Abstract
Mouse Trmt112, the homologous gene of yeast Trm112 (tRNA methyltransferase 11-2), was initially cloned from RIKEN with uncertain function. The yeast TRM112 is now known to play important roles in RNA methylation. Here, we studied the expression of Trmt112 by in situ hybridization and quantitative real-time RT-PCR (QRT-PCR). A higher expression level of Trmt112 was observed in the brain and nervous system by whole mount in situ hybridization from embryonic day 10.5 (E10.5) to E11.5. At later developmental stages E13.5 and E16.5, abundant expression was prominently found in various organs and tissues including developing brain, nervous system, thymus, lung, liver, intestine, kidney, and cartilage. Furthermore, Trmt112 was persistently expressed from E9.5 to E18.5 on whole embryos and highly expressed in multiple organs at E12.5, E15.5 and E18.5 by QRT-PCR. These results showed that Trmt112 gene was highly and ubiquitously expressed during mouse embryonic development, implying that it might be involved in the morphogenesis of diverse organs and tissues and numerous physiological functions.
Keywords: TRM112, mouse development, in situ hybridization, QRT-PCR
I. Introduction
In transfer RNA (tRNA), base and ribose methylations are the most frequent modifications [9, 20]. The majority of methylated modifications are catalyzed by S-adenosylmethionine-(SAM)-dependent methyltransferases (MTases) [1], whose larger family is Rossman fold MTase (RFM) containing a seven-stranded β-sheet surrounded by helices on each side [25]. Yeast tRNA MTase (Trm) has been broadly identified by now [6]. Yeast Trm112 exhibits no sequence similarity to known RFM superfamilies, implying that it lacks a methyltransferase domain and does not catalyze methylation. TRM112 is identified as a small zinc finger protein, that is required for tRNA methylation in vivo, but this family function is still uncertain [5, 23, 26].
The mouse gene Trmt112 was initially cloned from RIKEN [10]. The full-length of Trmt112 cDNA is 1047 bp, encoding a 125-amino-acid protein that is named yeast tRNA methyltransferase 11-2 (TRM112) homolog. A BLAST search in GenBank reveals that mouse Trmt112 shares a higher similarity with archaea and eukaryotes, implying that TRM112 is an evolutionarily conserved protein. Biochemical analysis indicates that TRM112 interacts with and activates both TRM11 and TRM9, two tRNA methyltransferases necessary for the formation of 2-methylguanosine at position 10 and modification of anticodons at the wobble uridine (U34) position, respectively [3, 18, 23, 26]. In addition, TRM112 was further found to be a cofactor of eukaryotic release factor 1 (eRF1) methyltransferase [5]. Translation termination is a very essential step, which is catalyzed by protein release factors (RFs) [11]. Methylation of RFs ensures efficient translation termination and release of newly synthesized peptides from the ribosome [19, 22]. Methylation of eRF1 in yeast is performed by the heterodimeric methyltransferase (MTase) Mtq2/Trm112 at the glutamine residue of the GGQ motift (ripeptide Gly-Gly-Gln) [4, 22], and Trm112 is necessary for the solubility and activity of the catalytic subunit of the protein methylase Mtq2p [5, 26]. More recently, the same mechanism was also found in human and mouse, the holoenzyme homologues of which are annotated as HEMK2α/hTRM112 and Pred28a (N6amt1)/mTrm112, respectively [2, 13, 14, 17, 24]. Furthermore, TRM112 has been regarded as a complex network of enzymatic activities [15], while deletion of Trm112 gene would lead to severe growth defect in yeast [15]. In parallel, in another TRM112 homolog, arabidopsis SMO2, inactivation of SMO2 leads to a defect in progression of cell division and organ growth [7]. Besides, TRM112 might also interact with LYS9, an alcohol dehydrogenase that possesses a near Rossman fold domain that lacks the seventh β-fold [23], and other proteins with unknown function, implying that TRM112 possibly plays roles in modification or regulation of additional cellular processes [12, 15, 28].
