screening entrainment factors for circadian clocks under serum- and hormone-free conditions. These results allow us to infer that insulin is a major synchronizer for the liver circadian clock.
In addition to using neuronal and humoral signals, the SCN controls peripheral oscillators through a more indirect route by driving daily rest-activity cycles, and consequently, feeding-fasting rhythms. Indeed, feeding rhythms appear to be a dominant zeitgeber for the entrainment of peripheral oscillators, and daytime-restricted feeding of nocturnal rodents completely inverts the phase of gene expression in peripheral tissues such as the liver, pancreas, and heart33,34. The signals that arise from feeding and entrain the liver clock and other peripheral oscillators could be food itself, food-induced metabolites, or hormones whose secretion is controlled by feeding. Hyperglycemia can induce the secretion of insulin. We, therefore, suggest that insulin acts as a molecular synchronizer for the liver clock that is associated with feeding behavior.
It is now recognized that genetically- and environmentally-induced dysfunctions of the circadian system contribute to the development of various pathological conditions15,16,35. Indeed, we have reported that abnormal rhythmicity of a metabolic enzyme transcript such as the Cyp7a1 gene deteriorates hypercholesterolemia in rats with irregular feeding16. We supposed that the phase advance of gene expression of cholesterol metabolism and other lipid metabolism in STZ-diabetic rats would aggravate hypercholesterolemia and other metabolic abnormalities. It was actually observed that STZ-induced rats exposed to a high-cholesterol diet (1% cholesterol) developed a severe hypercholesterolemia and hyperlipidemia (Supplementary Fig. 11, Supplementary Table 3). As part of the circadian rhythm in physiology, drug efficacy and toxicity can vary with time36. This implies the possibility that pharmacological modulators of the circadian rhythm might affect the outcome of pharmacotherapy. In diabetic subjects, insulin and combination therapies of anti-diabetic drugs are commonly used to ameliorate hyperglycemia. The data obtained in this study suggest that the timing of insulin administration plays an important role in resetting the circadian rhythm of the liver and subsequent pharmaceutical implications. Understanding the integration between the peripheral clock and metabolic pathways may ultimately contribute to the design of improved rational therapies for diabetes mellitus.
Methods
Preparation of hepatocytes
Adult male Wistar rats (150–200 g) were purchased from Japan SLC (Hamamatsu, Japan) and maintained; they had free access to animal chow and water for 3–10 days prior to use. Rat parenchymal hepatocytes were isolated by perfusing liver with collagenase, as described previously37. For the in vitro circadian variation experiment, the rat liver was routinely perfused at 11:00 and the cells were plated at 13:00. The hepatocytes were plated on dishes coated with a type I collagen (TIC), a type IV collagen (TIVC), a laminin, poly-N-p-vinylvenzyl-d-lactonamide (PVLA), and an Engelbreth-Holm-Swarm sarcoma gel (EHS gel), at a high density (1 × 107 cells/100-mm dish) or a low density (2.5 × 106 cells/100-mm dish). The cultures were maintained in a serum- and hormone-free Waymouth's MB 752/1 medium containing penicillin (5 IU/mL) and streptomycin (5 mg/mL). After 4 h, the cells were washed twice with PBS, and fresh warm medium was replaced. Thereafter, the medium was replaced daily with a fresh warm medium.
Preparation of culture dishes
TIC-coated dishes were prepared by adding 100 μg/mL of TIC (Nitta Gelatin, Japan) to plastic dishes (Falcon 1029). EHS gel-coated dishes were prepared as described previously20. TIVC and laminin-coated dishes were purchased from BD Biosciences (USA). PVLA-coated dishes were prepared by adding 100 μg/mL of PVLA25 to plastic dishes (Falcon 1029).
Treatment of hepatocytes
In studies to observe the effect of cytoskeleton-disrupting reagents, the cells were cultured for 45 h. Hepatocytes were then treated with the following cytoskeletal inhibitors: colchicine (1 × 10−6 M), paclitaxel (1 × 10−5 M), vinblastine sulfate (1 × 10−5 M), nocodazole (1 × 10−4 M), cytochalasin B (2 × 10−6 M), or 3,3′-iminodipropionitrile (IDPN, 1%) for additional 12 h.
RNA analysis
mRNA levels were analyzed using quantitative real-time PCR38 and northern blot analysis39. The sequences of the primer sets used are shown in Supplementary Table 4.
