Abstract
A gene encoding a manganese superoxide dismutase (MnSOD) enzyme (Mi-mnsod) was identified and characterized in second-stage juveniles of the root-knot nematode Meloidogyne incognita. The Mi-mnsod gene was found to possess five exons and four introns with (GT/AG) consensus splice-site junctions. The deduced amino acid sequence of Mi-mnsod encodes a putative 25 KDa protein, with conserved amino acid residues of the MnSOD family, including the Parker-Blake signature and four metal-binding sites. The derived amino acid sequence showed high similarity to other eukaryotic MnSODs, including a 23 amino acid N-terminal putative mitochondrial transit peptide. Gene expression was observed throughout the posterior nematode body region with elevated signal intensities at the anterior portion of the intestine. DNA blot analysis and sequencing data showed the occurrence of three putative copies of the MnSOD gene with nucleotide polymorphisms found at the fourth exon and the 3' un-translated region.
Keywords: Meloidogyne incognita, root knot nematode, manganese superoxide dismutase, anti-oxidant defense
Root-knot nematodes (Meloidogyne spp.) are root endoparasites of most cultivated crops that cause significant economic losses worldwide (Sasser, 1980). The infective second-stage juveniles (J2) penetrate the roots at the elongation zone and then migrate intercellularly, establishing feeding sites in the developing vascular tissue (Wyss et al., 1992). Although tomato (Solanum lycopersicon) is an excellent host for these nematodes, some varieties are resistant to M. incognita because of the presence of the dominant gene Mi. Resistance conferred by Mi-gene is exerted through hypersensitive response (HR) by the cells surrounding the feeding site of the nematode,which ultimately leads to a localized necrosis of the challenged host tissues (Droopkin et al., 1969; Paulson et al., 1972).
Hypersensitive response is triggered by a recognition event and consists of a complex response network which results in a programmed cell death, preventing further nourishment of the parasite and the establishment of a feeding site. One key feature of HR is the generation of reactive oxygen species (ROS), the oxidative burst. there are several proposed functions for ROS in the plant-pathogen interaction, such as signaling for local and systemic defense reactions, antimicrobial activity and cell-wall strengthening (Mehdy, 1994; Baker and Orlandi, 1995).
Toxic oxygen species can damage nucleic acids, proteins, and membrane lipids and therefore can be lethal (Fridovich et al., 1986; Hunter et al., 1997). To protect against these potentially damaging oxygen species, aerobic organisms evolved a multi-layered anti-oxidant defense system in which the anti-oxidant enzymes superoxide dismutase (SOD), glutathione peroxidase, and catalase, as well as non-enzymatic anti-oxidants, play a pivotal role (McCord and Fridovich, 1969; Henkle-Dührsen et al., 1995).
Anti-oxidant enzymes have been postulated to protect parasitic nematodes from host-induced oxidative damage in addition to the cellular scavenging functions of their own metabolism (Callahan et al., 1988). For instance, it has been found that manganese superoxide dismutase (MnSOD) activity was markedly higher in J2 extracts from virulent nematodes, selected from avirulent populations by repeated inoculations on resistant tomato, than in avirulent counterparts. This observation suggested a possible relationship between enhanced MnSOD activity and the ability of nematodes to grow on resistant tomato plants (Molinari et al., 2005).
In eukaryotes, free radicals have directly and indirectly been implicated in the aging process (Hartman et al., 1995) and cellular activities that eliminate free radicals have been associated with longer lifespan. For example, a transgenic strain of Drosophila melanogaster over-expressing both SOD and catalase exhibited a life-extension of up to one-third that of the wild-type (Orr and Sohal, 1994). Long-lived genetic variants of Caenorhabditis elegans also provided evidence for a positive correlation between SOD cellular levels and extended lifespans (Honda and Honda, 1999).
As originally formulated, the superoxide theory of oxygen toxicity ascribed the damaging effects of high oxygen tension to the formation of the superoxide radical (O2-•), although much of the cellular damage now appears to be the consequence of more reactive species (Halliwell and Gutteridge, 1989). SODs protect cells by catalyzing the dismutation of O2-• to hydrogen peroxide (H2O2) and molecular oxygen (O2). Three major classes of SODs have been described on the basis of the metal composition in the active site, i.e., Fe-Mn, and Cu-Zn SODs. FeSOD is found primarily in prokaryotes, MnSOD is found in both prokaryotes and eukaryotes, while the presence of Cu-Zn isoenzymes is restricted to eukaryotes (Halliwell and Gutteridge, 1989).