Above all, TRM112 has been involved in multiple biological activities as an important plurifunctional factor. Nevertheless, the expression patterns of mouse Trmt112 gene has not been reported thus far, hence we performed detailed expression analysis of Trmt112 gene during mouse embryonic development, the results from which would offer an excellent foundation for further studies in functional analyses. The results showed that Trmt112 was highly and ubiquitously expressed in zones of the brain, nervous system, and diverse tissues and organs, suggesting that Trmt112 may play significant roles during embryonic development.
II. Materials and Methods
Mouse and tissue preparation
DBA/2J and C57BL/6J mice were time-mated overnight. At noon the next day the presence of a vaginal plug was considered to be embryonic day 0.5 (E0.5). All procedures were carried out following the “Rules for experiments animals” published by the Chinese Government (Beijing, China).
Whole mount in situ hybridization (WISH) and in situ hybridization (ISH)
E10.5, E11.5, E13.5 and E16.5 mouse embryos were isolated at different developmental stages and fixed in 4% paraformaldehyde in PBS at 4°C overnight. E10.5 and E11.5 were prepared for WISH. E13.5 and E16.5 embryos were paraffin embedded and sectioned at 9 µm thickness. To obtain the RNA probes, cDNA fragment of Trmt112 (NM_026306, nt. 92-921, sense: 5'-ttcaatcgcagaaagagg-3', antisense: 5'-gtgtttctgttgtacggtct-3') were amplified by RT-PCR and inserted in pBluescript II KS(+) (Stratagene, La Jolla, CA, USA). Reaction for RT-PCR were performed for 32 cycles at 95°C for 2 min, 55°C for 50 s and 72°C for 45 s. RNA probe for the ISH were prepared using a DIG RNA labeling kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. WISH and ISH was performed by standard procedures as described previously [8, 16, 21, 27]. Briefly, we performed the standard procedures of the ISH experiments at least in triplicate at one time point. The experiments were pretreated by proteinase K with proper concentration and enough time to make holes on embryos prior to hybridization, and then, each parallel control was added with the sense RNA probe in contrast to the antisense probe and was hybridized at 65°C overnight. After adequate washing at 65°C by 2×SSC, 1% SDS and at room temperature by 0.2×SSC, the samples were blocked for 1 hr at room temperature and incubated with anti-digoxigenin-alkaline phosphatase conjugated antibody (Roche, 1:5000) at 4°C overnight. The slides were washed with several changes, developed with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP).
RNA analyses
Total RNA was isolated from E9.5 to E18.5 whole embryos and embryonic tissues (brain, tongue, heart, lung, liver and kidney) at E12.5, E15.5 and E18.5 using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and then were treated with DNaseI (Roche). cDNAs were synthesized using a SuperScriptTMIII RNase H– Reverse Transcriptase kit (Invitrogen, Eugene, OR, USA). Quantitative analysis of Trmt112 with specific primers (NM_026306, nt. 271-492, sense: 5'-cgtaagcctttgcagtctccc-3', antisense: 5'-ttcatgttgtca cagcggagc-3') was performed using the ABI Prism 7500 Real-Time PCR System, and reactions were done using the SYBR Green PCR master mix kit (Applied Biosystems, Foster City, CA, USA) following manufacturer’s instructions, and relative quantification was achieved following a standard curve method. Reactions for Trmt112 were performed for 36 cycles at 95°C for 15 s, 60°C for 1 min, and then followed by a dissociation stage analysis. β-actin (NM_007393, nt. 520–717, sense: 5'-taccacaggcattgtgtag gact-3', antisense: 5'-ttgatgtcacgcacgatttccct-3') was used as a reference gene. All reactions, including the standards and non-template control (H2O), were run in quartet from different embryos and organs. One-way analysis of variance (ANOVA) was used to analyze the significance of difference, followed by the Student-Newman-Keuls post-test. Students’ t-test and a P value less than 0.05 was considered statistically significant.
