Abstract
Hydrogels are networks of hydrophilic polymer chains that are swollen with water, and they are useful for a wide range of applications because they provide stable niches for immobilizing proteins and cells. We report here the marriage of hydrogels with digital microfluidic devices. Until recently, digital microfluidics, a fluid handling technique in which discrete droplets are manipulated electromechanically on the surface of an array of electrodes, has been used only for homogeneous systems involving liquid reagents. Here, we demonstrate for the first time that the cylindrical hydrogel discs can be incorporated into digital microfluidic systems and that these discs can be systematically addressed by droplets of reagents. Droplet movement is observed to be unimpeded by interaction with the gel discs, and gel discs remain stationary when droplets pass through them. Analyte transport into gel discs is observed to be identical to diffusion in cases in which droplets are incubated with gels passively, but transport is enhanced when droplets are continually actuated through the gels. The system is useful for generating integrated enzymatic microreactors and for three-dimensional cell culture. This paper demonstrates a new combination of techniques for lab-on-a-chip systems which we propose will be useful for a wide range of applications.
INTRODUCTION
Hydrogels are hydrophilic polymers with an open network structure that are swollen with water.1, 2, 3, 4 Hydrogels are uniquely useful for a wide range of applications because they can serve as stable niches for the immobilization of proteins and cells.5 For the former case, protein-bearing hydrogels are often formed to serve as microreactors for use in enzymatic digestion,6 catalysis,7 and biosensors.8 For the latter case, hydrogels are often used as scaffolds for tissue engineering in two-dimensional9, 10, 11, 12 (2D) and three-dimensional5, 13, 14 (3D) formats. In addition to being useful for encapsulating proteins and cells, hydrogels are useful for sensing operations involving temperature,15pH,16 and quantitation of analytes17 and can be used in a wide range of applications including drug delivery,18, 19 cosmetics,20, 21 and food.22, 23, 24
There has been great enthusiasm for combining hydrogels with microchannel-based lab-on–a-chip systems.25, 26, 27, 28, 29, 30, 31, 32, 33, 34 For example, microfluidic systems are useful for reagent delivery and collection of products from hydrogel-based enzyme microreactors.25 Likewise, microfluidic systems for culturing cells encapsulated in hydrogel structures are useful because fluidic ports are easily incorporated for nutrient delivery and waste removal.28 These advances are promising and useful; however, the plumbing complexities incurred with controlling many different reagents simultaneously make the combination of microchannels and hydrogels an imperfect match for all applications. Moreover, in continuous flow microchannel systems, analytes generated by enzymes or cells immobilized in hydrogels can diffuse away from the gels, complicating the analysis.
Digital microfluidics (DMF) is an alternative to microchannels for controlling fluids in miniaturized devices. In DMF, discrete droplets are manipulated by applying a series of electrical potentials to an open array of electrodes (with no channels or walls) covered by a hydrophobic insulator.35, 36, 37 In such systems, droplets are controlled by electromechanical forces.38 DMF enables facile control over many different reagents for multi-step processes, a property that has been useful for enzyme assays27, 39, 40, 41 and for applications involving cells.42, 43, 44, 45, 46, 47, 48, 49, 50, 51 In these initial applications, enzymatic bioreactors and cell culture and assays were implemented in homogeneous aqueous droplets manipulated by DMF. We propose that the marriage of DMF with hydrogels represents a welcome combination for forming miniaturized enzyme bioreactors and 3D cell culture scaffolds.
We report here the first integration of hydrogel structures onto DMF devices for applications involving encapsulated proteins and cells. A review paper published in 2007 (Ref. 52) included a short description of a procedure to mix two reagents to form a hydrogel in a DMF device. But beyond this paper, we are not aware of any other reports of combining hydrogels with DMF. Thus, this paper is the first to report digital microfluidic applications of hydrogels including reagent exchange, enzyme microreactors, and 3D cell culture. We propose that the combination of digital microfluidics and hydrogels will be useful for a wide range of applications.
METHODS AND MATERIALS
Reagents and materials
Agarose (two forms: low gelling temperature and ultra-low gelling temperature) and all other reagents (unless noted otherwise) were from Sigma Aldrich Canada (Oakville, ON) and were used as received. Cell culture reagents were from Invitrogen/Life Technologies (Burlington, ON), and Pluronic P105 was generously donated by BASF Brenntag Canada (Toronto, ON). Parylene-C dimer was from Specialty Coating Systems (Indianapolis, IN), and Teflon-AF was purchased from DuPont (Wilmington, DE). Deionized (DI) water had a resistivity of 18 MΩ cm at 25 °C.