An increase in MnSOD activity was observed in virulent compared to avirulent populations of M. incognita which led us to investigate the sequence and expression of MnSOD in M. incognita (Molinari et al., 2005). Available genomic data for C. elegans SOD and partial SOD EST sequences from M. incognita were integrated with data from the M. incognita genomic project which revealed the existence of three SOD genes in the M. incognita genome strain Morelos, two copies encoding the Cu-Zn enzyme and one copy encoding the Fe-Mn enzyme (Abad et al, 2008). In this study using available genomic data, it was possible to identify and characterize a gene, herein indicated as Mi-mnsod, encoding a MnSOD enzyme from J2 of a population of M. incognita.
Materials and Methods
Nematode population: To obtain M. incognita J2, the nematode population (MILEV-L4) originally from Leverano (Italy) was maintained in a greenhouse on susceptible tomato (cv. UC82) under controlled conditions (24-26°C). Galled roots were removed from soil and rinsed, then egg masses were collected with a scalpel. Harvested eggs were incubated at 26°C for 5 days on moist filter papers. The hatched J2, which migrated through the paper, were collected in a water-filled Petri dish. Afterwards, J2 were counted under a stereoscope before collection with a sterile glass pipette tip.
Nucleic acids isolation: For DNA extraction, J2 were placed in a 1.5 ml tube and incubated at −80°C for 10 minutes. Subsequently, 100 μl of extraction buffer and 50 mg of acid-washed glass beads (425-600 μm diameter, Sigma, St. Louis MO) were added and tissues disrupted by vortex for 5 minutes. The lysate was mixed with 50 μl phenol and incubated for 10 minutes at 60°C; then, 50 μl chloroform/isoamyl alcohol (24:1) were added and the suspension mixed by inversion. The aqueous layer was separated by centrifugation for 10 minutes at 11,000 rpm. DNA was precipitated by addition of 4 μl 5M NaCl and 200 μl 100% ethanol at −20°C for one hour. After centrifugation at 12,000 rpm for 10 minutes, the pellet was washed twice with 70% ethanol and dissolved in sterile distilled water (SDW).
Total RNA was extracted from approximately 3000 J2 nematodes. Extraction was carried out by improving the single-step RNA isolation method with a monophasic solution of phenol and guanidine isothiocyanate (TRIzol, Invitrogen Carlsbad, CA USA), according to the manufacturer's instructions. For nematode disruption, 0.1 g of glass beads was added to 250 μl J2 suspension in TRIzol and tubes vortexed for 5 minutes. The RNA pellet was dissolved in 15 μl nuclease-free water and treated with DNAse (Roche Applied Science, Indianapolis, IN, USA) to remove possible contaminating genomic DNA. The RNA pellet was dissolved in 10 μl of nuclease-free water and stored at –70°C.
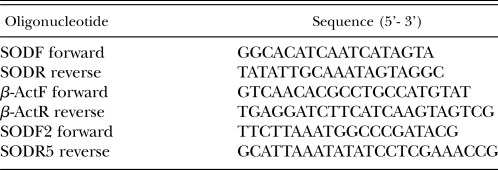
Amplification and sequencing analysis: cDNA synthesis was performed with 1 μg of total RNA in two steps with Enhanced avian HS RT-PCR Kit (Sigma, St. Louis MO, USA). For first strand synthesis, an anchored-oligo dT primer and AMV reverse transcriptase was used according to the manufacturer's instructions. Meloidogyne incognita ESTs CF980963 (Mc Carter J. P.) and BI773441 (Bird-Rao library, Mc Carter, J. P.) showing a high degree of similarity to the C. elegans Mn-SOD (CAEEL P31161) were used as a starting point for primer design. Specific internal primers were used in PCR on reverse-transcribed M. incognita poly (A)+ RNA. The 5' end of the Mi-mnsod cDNA was obtained using forward random primers and specific SODR reverse, whereas the 3' end was obtained using specific SODF forward and reverse anchored primer (Table 1). For PCR amplification 2 μl cDNA template were used directly in a 25 μl reaction mixture consisting of 1x PCR Buffer (Roche, Applied Science, Indianapolis, IN, USA), 100 μM each dNTPs, 1 unit Taq polymerase, and 500 nM primers. Amplification was carried out in a Thermocycler (iCycler, BioRad, Hercules CA, USA) programmed with an initial denaturation at 94°C for 4 minutes, 35 cycles at 94°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds, with a last step at 72°C for 5 minutes with a 4°C hold. The PCR products were analyzed by agarose gel electrophoresis and cloned into a pGemT easy vector (Promega, Madison WI, USA).
Table 1.
Acronyms and sequences of the oligonucleotides used in this study.
Using partial sequences generated by cDNA amplification, specific primers were designed to obtain the full-length genomic DNA sequences for M. incognita Mi-mnsod. PCR, cloning, and sequencing were performed using 40 ng of DNA and forward SODF2 and reverse SODR5 primers (Table 1) following amplification conditions described for the cDNA amplification.
