Abstract
The stubby root nematode, Paratrichodorus allius, is important to the potato industry in the Pacific Northwest of USA, because it vectors Tobacco rattle virus (TRV), the causal agent of corky ringspot disease. The current method for determining if nematodes are viruliferous for TRV takes several weeks, requiring a glasshouse bioassay followed by a serological test. To overcome this drawback, a rapid and affordable molecular test was developed using reverse transcription polymerase chain reaction (RT-PCR) to identify viruliferous P. allius nematodes within 48 hours. Primers from the 16 kDa gene of TRV were used to detect TRV in both greenhouse-reared and field collected P. allius. TRV RNA can be detected consistently in nucleic acids equivalent to one quarter of a viruliferous adult nematode reared in the greenhouse. In order to reduce the time and expense of processing individual nematodes from field samples, viral RNA was consistently and affordably detected in extracts from 5 field-collected adult P. allius.
Keywords: Detection, Reverse Transcription PCR, Paratrichodorus allius, viruliferous stubby root nematode, tobacco rattle virus
Nematodes of the genera Paratrichodorus and Trichodorus are polyphagous migratory ectoparasites that feed on roots of annual and perennial plants causing stunting and are referred to as the stubby root nematodes. Nine stubby root nematode species are currently known to vector tobraviruses that cause economically important diseases of potatoes and ornamental bulbs (Harrison and Robinson, 1986; Mumford et al., 2000; Taylor and Brown, 1997). The most prevalent trichodorid vector of Tobacco rattle virus (TRV) (genus Tobravirus) in the Pacific Northwest (PNW) of USA is Paratrichodorus allius (Jensen ,1963) Siddiqi 1974, and approximately 10% of field populations are viruliferous (Mojtahedi et al., 2000). In potato (Solanum tuberosum L), TRV causes corky ringspot disease (CRS) (Mojtahedi et al., 2000) which is increasing in importance in Washington State (Pelter, 1997), and has been reported in several other states (Kirk et al., 2008).
Existing biological tests that screen for viruliferous P. allius are time consuming, and inaccurate. These assays require indicator plants, such as tobacco (Nicotiana tabacum L.) ‘Samsun NN’, for detecting TRV in live nematodes, often followed by an ELISA test. However, this assay requires approximately 2 months. TRV can be detected in potato tubers using reverse transcription-polymerase chain reaction (RT-PCR) but has not been reported for the detection of TRV in P. allius. A real-time fluorogenic 5′ nuclease PCR assay has been developed for TRV detection in two stubby root nematodes P. pachydermus and Trichodorus similis, which are not found in the PNW (Holeva et al., 2006).
Stubby root nematodes are relatively common in the PNW and have been found more often in potato-growing areas than TRV (Mojtahedi et al., 2000). Therefore, the presence of P. allius in a field does not necessarily imply that nematode control measures are necessary. The development of rapid, reliable and affordable diagnostic tools to identify TRV-infected P. allius would benefit the potato industry by allowing growers to make early decisions in terms of P. allius management practices. In this study, we developed a molecular diagnostic test for identification of viruliferous P. allius nematodes that can be completed within 48 hours in order to meet this need. Furthermore, our test would determine how many field stubby root nematodes could be used in one PCR reaction in order to detect as few as one viruliferous nematode.
Materials and Methods
Nematode and virus isolates: A viruliferous P. allius population was isolated from a CRS-infested field in Pasco, WA and maintained on Nicotiana tabacum L. var. Samsun NN, a good host for both TRV and the stubby root nematode (Mojtahedi and Santo, 1999). Nematode-infested grown on tobacco plants under greenhouse conditions exhibited typical TRV symptoms, such as light brown necrotic ring spots on the leaves. The presence of the virus was confirmed by antigen-coated plate indirect enzyme-linked immunosorbent assay (ELISA) (Converse and Martin, 1990). Paratrichodorus allius was also collected from three commercial potato fields (Pasco and Prosser, WA; and Klamath Falls, OR) with reported incidence of CRS disease; a total of 22 different soil samples (250 cm3 each) were collected from the above fields; a total of 185 nematodes were collected from the field samples.