DMF device fabrication and operation
Digital microfluidic devices were fabricated in the University of Toronto Emerging Communications Technology Institute (ECTI) cleanroom facility, using a transparent photomask printed at Pacific Arts and Design (Markham, ON). Digital microfluidic device bottom plates bearing patterned chromium electrodes were formed by photolithography and etching as described previously53, 54 and were coated with 15 μm of Parylene-C and 200 nm of Teflon-AF. Parylene-C was applied using a vapor deposition instrument (Specialty Coating Systems), and Teflon-AF was spin-coated (1% wt/wt in Fluorinert FC-40, 1600 rpm, 60 s) followed by post-baking on a hot-plate (160 °C, 10 min). The polymer coatings were removed from contact pads by gentle scraping with a scalpel to facilitate electrical contact for droplet actuation. In addition to patterned DMF device bottom plates, unpatterned glass microscope slides and DMF device top plates formed from indium tin oxide (ITO) coated glass substrates (Delta Technologies Ltd, Stillwater, MN) were coated with Teflon-AF (200 nm, as above).
As shown in Figure 1, the device used here featured an array of 68 square actuation electrodes (2 × 2 mm ea.) connected to 10 reservoir electrodes (5 × 5 mm ea.), with an average inter-electrode gap size of 50-100 μm. Devices used for imaging cells incorporated 0.8 × 0.8 mm transparent windows (i.e., regions with no chromium) at the corners of some electrodes to assist with imaging.44 Devices were assembled with an unpatterned ITO–glass top plate and a patterned bottom plate separated by a spacer formed from 2 pieces of double-sided tape (total spacer thickness 140 μm). To actuate droplets, driving potentials (200–250 Vpp) were generated by amplifying the output of a function generator (Agilent Technologies, Santa Clara, CA) operating at 18 kHz. Liquid droplets were initially pipetted onto the device such that they were sandwiched between two plates, and then were actuated by applying driving potentials between the top electrode (ground) and sequential electrodes on the bottom plate via the exposed contact pads. No heating was observed as a consequence of droplet actuation. Droplet actuation was monitored and recorded by a charged couple device (CCD) camera mounted on a lens.
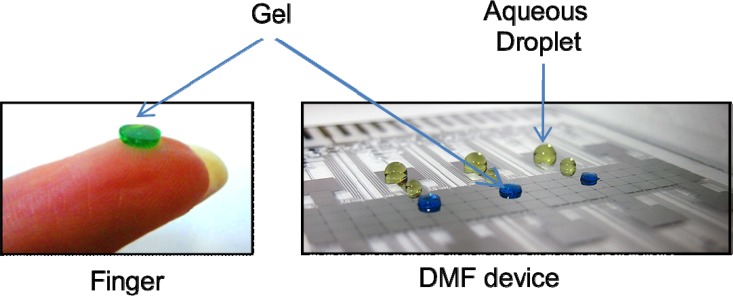
Figure 1.
Images of gel discs on a finger (left) and on a digital microfluidic device with no top plate (right). Aqueous droplets (yellow) are included in the image on the right to demonstrate the differences in geometry (i.e., the gel discs are cylinders, and the droplets have a domed shape). The gels and droplets contain dye to make them visible.
Formation of gel discs
Agarose was dissolved in DI water to a concentration of 0.5–4.0 wt. % by incubating the mixture in an oven at 70 °C for 1 h. Low gelling temperature agarose was used for the experiments not involving cells, and ultra-low gelling temperature agarose was used for the experiments involving cells. Some mixtures included food coloring dyes or 2 μm diameter polystyrene beads (Sigma Aldrich Canada) to make the gels easier to visualize. After removing the mixture from the oven, 0.4–20 μl aliquots were pipetted and sandwiched between two unpatterned Teflon AF-coated glass slides held together with 2 pieces of double-sided tape as a 140 μm spacer. These assemblies were placed on a cold pack to allow the droplets to gel (2 min). The resulting gel discs were ∼1 to 5 mm in diameter and 140 μm tall and could be retrieved from the glass-slide assembly using tweezers. Figure 1 shows the picture of gel discs and droplets of water, which demonstrates that the gel disc geometry is fixed in a cylindrical shape (in comparison to the water droplets, which are domed). For use with digital microfluidics, gel discs were sandwiched between a bottom plate and a top plate, with each disc positioned such that it straddled the interface between two electrodes on the bottom plate.