III. Results
Gene structure and homology analysis of the mouse Trmt112
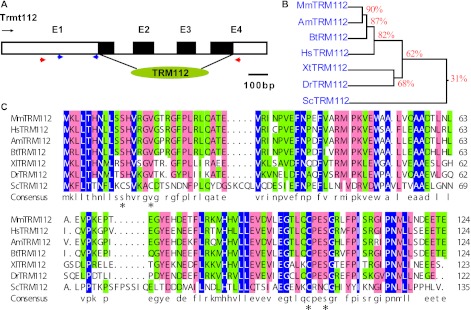
Trmt112 (gene ID: 67674) full-length cDNA has been initially identified from RIKEN [10]. Comparison with the genomic sequences showed that Trmt112 is assembled from 4 exons spanning 1.53 kb on mouse chromosome 19E1 (Fig. 1A), the open reading frame of which starts within exon 2, ends within exon 4, and encodes a 125 amino acid protein considerably homologous with the TRM112 function domain (Fig. 1A). Amino acid sequence alignment analysis showed high similarity of mouse Trmt112 protein with other TRM112 homologues including human (Homo sapiens), panda (Ailuropoda melanoleuca), cows (Bos taurus), zebra fish (Danio rerio), clawed frog (Xenopus tropicalis) and yeast (Saccharomyces cerevisiae) (Fig. 1B), implying that TRM112 is a functionally conserved protein in the phylogenetic evolution. However, the putative zinc-binding domain of TRM112s in yeast was not present in other multicellular organisms (Fig. 1B) [5].
Fig. 1.
Genomic organization and homology alignment of Trmt112. (A) Arrow denotes transcriptional orientations. Exons are represented as numbered boxes, introns as connecting lines, black boxes indicate coding protein, and conserved and function domain TRM112 (green oblate ellipsoid) is indicated below. Red and blue triangles were labeled the primers for in situ hybridization and QRT-PCR, respectively. Scale of genomic bar represents 100 bp. (B) Alignment of mouse TRM112 with other eukaryotes homologues. Shading colors represent highlight levels of amino acid conservation, and asterisks refer to the Cys residues for zinc binding in yeast TRM112. Mm, Mus musculus; Am, Ailuropoda melanoleuca; Hs, Homo sapiens; Bt, Bos taurus; Dr, Danio rerio; Xt, Xenopus tropicalis; Sc, Saccharomyces cerevisiae.
Expression patterns of Trmt112 in mouse embryos
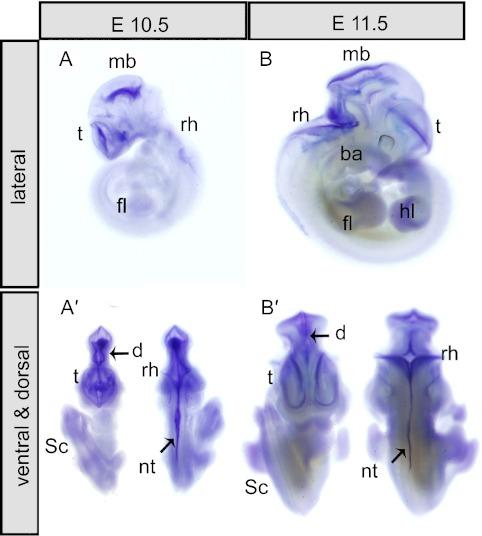
The expression profile of Trmt112 was analyzed in different tissues and at different developmental stages. Trmt112 was firstly detected by whole mount in situ hybridization (WISH) at E10.5 and E11.5. At E10.5, from the lateral view, strong expressional signals of Trmt112 were detected in the cortical anlage of the telencephalon vesicle (t), alar plate of midbrain (mb), and nervous plate of rhombencephalon (rh) (Fig. 2A). Dorsal view of the same embryos showed that the major signal was along the dorsal midline from the diencephalon (d) through the mid-hindbrain region to spinal cord (Sc) and neural tube (Nt) (Fig. 2A’). At E11.5, a similar expression pattern was maintained as E10.5, especially in brain and neural tube (Fig. 2B–B’). In addition, Trmt112 expression was detected both in the branchial arches (ba), forelimb (fl), and hindlimb (hl) (Fig. 2A–A’ & B–B’) at E10.5 and E11.5. As a negative control, there were no signals detected using the Trmt112 sense probes (data not shown).
Fig. 2.