Isolation of RNA from P. allius and reverse transcriptase reaction: Nematodes were collected from greenhouse-grown tobacco, and field soil samples by sieving infested soil followed by centrifugal flotation methods (Jenkins, 1964). Individual adult female P. allius were hand picked using a probe with an eyelash attached at its end, and total RNA was isolated based on a protocol obtained from Dr. R. Martin (USDA-ARS, Corvallis, OR) and modified as follows: RNA was extracted from single female individuals from greenhouse raised nematodes, and from soil samples from the field, RNA was extracted from sets of five adult female nematodes. Nematodes were handpicked and placed in 1.5 ml microcentrifuge tubes containing 100 μl sterile distilled H2O; 100 μl of collagenase (Sigma-Aldrich , St Louis, MO) solution (10mg/ml in 50mM Tris, l mM CaCl2, pH 7.4) was added to each tube and incubated at 37°C for 1 hour. Then 50 mg of glass beads (Acid-washed, 425-600 microns, Sigma) were added followed by 200 μl of 2X RNA Extraction Buffer (400 mM Tris pH 8.5; 600 mM LiCl; 20mM EDTA; 3% lithium dodecyl sulfate; 2% deoxycholic acid; 2% tergitol; 2% 2,β-mercaptoethanol (Sigma-Aldrich Milwaukee, WI). Tubes were vortexed for 1 min, followed by centrifugation for 30 sec at 21,000 x g. The supernatant was discarded and 400 μl 6 M potassium acetate pH 6.5 were added to the pelleted material; thoroughly mixed, and incubated on ice for 15 min. Tubes were centrifuged at 21,000 x g for 5 min and the supernatant was transferred to a new tube and incubated for 15 min at −20°C, and then centrifuged again for 5 min at 21,000 x g. The supernate was transferred to a new tube, and 1 μl glycogen at a concentration of 20μg/μl (Invitrogen, Carlsbad, CA) was added to each tube, followed by an equal volume of isopropanol. The tubes were incubated for 15 min at −20°C and centrifuged for 20 min at 21,000 x g. The supernate was discarded without disturbing the pellet and the RNA pellet was washed twice with 500 μl 70% ethanol. The pellet was air dried at room temperature for 10 min at 20°C. Pellets were dissolved in 20 μl diethylpyrocarbonate (DEPC) H2O (Sigma-Aldrich, St Louis, MO) and placed on ice for immediate use or stored at −20°C.
First strand synthesis from TRV RNA in extracts was primed with primer TRV1 (5′ CAGTCTATACACAGAAACAGA 3′) complementary to sequences near the 3′ terminus of the 16 kDa gene on TRV genomic RNA 1 (Robinson, 1992). A 10 μl aliquot of the RNA was added to 2.0 μl of TRV1 primer (20 pmol) and 1.8 μl of sterile distilled water and incubated for 5 min at 65°C then placed on ice. To each tube, 4 μl of M-MLV RT 5X Reaction Buffer (Promega, Madison, WI) 2.0 μl of a mixture containing 10 mM each of dATP, dCTP, dGTP, and dTTP (Invitrogen) and 0.2 μl (200u/μl) of M-MLV reverse transcriptase (Promega) were added. The mixture was incubated at 37°C for 1 hour followed by 65°C for 5 min to inactivate the enzyme.