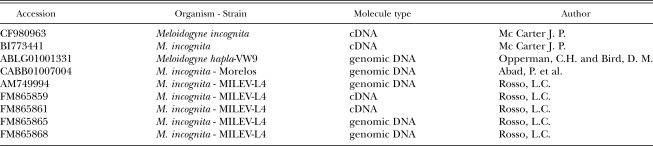
The cloned PCR products from cDNA and genomic DNA were sequenced by the MWG Biotech (Ebersberg, Germany) sequencing service and deposited in GenBank (Table 2). The SOD encoded protein sequence was analyzed for an N-terminal mitochondrial target domain by the MitoProt (Claros and Vincens, 1996) software. To compare MnSOD across species, related sequences were obtained from public databases (EMBL and NCBI). Where protein sequences were not available, homology searches were performed on genomic databases using the tblastn search program and the predicted proteins were assembled from identified translated exons. Sequences were aligned with Clustal W (Thompson et al., 1994).
Table 2.
Identification of sequences mentioned in this study.
DNA blot analysis: Genomic DNA from approximately 100,000 J2 was isolated and 8 μg of DNA were digested with Eco RI and Hind III restriction enzymes at 37°C for 18 hours and resolved on 0.7% agarose gel. After denaturation and neutralization, the DNA was capillary transferred onto a nylon membrane (Roche). Mi-mnsod chemiluminescent probe, produced by PCR using the PCR Dig probe synthesis kit (Roche, Applied Science, Indianapolis, IN, USA) was synthesized using, as a template, a plasmid DNA containing a fragment of the Mi-mnsod gene and SODR and SODF primers (Table 1). The Mi-mnsod probe sequence was confirmed by local BLAST against contigs resulting from the M. hapla strain VW9 available on line (http://www.hapla.org) and M. incognita strain Morelos genome assembly, deposited in the EMBL GenBank databases (Table 2). An 88% and a 97% similarity was observed with M. hapla ABLG01001331 (nucleotides 13873-14098) and M. incognita CABB01007004 (nucleotides 902-1246), respectively (Table 2). The hybridization temperature (45°C) was 2°C higher than the optimal temperature calculated according to both GC content and percent homology of probe to target indicated on the DIG Easy Hyb Granules (Roche) information sheet. Membranes were pre-hybridized at 45°C for 1 hour and hybridized at the same temperature overnight in DIG Easy Hyb Granules solution. After hybridization, the nylon membrane was washed twice at room temperature in 2 × SSC (NaCl, 3M; sodium citrate 0.3M; pH 7), 0.1% (w/v) sodium dodecyl sulfate (SDS) solution, and twice for 15 minutes with 1 × SSC, 0.1% (w/v) SDS at 65°C. Hybrid molecules were detected by the Dig luminescent detection kit (Roche) according to manufacturer's instructions. Hybridization events were detected by exposure to an x-ray film for 4 and 16 hours.
MnSOD gene induction assay: Paraquat dichloride (N,N-dimethyl-4,4-bipyridium dichloride, C12H14Cl2N2 · xH2O Sigma St. Louis MO, USA) solution was used to induce Mi-mnsod gene expression. A test to determine the threshold concentration of paraquat tolerated by J2 was performed by placing nematodes in 1.5 ml tubes containing paraquat solutions at 10, 50, and 100 mM final concentrations. The J2 were incubated in darkness at room temperature for 1, 3, 7, and 12 hours and viability checked under a stereoscope. Nematodes in 10 and 50 mM concentrations survived after 12 hours, whereas the J2 exposed at 100 mM died. The 50 mM concentration was selected for gene expression analysis.
Approximately 3,000 M. incognita J2 were placed in 1.5 ml tubes containing 50 mM paraquat and incubated in darkness at room temperature for 1, 3, 7, and 12 hours. After incubation, J2 were harvested by centrifugation at 2600 rpm for 3 minutes and washed twice in cold 0.1 M NaCl. The samples were frozen as a pellet in liquid nitrogen and stored at -80°C for subsequent RNA extraction. Sterile distilled water was added in control tubes. Transcript levels were measured by real-time PCR quantification using SYBR Green I Master Mix (Roche). Specific primers pairs SODF/SODR and βActF/βActR were used in the PCR reactions. β–actin primers were designed from M. incognita β–actin sequence, accession number BE225475 (amplicon 198 bp). The quantitative PCR reactions (20 μl) consisted of 2 μl first strand reverse transcriptase product, gene specific primers (500 nM), and fluorescent dye SYBR Green I (1×). The thermal profile was performed in a Mx3000P (Stratagene) 1 cycle 94°C for 4 minutes and 40 cycles at 94°C for 30 seconds, 49°C for 30 seconds, and 72°C for 20 seconds. Melting curve analysis was performed after the PCR reaction, to confirm that the signal was the result of a single product amplification. Cycle threshold (Ct) values were determined for three biological assays, with two or three replicates each. Amplification efficiencies were 101% for MnSOD and 109% for β–actin products. The corresponding Ct values were applied to estimate the ratio of starting amounts DNA according to the modified Pfaffl equation (Pfaffl, 2001): normalized treated (with paraquat)/normalized control (without paraquat) = (2-ΔCt target)/(2-ΔCt norm). Data were statistically analyzed by unpaired Student's t-test.