Gel disc dispensing measurements
In each experiment, a 2.5 μl DI water droplet was dispensed from a reservoir and then merged onto a 2 mm diameter 4 wt. % agarose gel disc. A sub-droplet was then dispensed away from the gel disc. Pictures were collected throughout the process, and volumes were estimated using image j software. The amount of water that remained on a gel after dispensing was calculated and averaged for 10 droplets on 10 different gels.
Gel disc solution exchange assay
In each experiment, a 5 μl droplet of 10 μM fluorescein in 100 μM borate buffer (pH 9.2) was dispensed from a reservoir and merged with a 2 mm dia. 4 wt. % agarose gel disc for passive or active exchange. For passive exchange, no further action was taken. For active exchange, the droplet was repeatedly actuated through the gel at ∼2.4 mm/s by alternately charging the two electrodes on either side of the disc. Pictures of the gel were collected using a microscope equipped with a fluorescent imaging module for 480/40 nm (Leica DM2000, Illinois, USA) every 5 (passive) or 2.5 (active) min. Each condition was repeated three times on three different gel discs.
The images were analyzed to measure the fluorescence in the gel using image j software. Briefly, in each image, the fluorescence intensity of the area of the inner 250 μm radius of the gel was divided by the average intensity of the same area outside the gel (in the droplet-at-large) to compensate for photobleaching. These corrected data were then plotted as a function of incubation time and then normalized to the intensity value of the last time point in each graph. Lines were fit to this data to determine the inflection point, correlating with saturation of the gel with fluorescein. The diffusion coefficient (passive) and apparent transport rate (active) were calculated as the square of the radius of the gel over the time it took to saturate the gel.
Enzyme microreactor assay
Enzymes were covalently attached to agarose gel discs using methods similar to those developed by Guisan and coworkers.6, 55, 56 To prime the gels for attachment, 2 mm diameter 4 wt. % agarose gel discs were isolated on Teflon AF-coated glass slides. A 3.5 μl aliquot of DI water was pipetted onto each gel, followed by 1 μl of 1.7 N NaOH containing 68 μg of NaBH4. The slide was placed on a cold pack and 0.7 μl of glycidol was pipetted onto the gel. After 5 min, the gel was removed from the cold pack and placed in a humidifier for 18 h at 25 °C. After incubation, the gel was washed 5 times with 10 μl of DI water. The gel was then oxidized by adding 30 μl of DI water and 6 μl of 2 M aqueous NaIO4 and then incubated in a humidifier for 2 h at 25 °C. Finally, the gel was washed 5 times with 10 μl aliquots of DI water.
After priming, gel discs were conjugated with alkaline phosphatase. A working buffer was formed from sodium borate in DI water (0.1 M, pH 10.0). 20 μl of 0.05 mg/ml alkaline phosphatase in working buffer was pipetted onto each primed gel, which was then incubated in a humidifier for 72 h at 25 °C. The gel was washed 5 times with 10 μl aliquots of working buffer and then stabilized by adding 20 μl of working buffer containing 20 μg NaBH4 and incubating for 30 min at 25 °C. Finally each gel was washed 5 times with 10 μl aliquots of sodium phosphate buffer (25 mM, pH 7.0). In the related work,57 we have demonstrated that the gel discs formed in this manner have enzymes distributed throughout the 3D gel structure.
For heterogeneous enzyme assays, a reaction buffer was formed from 10 mM diethanolamine and 1 mM MgCl2 in DI water (0.1 M, pH 10.0). Gels functionalized as described above were positioned into DMF devices and conditioned by dispensing 10 μl droplets of reaction buffer from reservoirs and merging them onto the gels. A 2.5 μl droplet of fluorescein diphosphate (0 to 40 μg/ml) in reaction buffer was then dispensed and merged onto each conditioned gel and incubated for 3 min. Droplets were then dispensed from the gel and 2 μl of each dispensed droplet was collected and mixed with 18 μl DI water in a well in a 384-well plate. Fluorescence was measured using a PheraStar multiwell plate reader with λex = 485 nm; λem = 520 nm; focal height = 9.0 mm; gain = 1). Each concentration of fluorescein diphosphate was evaluated three times on three different gels.
3D cell culture
Complete cell culture medium was formed from Dulbecco’s modified eagle medium (DMEM) (Sigma-Aldrich) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. NIH 3T3 cells were grown to near confluency in complete medium in T-25 flasks in an incubator at 37 °C with 5% CO2. At the beginning of each experiment, cells were detached using a solution of trypsin (0.25% w/v) and ethylenediaminetetraacetic acid (EDTA) (1 mM) for 5 min and then centrifuged at 173 × g for 5 min. The supernatant was removed and the cells were resuspended in complete medium at 107 cell/ml.