PCR amplification for virus detection: PCR amplification was performed using a thermocycler (Biometric T Gradient 96, Whatman, Gottingen, Germany). The 50 μl reaction mixture contained 10 μl first strand cDNA, 5 μl of 10X Buffer (Promega), 3.0 μl of 25 mM MgCl2 (Promega), 1.5 μl of 10mM dNTPs (Invitrogen), 1.0 μl of primers TRV1 and TRV2 (5′ GACGTGTGTACTCAAGGGTT 3′) (20 pmol), 0.2 μl (1 unit) of Taq Polymerase (Invitrogen, Madison, WI) and 28.3 μl sterile distilled water. After an initial denaturation for 2 min at 94°C, the amplification consisted of 40 cycles of the following: denaturation for 15 seconds at 94°C, annealing for 2 min at 58°C, extension for 1 min at 72°C; and a final extension step for 10 min at 72°C. PCR products were resolved by electrophoresis in a 1.5% agarose gel and 1X TAE Buffer (1mM EDTA; 40mM Tris; 228.4 μl/l of glacial acetic acid; pH 8.0) for 80 min at 40 volts. Caenorhabditis elegans cDNA and nonviruliferous P. allius cDNA (reared on spearmint to ensure that the virus will not be present according to Mojtahedi et al., 2002) were used as negative control in PCR reactions. In addition, 2 μl of TRV nucleic acid extracted from a symptomatic potato tuber of the cultivar Russet Ranger collected in Pasco, WA was used for the positive control; the method used to obtain TRV from potatoes was based on Dellaporta (1993). The expected size of the PCR-amplified TRV fragment was 463 bp (Robinson, 1992).
Results and Discussion
We used adult females of P. allius to assay for the detection of TRV particles because they are most likely to carry the virus. According to MacFarlane (2003), tobravirus particles can be acquired by both juvenile and adult nematodes and retain the virus particles for long periods. However, the virus can be lost from the juveniles after molting, therefore, in our study only adult female nematodes were used from both the greenhouse and field samples.
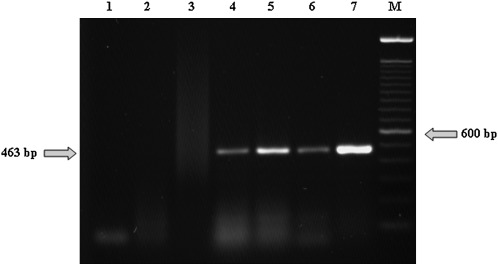
Our diagnostic assay routinely detected TRV RNA in nucleic acid from the equivalent of one quarter adult female viruliferous P. allius from greenhouse reared nematodes which are grown in 10 liter pots containing a tobacco plant infected with TRV to assure that most of the adult females will acquire TRV (Fig. 1). Nucleic acid extracted from single P. allius was resuspended in 10 μl of DEPC-treated water, which was used entirely for the RT-PCR reaction (a 20 μl reaction). The subsequent PCR reaction contained 10 μl of the reverse transcription reaction, which represents nucleic acids from one quarter of a nematode. RT-PCR produced an amplicon of the expected size (463 bp) from viruliferous P. allius nematodes. The 463 bp amplicon has been sequenced and was found to be identical to the homologous sequence of a Florida TRV isolate (AF055912) (Crosslin, unpublished data). TRV comes from viruliferous nematodes reared in the greenhouse and originally obtained from a CRS-infested field in Pasco, WA (Fig.1). The non-viruliferous stubby root nematodes which were used as controls were reared on spearmint to assure absence of TRV (Mojtahedi et al., 2002).
Fig. 1.
Agarose gel (1.5%) stained with ethidium bromide showing amplicons of the expected size (463 bp) from RT-PCR reactions with nucleic acids extracted from the equivalent of 1/4 viruliferous adult female Paratrichodorus allius (lane 4-6), reared in the greenhouse. Lane 7 is TRV isolated from a symptomatic potato tuber cv. Ranger Russet as the positive control. Lane 1 is water; lane 2 is cDNA from Caenorhabditis elegans as negative control; and lane 3 in cDNA from non-viruliferous P. allius from spearmint plants, as negative control. Lane (M) is a 100 bp DNA ladder (Invitrogen).
In order to determine detection limits, nucleic acids extracted from a single viruliferous P. allius were diluted in water and TRV was detected in 25%, 15%, and 5% of nucleic acids extracted from a single viruliferous female (data not shown). However, the detection of TRV was not consistent at higher dilutions. Nucleic acids were not extracted from a mixture of greenhouse tobacco reared viruliferous and greenhouse spearmint reared non-viruliferous nematodes as we do not know exactly how many of the tobacco reared nematodes were viruliferous at the time of the assay.