In situ hybridization: Primer SODR and SODF were used to synthesize digoxigenin-labeled sense and anti-sense single-stranded DNA by asymmetric PCR from the M. incognita Mi-mnsod cDNA. In situ hybridization was performed as described by De Boer et al. (1998). Approx 40,000 J2 of M. incognita were treated with 50 mM paraquat for 7 hours and washed three times with 1 ml SDW. Juveniles incubated in SDW were used as control. After treatment, J2 were fixed in 2% formalin buffered in M9 (pH 7.0) for 18 hours at 4°C and 4 hours at room temperature. Nematodes were randomly cut on glass slides with a razor blade. Partial digestion was performed with 500 ng/ml proteinase-K (Roche) at room temperature for 40 minutes. Denatured PCR digoxigenin-labeled DNA probes (approximately 20 ng) were added to each tube and hybridization was performed overnight at 45°C. cDNA was detected by alkaline phosphatase immunostaining at 4°C for 16 hours and sections were examined by light microscopy. Images were captured using a Nikon digital camera.
Results
cDNA sequence analysis: Using M. incognita cDNA as a template and the primers pairs SODF forward and anchored oligo dT, or random primers and SODR reverse, yielded amplicons that were 98% identical to CF980963. Assembling these amplicons resulted in a putative full-length open reading frame (ORF) of 660 bp within a total of 820 bp cDNA (Fig. 1). This cDNA sequence also contained a partial 5' untranslated region (UTR) and a 3' UTR of 30 bp and 119 bp, respectively. Six adenylate-uridylate-rich elements (ARE) were observed in the sequence, three of which were found in the transit peptide coding region, two were positioned in the mature peptide coding region, and one in the 3' UTR. The 3' UTR included a poly-(A) tail and the polyadenylation signal sequence 5'-AATAAA-3' at position 702. Mi-mnsod mRNA contained also two polypyrimidine tracts at the 3' UTR (Fig. 1).
Fig. 1.
Nucleotide and deduced amino acid sequence of M. incognita Mi-mnsod sequence, AM749994. Numbering is given for the gene (including introns) and amino acid sequences. Primers sequences (SODF2 forward and SODR5 reverse) are indicated in bold. Introns and flanking regions are showed in lowercase letters and exon in uppercase letters. Underlined motifs represent potential regulatory elements (see text). The putative mitochondria transit leader sequence of 22 amino acids in length is shown in italics. The Parker and Blake signature amino acids of MnSODs are marked with grey boxes; conserved metal-binding residues are identified by asterisks.
The translational start site was a typical ATG codon of eukaryotic mRNAs (Kozak, 1991). BlastP (Altschul et al., 1997) of the translated sequence, consisting of 220 amino acids with a predicted molecular mass of 25 kDa, revealed a high degree of similarity to the SOD family member (sod-2) of C. elegans (amino acid identities 74%, positives 83%), C. briggsae (amino acid identities 73%, positives 82%), and the MnSOD from the giant freshwater prawn Macrobrachium rosenbergii (amino acid identities 73%, positives 85%).
DNA Sequence analysis: Sequences of the entire Mi-mnsod gene, as well as 30 bp of 5' flanking sequence (not shown in Fig. 1) and 86 bp of 3' sequence were determined by amplification and sequencing from M. incognita genomic DNA with primers SODF2 forward and SODR5 reverse, located in the 5' and 3' UTR, respectively (Fig. 1). The genomic sequence was deposited in GenBank (EMBL) as accession number AM749994. The Mi-mnsod genomic sequence showed 96% similarity to M. incognita strain Morelos CABB01007004 (nucleotides 474-1417) and 82% with M. hapla ABLG01001331 (nucleotides 13431-14364).
Sequence comparisons of the cDNA and genomic clones indicated that Mi-mnsod gene is composed of five exons encoding 12, 41, 20, 98, and 49 amino acids and four introns 42, 42, 39, and 48 bp long each. The intron/exon boundaries of the Mi-mnsod gene had a conserved 5'-GT – AG-3' intron splice–site junctions (Mount, 1982) (Fig. 1).