Expression pattern of mouse Trmt112 at E10.5 and E11.5. (A–B) Lateral view of embryos, Trmt112 was expressed in the telencephalon vesicle (t), midbrain (mb), rhombencephalon (rh), diencephalon (d) and branchial arches (ba). (A’–B’) Dorsal or ventral view of E10.5 and E11.5 showed Trmt112 expression was detected through the mid-hindbrain region to rhombencephalon (rh) and spinal cord (sc).
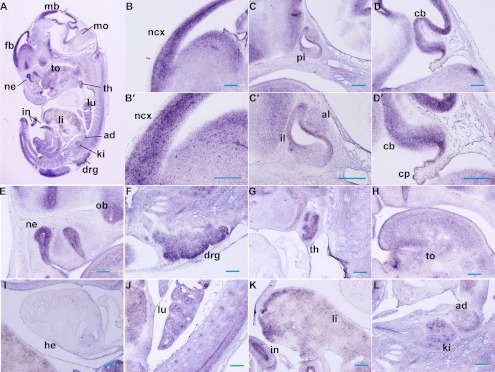
At E13.5, the spatial expression of Trmt112 was detected by in situ hybridization on histological sections. The expression was further expanded in the brain and spinal cord (Fig. 3A), and found in multiple developing organs. In the developing central nervous system (CNS), strong signals were detected in the lateral ventricular zone of the neocortex (ncx), anterior lobe (al), and intermediate lobes (il) of pituitary gland, cerebellum (cb), nasal epithelium (ne), olfactory bulb (ob), and dorsal root ganglia (drg) (Fig. 3B–F & B’–D’). In developing organs, prominent hybridization signals were observed within the thymus (th), tongue (to), epithelium of the lung (lu) and intestine (in), liver (li), kidney (ki), adrenal gland (ad) (Fig. 3G–L), but the expression in the heart (he) was low (Fig. 3I).
Fig. 3.
Expression pattern of Trmt112 at E13.5 by section in situ hybridization. (A) Sagittal sections of complete mouse embryos at E13.5 showed abundant expression of Trmt112. (B–D & E–F) Trmt112 expression signals were detected in CNS. (B’–D’) Magnifications of observations of (B–D). (G–L) Trmt112 prominent expression signals were detected in the main organs including thymus gland, lung, liver, intestine, kidney, and adrenal gland at E13.5. fb, forebrain; mo, medulla oblongata; ncx, neocortex; al, anterior lobe; il, intermediate lobes; pi, pituitary gland; cb, cerebellum; cp, choroid plexus; ne, nasal epithelium; ob, olfactory bulb; drg, dorsal root ganglia; th, thymus; to, tongue; h, heart; lu, lung; li, liver; in, intestine; ki, kidney. Bars=100 µm.
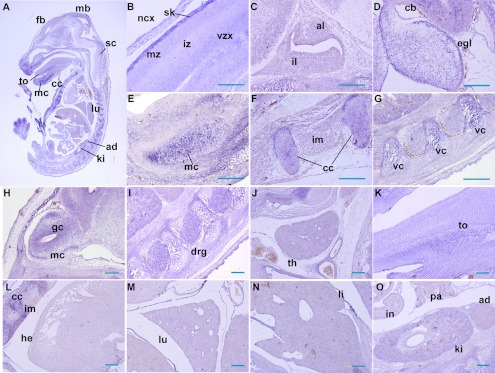
By E16.5, Trmt112 was ubiquitously expressed in tissues. In the developing CNS, strong signals were detected in the regions of the neocortex (ncx), cerebellum (cb), olfactory bulb (ob), spinal cord (sc), and dorsal root ganglia (drg) (Fig. 4A–D & H–I). For instance, in the neocortex, the signals were detected in the marginal zone (mz) and ventricular zone (vz), with low expression in the intermedial zone (iz) (Fig. 4B). In cerebellum, the external granular layer (egl) exhibited clear signals (Fig. 4D). In the olfactory bulb, Trmt112 mainly distributed in the granule cell layer (gc), with weaker signals in the mitral cell layer (mc) (Fig. 4H). Alternatively, clear signals were also visible in proliferating chondrocytes and perichondrium, including skull (Fig. 4B), Meckel’s cartilage (mc) (Fig. 4E), costal cartilage (cc) (Fig. 4F), and cartilages of vertebral column (vc) (Fig. 4G), which signals were not that evident at E13.5. Furthermore, Trmt112 was moderately expressed in multiple organs like the thymus (th), tongue, heart, lung, liver, and kidney (Fig. 4G–O). As a control, sense probes were performed on adjacent tissue sections and showed no detectable labeling (data not show).