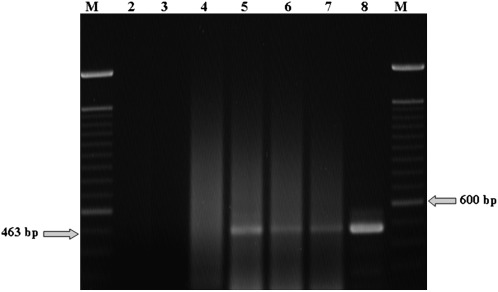
To provide growers with a timely, affordable and reliable assay, five P. allius were used per nucleic acid extraction; it would not be efficient or economical to use single nematodes. We have been able the reliably detect TRV in a mixture of 5 stubby root nematodes containing at least one viruliferous individual (Fig. 2); also TRV can be detected in samples containing less than 5 nematodes. None of the 12 soil samples from Klamath Falls containing 80 nematodes in total yielded a detectable amplified product. One of the 3 soil samples from Pasco containing a total of 75 stubby root nematodes produced a detectable amplified product. Five of the 7 soil samples from Prosser containing 30 nematode samples produced a positive PCR amplification product. However, we were not able to detect TRV in mixtures of one viruliferous nematode in 10, 20 and 30 non-viruliferous greenhouse reared nematodes (data not shown). It is possible that the presence of nematode nucleic acids in large amounts interfere with the ability to amplify a detectable product. Therefore, five female adults are used for routine diagnostic tests from field samples. It is possible that the brightness of the bands seen in Fig. 2 indicates that the titer (or viral RNA recovered) varies from nematode to nematode.
Fig. 2.
Amplification products of RNA isolated from 5 field adult female Paratrichodorus allius using TRV specific primers (TRV1 and TRV2) in lanes 5 (Pasco field), and 6 and 7 (Prosser field); viruliferous P. allius shown here were collected from two of the three potato fields; no viruliferous nematodes were detected in the Klamath Falls field (not shown). Lane 2 is water control, lane 3 is cDNA from non-viruliferous P. allius from spearmint plants, and lane 4 is cDNA from Caenorhabditis elegans as negative controls. Lane 8 is TRV isolated from a symptomatic potato tuber, cv. Ranger Russet, as the positive control. Lanes (M) are the size marker, 100 bp DNA ladder (Invitrogen).
This is the first report on the detection of TRV in adult females of P. allius. Additionally, the use of molecular based assays using RT-PCR produced results in approximately 48 hours. This assay is affordable for routine diagnostic tests in pre-plant situations.
To date, there are few reports on the direct detection of TRV from trichodorid nematodes. Van der Wilk et al. (1994) developed an RT-PCR assay that was able to detect TRV in viruliferous Trichodorus primitivus nematodes. More recently, Boutsika et al. (2004) developed an RT-PCR assay that was able to amplify TRV in P. anemones and P. pachydermus, two species related to P. allius but not found in the PNW. Holeva et al. (2006) reported TRV detection in P. pachydermus and T. similis using real-time PCR assay (TaqMan) which is a very reliable assay but expensive for routine testing because of the cost of the labeled probe and the need for the more expensive real-time PCR instrumentation.
The results reported here are consistent with recent advances on the direct detection of TRV-infected trichodorid nematodes and the technique will allow the rapid, reliable and affordable detection of viruliferous P. allius from field samples. As a result, the PNW potato growers will be able to make timely decisions on stubby root nematode control practices.
This research is part of an ongoing project aimed to provide scientific basis for a field management strategy to control TRV epidemics in commercial potato fields. It includes a comprehensive assessment of TRV infection rates of field collected nematodes (to determine sample size at different levels of confidence); seasonal variations of the nematode population in the field and its correlation with the rate of TRV-infected individuals in the population (to determine the optimal timing to assess the risk for economical damage caused by TRV); and the patterns of distribution of the nematode in different types of soil (to determine proper sampling methods). The results reported here represent an important step to develop an affordable and consistent risk assessment process for a management system to control TRV in commercial potato fields in the Pacific Northwest of USA.
Footnotes
E. Riga is supported by the College of Agricultural, Human, and Natural Resource Sciences Agricultural Research Centre, PPNS No. 0450, Department of Plant Pathology and IAREC WSU-Prosser, Project No.WNPO0542, Washington State University, Pullman, WA 99164-6430, USA. This research was funded by the Washington State Potato Commission and the Washington State Commission on Pesticide Registration.