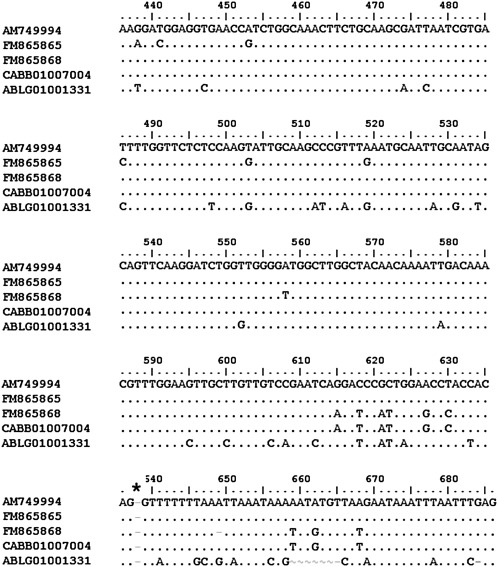
Analyzing partial sequences obtained from cDNA (accession number FM865859 and 61) and genomic DNA (accession number FM865865 and 68) of M. incognita MILEV-L4, nucleotide polymorphisms were observed. AM749994 sequence showed a 97% similarity with FM865865, CABB01007004 and FM865868 in the exon 4 and intron 4 regions (Fig. 2). The AM749994 3' UTR region showed 93% similarity with FM865859 and CABB01007004 and 79% with FM865861 (Fig. 3).
Fig. 2.
Comparison of Mi-mnsod sequences of exon and intron number four from M. incognita strain MiLEV-L4 (AM749997, FM865865, FM865868), strain Morelos (CABB01007004), and M. hapla (ABLG01001331). Identity between sequences is indicated by a dot, asterisk indicates the start of the intron site.
Fig. 3.
Comparison of Mi-mnsod 3'UTR sequences from M. incognita strain Morelos (CABB01007004), strain MiLEV-L4 (AM749997, FM865859, FM865860), and M. hapla (ABLG01001331). The stop codon is indicated in a box.
Amino acid sequence analysis: The deduced amino acid sequence of AM749994 (CAO78623) displayed the consensus region DXWEH[STA][FY]. In addition, it presented the four residues known to be responsible for the Mn binding (His-48, His-96, Asp-180 and His-184) and the Parker and Blake signature MnSOD (Gly-91, Gly-92, Phe-99, Gln-164 and Asp-165) (Parker and Blake, 1988) (Fig. 1). The MitoProt analysis identified a putative mitochondrial localization signal encoded by amino acids 1-22. Removal of this 2.7 kDa mitochondrial transit peptide resulted in a 22.3 kDa mature enzyme. The transit peptide amino acid sequence was rich in positively charged groups (Arg-14, Arg-21, Lys-4, Lys-8 and Lys-20) and lacked acidic amino acids. The putative transit peptide of Mi-MnSOD also possessed an arginine residue in the penultimate position, present in all eukaryotic MnSODs. The N-terminal LPD region, which is conserved in all eukaryotic MnSODs, was also present in M. incognita (Fig. 4). When considering the nucleotide polymorphisms observed in coding regions, only one present at nucleotide 503 introduced an amino acid substitution at position 27, giving rise to glycine-to-valine substitution (Fig. 1 and 2).
Fig. 4.
Alignment of the translated AM749994 Mi-MnSOD mitochondrial transit peptide and the first seven amino acids of mature Mi-MnSOD protein with characterized MnSOD from other species. The predicted cleavage site is indicated by an arrow. Positively charged amino acid residues are shown in bold. the asterisks indicate conserved lysine and arginine residues at the beginning of the mature proteins and penultimate to the maturation cleavage site, respectively.
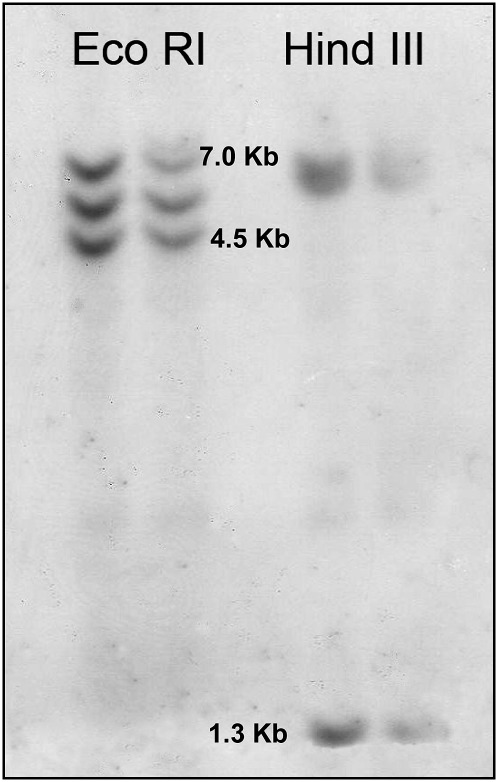
The DNA blot analyses of the digested DNA with Eco RI or Hind III, hybridized with the Dig-labeled Mi-mnsod probe, showed three hybridization signals for both restriction enzymes (Fig. 5). Cleavage sites for Eco RI or Hind III are not present in the sequence of the Mi-mnsod probe.