Fig. 4.
Expression pattern of Trmt112 at E16.5 by section in situ hybridization. (A) Sagittal sections of complete mouse embryos at E15.5 showed ubiquitous expression of Trmt112. (B–D & H–I) In developing CNS, strong signals were detected in the regions of the neocortex (ncx), cerebellum (cb), olfactory bulb (ob), and dorsal root ganglia (drg). (E–G) Trmt112 staining was significantly present in both proliferating chondrocytes and perichondrium. (G–O) Trmt112 was persistently and moderately expressed in multiple important organs at E16.5 like E13.5. fb, forebrain; sc, spinal cord; ncx, neocortex; sk, skull; mz, marginal zone; vz, ventricular zone; iz, intermedial zone; al, anterior lobe; il, intermediate lobes; cb, cerebellum; mc, Meckel’s cartilage; cc, costal cartilage; vc, cartilages of vertebral column; mc, mitral cell layer; gc, granule cell layer; drg, dorsal root ganglia; th, thymus; to, tongue; h, heart; lu, lung; li, liver; in, intestine; ki, kidney. Bars=100 µm.
Expression analysis of Trmt112 during embryonic development
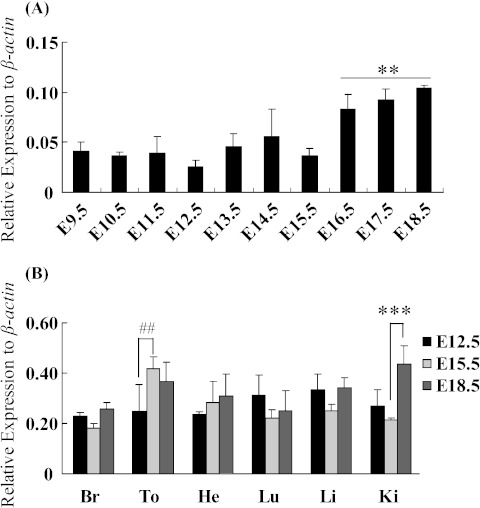
In order to further investigate the Trmt112 expressional pattern, we utilized QRT-PCR to confirm its spatiotemporal expression during mouse development. Results showed that Trmt112 expression was up-regulated from E9.5 to E18.5, especially up-regulated at E16.5, E17.5 and E18.5, but slightly down-regulated at E12.5 and E15.5 on whole embryos (Fig. 5A). Additionally, Trmt112 was highly expressed in multiple tissues at different developmental stages. Trmt112 expression was significantly up-regulated in tongue at E15.5 relative to E12.5, and in kidney at E15.5 relative to E18.5, but there no significant differences between tissues expression, expect for a litter lower expression of heart during embryonic development. (Fig. 5B).
Fig. 5.
Trmt112 expression during embryonic development by QRT-PCR analysis. (A) Expression of Trmt112 in whole embryos during mouse development from E9.5 to E18.5. **P<0.05 represents E16.5-E18.5 vs E9.5-E15.5 by one-way analysis of variance (ANOVA) followed by Tukey test. (B) Relative expression of Trmt112 in diverse tissues at E12.5, E15.5 and E18.5. Values (means±SEM) represent the expression level relative to that of β-actin expression level from four biological replicates. (n=4). Br, brain; To, tongue; He, heart; Lu, lung; Li, liver; Ki, kidney. ##P<0.05 represents tongue expressed at E12.5 vs E15.5; ***P<0.01 represents kidney expressed at E15.5 vs E18.5 by two-way ANOVA followed by Tukey test.