To prepare cell-laden gel discs, 4 wt. % ultra-low gelling temperature agarose in DI water was diluted with complete cell media to a final concentration of agarose ranging from 0.4 to 3.6 wt. %. The 107 cell/ml suspension in medium was added to these agarose solutions to obtain a suspension containing 106 cell/ml. The results of initial experiments indicated that 0.58 wt. % agarose in such suspensions was an appropriate concentration for 3D cell growth (see Results and Discussion section), and this concentration was used for all the subsequent experiments. Droplets of this agarose/cell mixture were pipetted onto DMF bottom plates (straddling an electrode bearing a transparent window, see the DMF Device Fabrication and Operation section), covered with top plates and then placed in a humidified chamber at 4 °C for 15 min to gel the discs. Devices were stored in an incubator at 37 °C/5% CO2 for 1 day, and gels were then visualized through the transparent windows using a Leica DM2000 microscope (Leica Microsystems Canada, Richmond Hill, ON).
In some cases, cells in gel discs were viability stained or fixed and stained for nuclei and F-actin. All steps were implemented by digital microfluidics—droplets of reagents were dispensed from reservoirs, merged onto gels, incubated, and then dispensed away from them. For the former (viability staining), a 2 μl droplet of 1 μM calcein-AM (Invitrogen/Life Technologies) in phosphate buffered saline (PBS) supplemented with 0.05% Pluronic F68 (Ref. 58) was delivered to each gel and incubated for 30 min at room temperature. Each gel was then washed twice with 2 μl droplets of PBS, after which cells were imaged by fluorescence microscopy (Leica DM2000). For the latter (cell fixation and staining), gel discs were washed with 2 μl droplets of PBS and then exposed to 2 μl droplets of 3.7% (wt./vol.) paraformaldehyde in PBS supplemented with 0.05% Pluronic F68 (Ref. 58) for 15 min. The discs were then washed three times with 2 μl droplets of PBS and then dehydrated by incubation in 2 μl droplets of cold acetone for 10 min at −20 °C. The discs were then rehydrated by exposure to 2 μl droplets of PBS for 10 min before incubation with 2 μl droplets of dye solution containing 50 μg/ml phalloidin, 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and 0.02% (wt/v) Pluronic P105 (Ref. 59) in PBS for 30 min at room temperature. The discs were then washed three times with 2 μl droplets of PBS and the gels were imaged using confocal imaging on a Leica TCS SP2 (Leica Microsystems).
RESULTS AND DISCUSSION
Design and behaviour of hydrogel discs for digital microfluidics
A previous report52 described a method for the preparation of hydrogels on DMF devices. In that initial work, droplets of alginate were combined with droplets of calcium chloride and the resulting mixture was observed to be non-movable (and thus was assumed to be gelled). In this initial work,52 it was not clear from the brief discussion whether the gels were subsequently addressable by liquid droplets for applications. Here, we introduce the use of thermoreversible agarose polymers on DMF devices and demonstrate the use of these gels for use with enzyme microreactors and 3D cell culture. We used two forms of agarose, low gelling temperature agarose which melts at 65 °C and gels at 30 °C, and ultra-low gelling temperature agarose which melts at ≥50 °C and gels between 8 and 17 °C. Figure 1 shows the morphology of a gel disc formed using this technique—when formed sandwiched between two plates, the gels maintain their shapes and can be used and re-used in different devices and experiments. In most of the work reported here, we used ∼2 mm-diameter discs to match the dimensions of the DMF actuation electrodes, but in practice, any desirable size can be formed by adjusting the volume of the agarose mixture prior to gelling.