Fig. 5.
DNA blot analysis of MnSOD genes. Meloidogyne incognita genomic DNA was digested with Eco RI or Hind III and hybridized with the Mi-mnsod probe in two replicates.
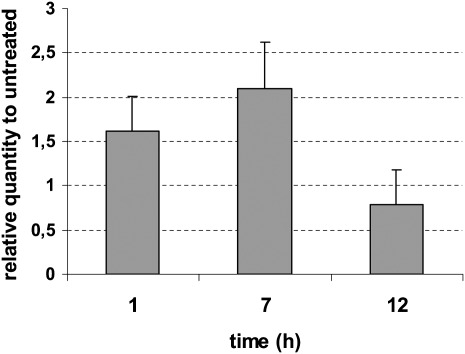
Expression analysis: Higher levels of antioxidant enzyme activity in response to paraquat have been demonstrated in several organisms, including Drosophila (Orr and Sohal, 1994) and C. elegans (Vanfleteren, 1993). Therefore, paraquat was used for SOD gene expression analyses in M. incognita. Exposures of M. incognita J2 to 50 mM paraquat was carried out successfully and no mortality was observed at any of the exposure times. Mi-mnsod expression was significantly higher (P < 0.05), than in untreated controls, at 1 (1.61-fold) and 7 hours (2.09-fold) exposures. After 12 hours, the Mi-mnsod mRNA quantity decreased (P < 0.05) 1.3-fold when compared to the 7 hours after exposure (Fig. 6).
Fig. 6.
Effect of paraquat on transcript levels of Mi-mnsod at different exposure times. Transcript levels were monitored with RT-PCR in three replicates and normalized against β-actin expression. Bars represent standard errors.
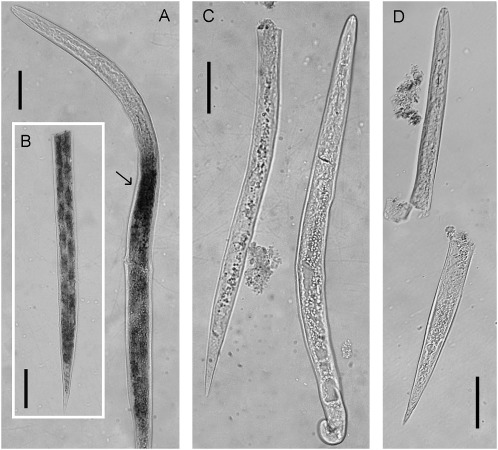
In situ hybridization: The digoxigenin labeled antisense probe for M. incognita Mi-mnsod showed a signal present throughout the posterior portion of the nematode body (Fig. 7A and B), with higher intensity at the anterior portion of the intestine (Fig. 7A). No signal was observed in controls hybridized with the sense strand probe (Fig. 7C) and a background level signal was observed in the anterior portion of the nematode (Fig. 7D).
Fig. 7.
In situ hybridization of digoxigenin labeled cDNA probe of Meloidogyne incognita Mi-mnsod. Juveniles were treated with 50mM paraquat for 7 hours. MnSOD expression is detected throughout the posterior body region A and inset B. Arrow A, anterior region of intestine. C, sense strand hybridization control. D, untreated control. Scale bars = 20 μm.
Discussion
This study allowed the identification and characterization of Mi-mnsod, a gene encoding a MnSOD enzyme from M. incognita. The complete nucleotide sequences of MnSOD genes have been previously characterized in C. elegans (Hunter et al., 1997) and Onchocerca volvulus (Henkle-Dührsen et al., 1995). Whereas the C. elegans genome encodes a 1.3 kb and 1.5 kb genes for Sod-2 and Sod-3 respectively, and the Ov MnSOD from O. volvulus is 1.9 kb in length, the Mi-mnsod is only 1 kb in length. However, there is a striking similarity in the overall gene organization and structure, with all genes being organized with five exons and four introns. In addition, the deduced M. incognita Mi-MnSOD mature enzyme molecular mass of 22.3 kDa is similar to that of other eukaryotic species, including O. volvulus 22.3 kDa, C. elegans 21.9 - 22.1 kDa, and Bombyx mori 24 kDa.
In the eukaryotic cells, MnSOD is encoded in the nucleus, synthesized in the cytosol, and imported post–translationally into the mitochondria matrix where 90% of the oxygen in the cells is consumed (Halliwell and Gutteridge, 1989). The amino acid sequence of Mi-MnSOD indicates the presence of a leader sequence at the amino-end of the deduced precursor protein, suggesting that the Mi-MnSOD might also be synthesized as a precursor in the cytosol and then transferred into the mitochondria (Fig. 4).