IV. Discussion
In this study, Trmt112 was prominently expressed in developing brain and neural tubes at E10.5 and E11.5, which were the proliferating zones of the nervous system, implying the possible role in the neural tube and nervous system. Section ISH results showed that Trmt112 was ubiquitously expressed at mid-later-gestation of organogenesis stages. At E13.5, Trmt112 was abundantly expressed in a variety of neural epithelium tissues including neocortex, cochlea, olfactory epithelium, Rathke’s pouch, medulla oblongata, and dorsal root ganglia (drg). Rathke’s pouch, one of the major neuroendocrine organs, which derives from the oral ectoderm and forms the anterior lobe (al) and intermediate lobes (il) of developed pituitary gland, is important for embryonic growth. At the stage of E16.5, organogenesis is largely complete, in accord with the signals detected in those tissues of brain and nervous system at E13.5, but abundant signals, that were also found in chondrocytes, where were not evident at E13.5, indicating that Trmt112 possessed spatiotemporal specific expression patterns in development progress. QRT-PCR results showed that Trmt112 was expressed in abundance throughout the developmental process from E9.5 to E18.5, and higher in multiple key organs. Thus, the Trmt112 might be considered as an essential gene which persistently maintains roles in diverse organs and tissues during the embryonic stages. Alternatively, we found that Trmt112 expression was much higher in parthenogenetic embryos than normal blastocysts of the same stage (E4.5) (data not shown). Thus we initially speculated whether Trmt112 was associated with imprinting, but the result showed it was a biallelic gene (data not shown), possibly implying Trmt112 expression in parthenogenetic embryos was functional at early development stage. TRM112 is an evolutionarily conserved protein, and the putative zinc-binding domain of TRM112 was present in yeast, bacteria, and some archaea but not in multicellular organisms [5]. However, disruption of the respective Trmt112 gene in yeast and arabidopsis resulted in the slow growth of both organisms [7, 23]. Additionally, we also found mouse Trmt112 had much higher homology with mammals than yeast, implying that Trmt112 protein is functionally conserved in multicellular organisms.
In conclusion, we reported the characteristic expression patterns of mouse Trmt112 during embryonic development. Our results suggested that Trmt112 could play broad roles in the developing brain, nervous system, and morphogenesis of diverse tissues. The present study will serve as a basis for future in vivo genetic, functional, and mutational analyses.
V. Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30971645 and 31100934), the Fundamental Research Funds for the Central Universities (Grant No.HIT.NSRIF.2010027).
VI. Conflict of Interest
All authors of this paper declare no conflict of interest.
VII. References
- 1.Bujnicki J. M. Comparison of protein structures reveals monophyletic origin of the adomet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of n4-cytosine and n6-adenine DNA methylation. In Silico Biol. 1999;1:175–182. [PubMed] [Google Scholar]
- 2.Figaro S., Scrima N., Buckingham R. H., Heurgue-Hamard V. Hemk2 protein, encoded on human chromosome 21, methylates translation termination factor erf1. FEBS Lett. 2008;582:2352–2356. doi: 10.1016/j.febslet.2008.05.045. [DOI] [PubMed] [Google Scholar]
- 3.Fu D., Brophy J. A., Chan C. T., Atmore K. A., Begley U., Paules R. S., Dedon P. C., Begley T. J., Samson L. D. Human alkb homolog abh8 is a trna methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heurgue-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Kisselev L. L., Buckingham R. H. The glutamine residue of the conserved ggq motif in saccharomyces cerevisiae release factor erf1 is methylated by the product of the ydr140w gene. J. Biol. Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- 5.Heurgue-Hamard V., Graille M., Scrima N., Ulryck N., Champ S., van Tilbeurgh H., Buckingham R. H. The zinc finger protein ynr046w is plurifunctional and a component of the erf1 methyltransferase in yeast. J. Biol. Chem. 2006;281:36140–36148. doi: 10.1074/jbc.M608571200. [DOI] [PubMed] [Google Scholar]