DNA microarray
The RNA samples were applied to a rat microarray (GeneChip Rat Genome 230 2.0 Array; Affymetrix Japan, Tokyo, Japan). Microarrays were hybridized and scanned, according to manufacturer instructions, with an Affymetrix fluidics station and Affymetrix GeneArray scanner. For normalization, GeneChip files were subjected to global scaling of each chip to a common mean target intensity using R (ver.2.7.2) and the MAS5 (Microarray Suite) function, which is included in the /affy/ package of the bioconductor. As the first step, probe sets called “absent” over all conditions and replicates were removed. As the second step, the 95th percentile of all the signals of the entire dataset that were flagged with an absent call was determined and used as a threshold to remove all the remaining probe sets whose expression values were consistently below this value. Bioinformatic analysis was performed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA), using differentially expressed gene expression values. Bio-functions were filtered by a p value of ≤0.005.
Cell culture experiments
Rat-1 fibroblasts cells were grown in Dulbecco's modified Eagle medium (DMEM)/10% fetal bovine serum (FBS). H4IIE cells were grown in minimum essential medium (MEM)/10% FBS/100 mM sodium pyruvate/10 mM non-essential amino acids. Cells were plated at density of 1.5 × 106 cells/60-mm dish. The cells reached confluency 2 days after plating, and they were incubated for 1 day in serum-free medium before initiating the experiment. At time 0, insulin (1, 5, and 50 nM) was added to the culture medium. For signaling experiments, LY2940020 (50 μM) and PD98059 (30 μM) were added to the medium 30 min before starting the 1-h insulin (50 nM) treatment. Cycloheximide (10 μM) was added to the culture medium 30 min before the insulin (50 nM) treatment. For insulin shock, the medium of the confluent cultures was replaced with 50 nM insulin (t = 0) and after 1 h, the cells were washed with phosphate-buffered saline (PBS) and incubated with serum-free medium. RNA was prepared at the indicated times. For the reporter gene assay, H4IIE cells were plated on a poly D-lysine-coated 24-well plate (1.0 × 105 cells/well) and transfected with 200 ng of the reporter constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. Construction of the reporter vectors (mPer1 and mPer2-dLuc) was described previously40. After 24 h of transfection, the medium was replaced with serum-free medium with insulin (50 nM). After 3 h, the cells were lysed using Passive Lysis Buffer (Promega, Madison, WI, USA) and the enzyme activity was measured using the Luciferase Assay System (Promega). Bioluminescence was measured with an LmaxII luminometer (Molecular Devices, Sunnyvale, CA, USA). To correct for variations in transfection efficiency, 20 ng pGL4.7 was cotransfected.
Real-time monitoring of bioluminescence
Per2-dLuc transgenic rats were generated as described previously26. Male Per2-dLuc transgenic rats, 8-week-old and weighing about 200 g, had free access to water and food. The animals were kept on a 12-h light/12-h dark cycle (light from 08:00–20:00). Rat parenchymal hepatocytes were isolated by perfusion of the liver with collagenase. Cells were plated on EHS gel-coated dishes (3.0 × 105 cells/35-mm dish). For 4 h after plating, the medium was washed, and then a serum- and hormone-free Waymouth's MB 752/1 medium supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and 0.1 mM luciferin was added. Bioluminescence was measured with photomultiplier tube (PMT) detector assemblies (Hamamatsu Photonics, Hamamatsu, Japan). At the indicated circadian time, the phase shift experiment was performed by replacing the medium with the one containing insulin (50 nM) or dexamethasone (100 nM), and 2 h later, the medium was replaced with the recording medium. The phase was calculated as described in previous studies40. Data sets were detrended by subtracting the 24-h running average from the raw data. The phase shift was calculated using the Origin 6.1 program (Origin Lab, Corporation Northampton, MA, USA).
Animals and treatment
Male rats of the Wistar strain (Japan SLC, Hamamatsu, Japan), 5-week-old and weighing ∼90 g, were kept on a 12-h light/12-h dark cycle (light from 08:00–20:00). Before being divided into 2 groups matched for body weight, they were fed 20% casein-containing diets for 5 days ad libitum. The compositions of the diets are shown in Supplementary Table 3. Diabetes was induced by a single intraperitoneal injection of STZ (60 mg/kg). Control rats were injected with saline. Blood glucose levels were checked 9 days after STZ injection. Food consumption was measured every 3 h for 63 h from day 11 to day 14 after STZ injection. Rats were killed by decapitation at 4 h intervals after 4-h fasting, 16 days after STZ injection. The tissues were immediately removed, frozen in liquid nitrogen, and stored at −80°C until assayed. Blood was collected from the cervical wound. Serum was prepared by centrifugation at 1500 × g for 10 min. The total cholesterol, phospholipid, triglyceride, and glucose in serum were enzymatically determined using a commercial kit (Cholesterol C–test, phospholipid C–test, Triglyceride E–test, and Glucose CII–test; Wako Pure Chemical, Osaka, Japan). Twenty days after the administration of STZ, the rats were injected intraperitoneally with insulin (Humalin R, 50 U·mg−1·kg−1) (ZT6, ZT10, ZT14, ZT18, and ZT2), and rats were killed by decapitation at 4-h intervals (ZT6, ZT10, ZT14, ZT18, and ZT2). The tissues were immediately removed, frozen in liquid nitrogen, and stored at −80°C until use. Animal studies were performed in compliance with the Rules and Regulations of the Guide for the Care and Use of Laboratory Animals, Nagoya University.