The conservation of the four residues responsible for the Mn binding present in all eukaryotic MnSODs, (amino acids His-48, His-96, Asp-180 and His-184 in the Mi-MnSOD) suggests the important role played by these residues in determining the catalytic activity of the enzyme and, consequently, redox balance in the cell. Regulation of the cell's redox state is critically linked to the levels of mitochondrial MnSOD, whose activity is regulated both at the transcriptional and post-translational levels (Davis et al., 2001). The genomic DNA sequence of Mi-mnsod revealed some features that also suggest a possible post-transcriptional control mechanism associated with mRNA stability, namely presence of ARE elements and polypyrimidine tracts.
The Mi-mnsod (-AUUUA-) ARE elements appear to belong to class I, one of the most ubiquitous and destabilizing elements found within rapidly degraded mRNA (Zhang et al., 2002). These elements are characterized by the presence of one to three pentamers that are distributed within the 3' UTR, coupled with a nearby U-rich region (Xu et al., 1997). At least three classes of AREs have been identified based on their composition, number of pentamer repeats, and subsequent effects of RNA deadenylation and decay (Xu et al., 1997). Even if M. incognita AREs were observed mainly in the coding region (only one is present at 3′ UTR), it is interesting to note the high frequency of these elements within a confined genomic region, which are not present (or occur in a low frequency) in nematode mRNA population (Vassella et al., 1994; De Giorgi et al., 1997). The polypyrimidine tracts found in the 3' UTR of Mi-mnsod have been described in the 5' and 3' UTR of procyclin-associated genes (PAGs) of Trypanosoma brucei (Vassella et al., 1994) and within a gene encoding a cuticlin protein of Meloidogyne artiellia (De Giorgi et al., 1997). This region binds proteins involved in the processing of primary transcripts to mature mRNAs (Singh et al, 1995). Further studies will be needed to test the role of these elements in any post-transcriptional regulatory processes.
The presence of three fragments in the DNA blot analysis, together with the occurrence of three different amplified products characterized by replicated nucleotide polymorphisms, suggest the occurrence of three putative copies of the MnSOD gene in M. incognita. This is similar to what has been found for C. elegans (Hunter et al., 1997) but not for O. volvulus, in which a single-copy MnSOD gene was observed (Henkle-Dührsen et al., 1995).
The Mi-mnsod transcript levels are significantly higher (1.61-fold) after 1 hour of paraquat treatment than in untreated control, suggesting a rapid response to oxidative stress. The decrease in the gene expression after 12 hours could be related to an irreversible toxic condition caused by the continuous exposure to an oxidative environment. The increase in the amount, and spatial location of Mnsod mRNA as shown by the in situ hybridization, could suggest a reaction to the oxidative stress generated by paraquat, according to the role of this enzyme in the ROS detoxification process. The presence of Mnsod mRNA in the intestine also suggests a putative detoxification function, active in this body region, where plant products are present due to feeding.
The ability of J2 to quickly react to the oxidative stress could play an important role in establishing and maintaining the nematode inside the plant tissue. Biochemical observations showed that tomato plants react to root-knot nematodes by mounting defense responses, implicating ROS production (Melillo et al., 2006). The superoxide induction is an early event during nematode infection which determines different patterns of ROS production in compatible and incompatible nematode-tomato interactions. A high concentration of ROS was detected as a result of the incompatible interaction, which can be related to a rapid induction of HR (Melillo et al., 2006). Present data showed that Mi-mnsod may play a key role in nematode survival under oxidative stress conditions. MnSOD activity, which scavenges superoxide anions and catalyzes their dismutation into hydrogen peroxide and oxygen, may represent the first defense of the J2 against ROS overproduction, occurring as early as 1 hour after exposure to oxidative stress. This early defense system may be important in determining the nematode's capability to organize a complex response to counteract the plant defense mechanisms in incompatible reactions.
Footnotes
The author thanks S. Molinari and A. Ciancio for helpful comments on the manuscript. D. Catalano is gratefully acknowledged for assistance with bioinformatic analysis. This work was supported in part by Regione Puglia PE040.
This paper was edited by Isgouhi Kaloshian.