- 6.Hou Y. M., Perona J. J. Stereochemical mechanisms of trna methyltransferases. FEBS Lett. 2010;584:278–286. doi: 10.1016/j.febslet.2009.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z., Qin Z., Wang M., Xu C., Feng G., Liu J., Meng Z., Hu Y. The arabidopsis smo2, a homologue of yeast trm112, modulates progression of cell division during organ growth. Plant J. 2010;61:600–610. doi: 10.1111/j.1365-313X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 8.Iijima N., Yokoyama T. Apoptosis in the medaka embryo in the early developmental stage. Acta Histochem. Cytochem. 2007;40:1–7. doi: 10.1267/ahc.06013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juhling F., Morl M., Hartmann R. K., Sprinzl M., Stadler P. F., Putz J. Trnadb 2009: Compilation of trna sequences and trna genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai J., Shinagawa A., Shibata K., Yoshino M., Itoh M., Ishii Y., Arakawa T., Hara A., Fukunishi Y., Konno H., et al. Functional annotation of a full-length mouse cdna collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 11.Kisselev L. L., Buckingham R. H. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 12.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., et al. Global landscape of protein complexes in the yeast saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 13.Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R. H., van Tilbeurgh H., Heurgue-Hamard V., Graille M. Mechanism of activation of methyltransferases involved in translation by the trm112 ‘hub’ protein. Nucleic Acids Res. 2011;39:6249–6259. doi: 10.1093/nar/gkr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P., Nie S., Li B., Yang Z. Q., Xu Z. M., Fei J., Lin C., Zeng R., Xu G. L. Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol. Cell. Biol. 2010;30:4245–4253. doi: 10.1128/MCB.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazauric M. H., Dirick L., Purushothaman S. K., Bjork G. R., Lapeyre B. Trm112p is a 15-kda zinc finger protein essential for the activity of two trna and one protein methyltransferases in yeast. J. Biol. Chem. 2010;285:18505–18515. doi: 10.1074/jbc.M110.113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorman A. F., Houweling A. C., de Boer P. A., Christoffels V. M. Sensitive nonradioactive detection of mrna in tissue sections: Novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- 17.Nakahigashi K., Kubo N., Narita S., Shimaoka T., Goto S., Oshima T., Mori H., Maeda M., Wada C., Inokuchi H. Hemk, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemk knockout induces defects in translational termination. Proc. Natl. Acad. Sci. U S A. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada K., Muneyoshi Y., Endo Y., Hori H. Production of yeast (m2g10) methyltransferase (trm11 and trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. (Oxf). 2009:303–304. doi: 10.1093/nass/nrp152. [DOI] [PubMed] [Google Scholar]
- 19.Petry S., Brodersen D. E., Murphy F. V., 4th., Dunham C. M., Selmer M., Tarry M. J., Kelley A. C., Ramakrishnan V. Crystal structures of the ribosome in complex with release factors rf1 and rf2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 20.Phizicky E. M., Hopper A. K. Trna biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piette D., Hendrickx M., Willems E., Kemp C. R., Leyns L. An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 2008;3:1194–1201. doi: 10.1038/nprot.2008.103. [DOI] [PubMed] [Google Scholar]
- 22.Polevoda B., Span L., Sherman F. The yeast translation release factors mrf1p and sup45p (erf1) are methylated, respectively, by the methyltransferases mtq1p and mtq2p. J. Biol. Chem. 2006;281:2562–2571. doi: 10.1074/jbc.M507651200. [DOI] [PubMed] [Google Scholar]
- 23.Purushothaman S. K., Bujnicki J. M., Grosjean H., Lapeyre B. Trm11p and trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast trna. Mol. Cell. Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratel D., Ravanat J. L., Charles M. P., Platet N., Breuillaud L., Lunardi J., Berger F., Wion D. Undetectable levels of n6-methyl adenine in mouse DNA: Cloning and analysis of pred28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006;580:3179–3184. doi: 10.1016/j.febslet.2006.04.074. [DOI] [PubMed] [Google Scholar]
- 25.Schubert H. L., Blumenthal R. M., Cheng X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Studte P., Zink S., Jablonowski D., Bar C., von der Haar T., Tuite M. F., Schaffrath R. Trna and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson D. G., Nieto M. A. Detection of messenger rna by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 28.Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]