Peak time analysis
We estimated the peak time of each cycling gene from the peak time of the best-fitted cosine curve, and defined it as the molecular peak time. We prepared cosine curves of 24-h periodicity with peaks from 0 to 24 h at increments of 10 min, yielding a total of 144 test cosine curves, and calculated the correlation value of the best-fitted cosine curve for each probe set. Estimation of molecular peak time was conducted as described by Ueda et al41.
Statistical analysis
The t-test assesses whether the means of two groups are statistically different from each other. Multiple comparisons among group mean differences were checked using Dunnett's test. We analyzed significance of the interaction between circadian time and diabetic / insulin effect by two-way ANOVA and then by Duncan's multiple range test. Values with asterisk indicate a statistically significant difference.
These studies were approved by Astellas Pharma and Nagoya University.
Author Contributions
D.Y. conceived, designed, and carried out experiments and had the primary role in writing the paper. T.I., T.H., S.O., Y.K., S.K., and K.I. carried out experiments and contributed to data analysis and data interpretation. T.L. carried out data analysis. T.K. and K.N. carried out experiments. S.H. supervised and advised on scientific aspects. H.O. conceived, designed, and supervised all studies, wrote the final draft of the manuscript, and acts as a guarantor for the studies.
Supplementary Material
Supplementary Information
Acknowledgments
The authors thank Y. Shibata for helpful data analysis. We are grateful to H. Okamura for the rPer1 and rPer2 probes, and U. Schibler for rDbp cDNA. This research was done as part of a research and development project of the Industrial Science and Technology Program supported by the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
The authors declare that they have no competing financial interests.
References
- Reppert S. M. & Weaver D. R. Coordination of circadian timing in mammals. Nature 418, 935–941 (2002). [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Damiola F. & Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998). [DOI] [PubMed] [Google Scholar]
- Yamazaki S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000). [DOI] [PubMed] [Google Scholar]
- Nagoshi E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S. & Kay S. A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K., Tamanini F., van Der Horst G. T. & Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292, 278–281 (2001). [DOI] [PubMed] [Google Scholar]
- Yoo S. H. et al. PERIOD2::LUCIFERASE real–time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101, 5339–5346(2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A. et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000). [DOI] [PubMed] [Google Scholar]
- Herichová I., Zeman M., Stebelová K. & Ravingerová T. Effect of streptozotocin-induced diabetes on daily expression of per2 and dbp in the heart and liver and melatonin rhythm in the pineal gland of Wistar rat. Mol. Cell. Biochem. 270, 223–229 (2005). [DOI] [PubMed] [Google Scholar]
- Kohsaka A. et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 6, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- Kudo T. et al. Night-time restricted feeding normalises clock genes and Pai-1 gene expression in the db/db mouse liver. Diabetologia 47, 1425–1436 (2004). [DOI] [PubMed] [Google Scholar]
- Kuriyama K., Sasahara K., Kudo T. & Shibata S. Daily injection of insulin attenuated impairment of liver circadian clock oscillation in the streptozotocin-treated diabetic mouse. FEBS Lett. 572, 206–210 (2004). [DOI] [PubMed] [Google Scholar]
- Oishi K., Kasamatsu M. & Ishida N. Gene- and tissue-specific alterations of circadian clock gene expression in streptozotocin-induced diabetic mice under restricted feeding. Biochem. Biophys. Res. Commun. 317, 330–334 (2004). [DOI] [PubMed] [Google Scholar]
- Yanagihara H., Ando H., Hayashi Y., Obi Y. & Fujimura A. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol. Int. 23, 905–914 (2006). [DOI] [PubMed] [Google Scholar]
- Lamia K. A., Storch K. F. & Weitz C. J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. USA 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamajuku D. et al. Regular feeding plays an important role in cholesterol homeostasis through the liver circadian clock. Circ. Res. 105, 545–548 (2009). [DOI] [PubMed] [Google Scholar]