This paper was edited by Brent Sipes.
Literature Cited
- Boutsika K, Phillips MS, MacFarlane SA, Brown DJF, Holeva RC, Blok VC. Molecular diagnostics of some trichodorid nematodes and associated Tobacco rattle virus. Plant Pathology. 2004;53:110–116. [Google Scholar]
- Converse RH, Martin RR. ELISA methods for plant viruses. In: Hampton R, Ball E, De Boer S, editors. Serological Methods for Detection and Identification of Viral and Bacterial Plant Pathogens. St. Paul, MN: APS Press; 1990. [Google Scholar]
- Dellaporta SL. Plant DNA miniprep and microprep: Version 2.1-2.3. In: Freeling M, Walbot V, editors. The Maize Handbook. New York, NY: Springer-Verlag; 1993. pp. 522–25. [Google Scholar]
- Harrison BD, Robinson DJ. Tobraviruses. In: van Regenmortel MHV, Fraenkel-Conrat H, editors. The Plant Viruses. New York: Plenum Press; 1986. pp. 339–369. [Google Scholar]
- Holeva R, Phillips MS, Neilson R, Brown DJF, Young V, Boutsika K, Blok VC. Real-time PCR detection and quantification of vector trichodorid nematodes and Tobacco rattle virus. Molecular and Cellular Probes. 2006;20:203–211. doi: 10.1016/j.mcp.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Jenkins WR. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Disease Reporter. 1964;48:692. [Google Scholar]
- Kirk WW, Gieck SL, Crosslin JM, Hamm PB. First report of corky ringspot caused by Tobacco rattle virus on potatoes (Solanum tuberosum) in Michigan. Plant Disease. 2008;92:485. doi: 10.1094/PDIS-92-3-0485B. [DOI] [PubMed] [Google Scholar]
- MacFarlane SA. Molecular determinants of the transmission of plant viruses by nematodes. Molecular Plant Pathology. 2003;4:211–215. doi: 10.1046/j.1364-3703.2003.00164.x. [DOI] [PubMed] [Google Scholar]
- Mojtahedi H, Santo GS. Ecology of Paratrichodorus allius and its relationship to the corky ringspot disease of potato in the Pacific Northwest. American Journal of Potato Research. 1999;76:273–280. [Google Scholar]
- Mojtahedi H, Santo GS, Handoo Z, Crosslin JM, Brown CR, Thomas PE. Distribution of Paratrichodorus allius and tobacco rattle virus in Pacific Northwest potato fields. Journal of Nematology. 2000;32:447. (Abstr.). [Google Scholar]
- Mojtahedi M, Santo GS, Thomas PE, Crosslin JM, Boydston RA. Eliminating tobacco rattle virus from viruliferous Paratrichodorus allius and establishing a new virus-vector combination. Journal of Nematology. 2002;34:66–69. [PMC free article] [PubMed] [Google Scholar]
- Mumford RA, Walsh K, Barker I, Boonham N. Detection of Potato mop top virus and Tobacco rattle virus using a multiplex real-time fluorescent reverse-transcription polymerase chain reaction assay. Phytopathology. 2000;90:448–453. doi: 10.1094/PHYTO.2000.90.5.448. [DOI] [PubMed] [Google Scholar]
- Pelter G. Corky ringspot disease of potatoes. Spud Topics. 1997;43:1. [Google Scholar]
- Robinson DJ. Detection of tobacco rattle virus by reverse transcription and polymerase chain reaction. Journal of Virological Methods. 1992;40:57–66. doi: 10.1016/0166-0934(92)90007-z. [DOI] [PubMed] [Google Scholar]
- Taylor CE, Brown DJF. Wallingford, UK: CAB International; 1997. Nematode Vectors of Plant Viruses. [Google Scholar]
- Van der Wilk F, Korsman M, Zoon F. Detection of tobacco rattle virus in nematodes by reverse transcription and polymerase chain reaction. European Journal of Plant Pathology. 1994;100:109–122. [Google Scholar]