Literature Cited
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EGJ, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok V, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier T, Markov GV, McVeigh P, Pesole P, Poulain J, Robinson-Rechavi M, Sallet E, Ségurens B, Steinbach D, Tytgat T, Ugarte E, Van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CJ, Orlandi EW. Active oxygen species in plant pathogenesis. Annual Review of Phytopathology. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- Callahan LH, Crouch RK, James ER. Helminth anti-oxidant enzymes: a protective mechanism against host oxidants? Parasitology Today. 1988;4:218–25. doi: 10.1016/0169-4758(88)90162-7. [DOI] [PubMed] [Google Scholar]
- Claros M, Vincens GP. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry. 1996;241:770–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Davis CA, Monnier JM, Nick HSA. Coding region determinant of instability regulates levels of manganese Superoxide Dismutase mRNA. Journal Biological Chemistry. 2001;276:37317–37326. doi: 10.1074/jbc.M104378200. [DOI] [PubMed] [Google Scholar]
- De Boer JM, Yan Y, Smant G, Davis EL, Baum TJ. In situ hybridization to messenger RNA in Heterodera glycines. Journal of Nematology. 1998;30:309–12. [PMC free article] [PubMed] [Google Scholar]
- De Giorgi C, De Luca F, Di Vito M, Lamberti F. Modulation of expression at the level of splicing of cut-1 RNA in the infective second-stage juvenile of the plant parasitic nematode Meloidogyne artiellia. Molecular and General Genetics. 1997;253:589–598. doi: 10.1007/s004380050361. [DOI] [PubMed] [Google Scholar]
- Droopkin VH, Helgeson JP, Upper CD. The hypersensitivity reaction of tomatoes resistant to Meloidogyne incognita: reversal by cytokinins. Journal of Nematology. 1969;1:55–60. [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. Biological effects of the superoxide radical. Advances in Enzymology and Related Areas of Molecular Biology. 1986;58:62–97. doi: 10.1002/9780470123041.ch2. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. 2nd ed. Oxford, UK: Oxford University Press; 1989. Free radicals in biology and medicine. [Google Scholar]
- Hartman P, Chidress E, Beyer T. Nematode development is inhibited by methyl viologen and high oxygen concentrations at a rate inversely proportional to life span. Journal of Gerontology. 1995;50:B322–B326. doi: 10.1093/gerona/50a.6.b322. [DOI] [PubMed] [Google Scholar]
- Henkle-Dührsen K, Tawe W, Warnecke C, Walter RD. Characterization of the manganese superoxie dismutase cDNA and gene from the human parasite Onchocerca volvulus. Biochemical Journal. 1995;308:441–446. doi: 10.1042/bj3080441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress reisitance and Mn – superoxide dismutase gene expression in Caenorhabditis elegans. FASEB Journal. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hunter T, Bannister WH, Hunter GJ. Cloning, expression and characterization of two Manganese Superoxide Dismutase from Caenorhabditis elegans. The journal of Biological Chemistry. 1997;272:28652–28659. doi: 10.1074/jbc.272.45.28652. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. The journal of Biological Chemistry. 1991;266:19867–70. [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) The journal of Biological Chemistry. 1969;244:6049–6055. [PubMed] [Google Scholar]
- Mehdy M. Active oxygen species in plant defense against pathogens. Plant Physiology. 1994;105:467–72. doi: 10.1104/pp.105.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo MT, Leonetti P, Bongiovanni M, Castagnone-Sereno P, Bleve Zacheo T. Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. New Phytologist. 2006;170:501–512. doi: 10.1111/j.1469-8137.2006.01724.x. [DOI] [PubMed] [Google Scholar]
- Molinari S, Rosso L, Ornat Longaron C. The role of antioxidant enzymes in the virulence of root-knot nematodes on resistant tomato. Nematropica. 2005;35:88. [Google Scholar]
- Mount SA. Catalogue of splice junction sequences. Nucleic Acids Research. 1982;10:459–72. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WF, Sohal SR. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Blake CF. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Letters. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- Paulson RE, Webster JM. Ultrastructure of the hypersensitivity reaction in roots of tomatoes, Lycopersicum esculentum L., to infection by the root-knot nematode, Meloidogyne incognita. Physiological and Molecular Plant Pathology. 1972;2:227–234. [Google Scholar]
- Roise D, Schatz G. Mitochondrial presequences. The journal of Biological Chemistry. 1988;263:4509–4511. [PubMed] [Google Scholar]
- Sasser JM. Root-knot nematodes: a global menace to crop production. Plant Disease. 1980;64:36–41. [Google Scholar]
- Singh R, Valcarcell J, Green MR. Distinct specificities and function of higher eukaryotic polypirymidine tract – binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;25:4876–82. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochemical Journal. 1993;292:605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Braun R, Roditi I. Control of polyadenilation and alternative splicing of transcripts from adjacent genes in aprocycling espression site: dual role for polypirymidine tracts in Trypanosoma. Nucleic Acids Research. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss U, Grundler FMW, Munch A. The parasitic behaviour of second-stage juveniles of Meloidogyne incognita in roots of Arabidopsis thaliana. Nematologica. 1992;38:98–111. [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Molecular Cell Biology. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kruys V, Huez G, Gueydan C. Au-rich element-mediated translational control: complexity and multiple activities of trans–activating factors. Biochemical Society Transactions. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]