- Yamada K. et al. Insulin induces the expression of the SHARP-2/Stra13/DEC1 gene via a phosphoinositide 3-kinase pathway. J. Biol. Chem. 278, 30719–30724 (2003). [DOI] [PubMed] [Google Scholar]
- Balsalobre A., Marcacci L. & Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr. Biol. 10, 1291–1294 (2000). [DOI] [PubMed] [Google Scholar]
- Kleinman H. K. et al. Basement membrane complexes with biological activity. Biochemistry 25, 312–318 (1986). [DOI] [PubMed] [Google Scholar]
- Oda H., Nozawa K., Hitomi Y. & Kakinuma A. Laminin–rich extracellular matrix maintains high level of hepatocyte nuclear factor 4 in rat hepatocyte culture. Biochem. Biophys. Res. Commun. 212, 800–805 (1995). [DOI] [PubMed] [Google Scholar]
- Oda H., Yoshida Y., Kawamura A. & Kakinuma A. Cell shape, cell-cell contact, cell-extracellular matrix contact and cell polarity are all required for the maximum induction of CYP2B1 and CYP2B2 gene expression by phenobarbital in adult rat cultured hepatocytes. Biochem. Pharmacol. 75, 1209–1217 (2008). [DOI] [PubMed] [Google Scholar]
- Schuetz E. G. et al. Regulation of gene expression in adult rat hepatocytes cultured on a basement membrane matrix. J Cell Physiol. 134, 309–323 (1988). [DOI] [PubMed] [Google Scholar]
- Singhvi R. et al. Engineering cell shape and function. Science 264, 696–698 (1994). [DOI] [PubMed] [Google Scholar]
- Oda H. Molecular mechanism of maintenance of differentiation function in cultured hepatocytes and application of the hepatocytes to molecular nutritional study. Nippon Nogeikagaku Kaishi 75, 1267–1274 (2001). [Google Scholar]
- Kobayashi A., Goto M., Kobayashi K. & Akaike T. Receptor-mediated regulation of differentiation and proliferation of hepatocytes by synthetic polymer model of asialoglycoprotein. J. Biomater. Sci. Polym. Ed. 6, 325–342 (1994). [DOI] [PubMed] [Google Scholar]
- He P. J., Hirata M., Yamauchi N., Hashimoto S. & Hattori M. A. The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real-time monitoring system. J. Endocrinol. 193, 413–420 (2007). [DOI] [PubMed] [Google Scholar]
- Young M. E., Wilson C. R., Razeghi P., Guthrie P. H. & Taegtmeyer H. Alterations of the circadian clock in the heart by streptozotocin-induced diabetes. J. Mol. Cell. Cardiol. 34, 223–231 (2002). [DOI] [PubMed] [Google Scholar]
- Koyanagi S. et al. Chronic treatment with prednisolone represses the circadian oscillation of clock gene expression in mouse peripheral tissues. Mol. Endocrinol. 20, 573–583 (2006). [DOI] [PubMed] [Google Scholar]
- Grechez-Cassiau A. et al. The transcriptional repressor STRA13 regulates a subset of peripheral circadian outputs. J. Biol. Chem. 279, 1141–1150 (2004). [DOI] [PubMed] [Google Scholar]
- Nakashima A. et al. Mol. Cell Biol. 28, 4080–4092 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen D. R. et al. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc. Natl. Acad. Sci. USA 103, 3393–3398 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka M. J., Shearman L. P., Weaver D. R & Reppert, S. M. Three period homologs in mammals: different light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20, 1103–1110 (1998). [DOI] [PubMed] [Google Scholar]
- Xu K., Diangelo J. R., Hughes M. E., Hogenesch J. B. & Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 13, 639–654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes. Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M. et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat. Med. 16, 67–74 (2010). [DOI] [PubMed] [Google Scholar]
- Paschos G. K., Baggs J. E., Hogenesch J. B. & FitzGerald G. A. The role of clock genes in pharmacology. Annu. Rev. Pharmacol. Toxicol. 50, 187–214 (2010). [DOI] [PubMed] [Google Scholar]
- Nagaoka S., Kamuro H., Oda H. & Yoshida A. Effects of polychlorinated biphenyls on cholesterol and ascorbic acid metabolism in primary cultured rat hepatocytes. Biochem. Pharmacol. 41, 1259–1261 (1991). [DOI] [PubMed] [Google Scholar]
- Ueda H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 (2005). [DOI] [PubMed] [Google Scholar]
- Chomczynski P. & Sacchi N. Single–step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 (1987). [DOI] [PubMed] [Google Scholar]
- Yamajuku D. et al. Identification of functional clock-controlled elements involved in differential timing of Per1 and Per2 transcription. Nucleic Acids Res. 38, 7964–7973 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. R. et al. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl Acad. Sci. USA, 101, 11227–11232 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information