A first question in our experiments was whether the agarose gel discs would remain stationary or would become mobile when exposed to moving droplets on DMF devices. We hypothesized that adhesion and friction would keep the gel discs stationary, and this turned out to be the case. In fact, over the course of hundreds of experiments, gel discs were never observed to move, which is similar to our observations of porous polymer monoliths on DMF devices.60
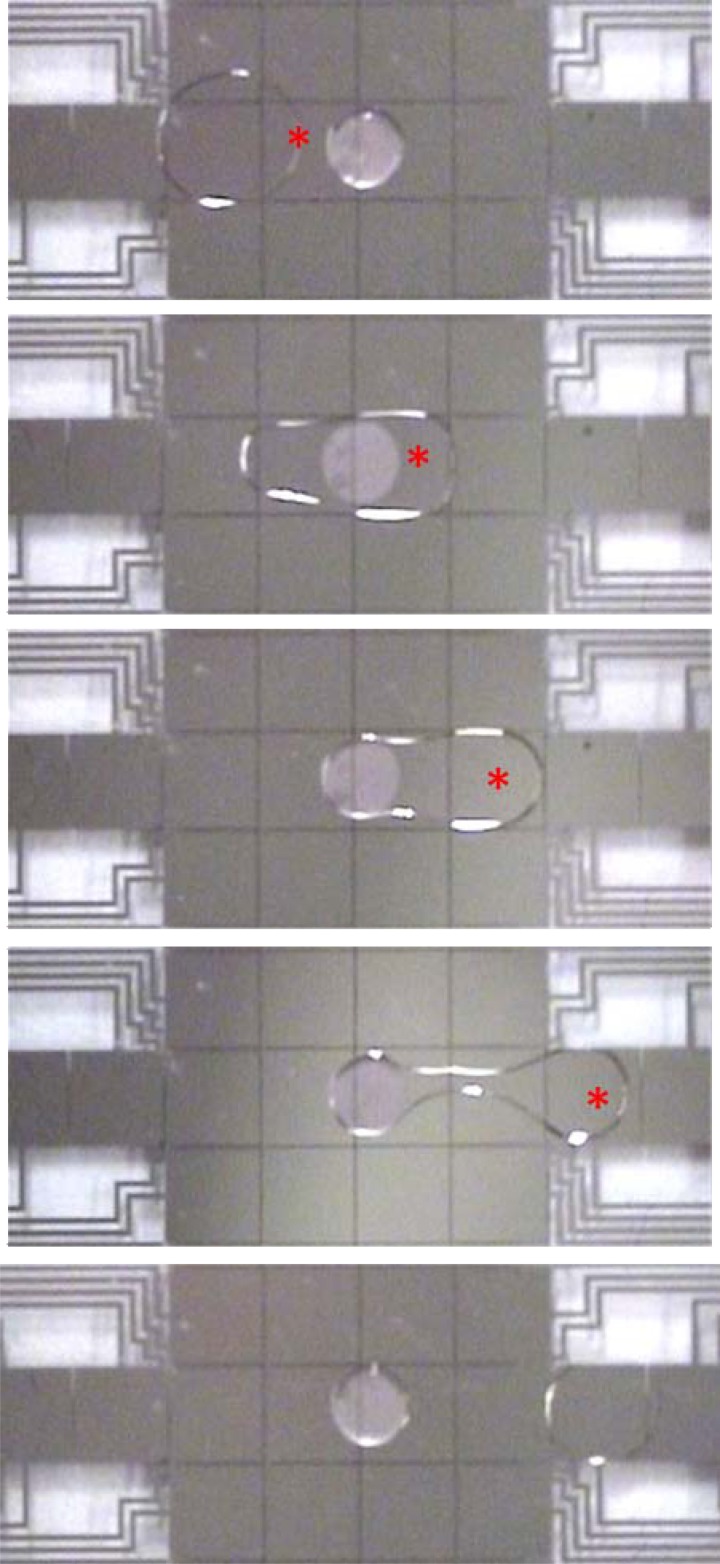
A second question about gel discs on digital microfluidic devices was whether aqueous droplets could be dispensed from the gels, or whether the droplets would remain permanently bound to the extremely hydrophilic gel discs. In fact, droplet dispensing was not only possible but quite straightforward. As shown in Figure 2, when the three electrodes adjacent to a gel disc are actuated in succession, a droplet (connected by a tail) is pulled away from the gel until it pinches off. This occurs reproducibly and predictably. After dispensing, a small amount of the original droplet volume remains on the gel. To characterize this quantitatively, ten 2.5 μl droplets of water were merged and dispensed from hydrogel discs, and on average, ∼7% of the volume (178 ± 13 nl) remains bound to the gel, and ∼93% is dispensed. Note that the droplets dispensed in most of the experiments reported here are “unit droplets” (as depicted in Figure 2)—that is, the dispensed droplets have a volume that corresponds to an area slightly larger than that of a single square electrode. Larger volumes can also be dispensed from gels by using a larger source droplet and actuating two or more destination electrodes in parallel (data not shown). We speculate that volumes smaller than unit droplets cannot be dispensed from gel discs (without special driving circuits designed for dispensing sub-unit volumes61), but this was not evaluated here. Future work exploring the relationship between gel disc size, droplet size, and electrode size is warranted.
Figure 2.
Droplet actuation and hydrogel discs in digital microfluidics: A series of images from a movie (top-to-bottom) depicting the merging of a 2 μl droplet with an immobilized gel disc, followed by dispensing of a portion of the volume. The red asterisks (*) denote which electrode was activated in each frame. The gel disc contains a suspension of 2 μm diameter beads to make it visible.
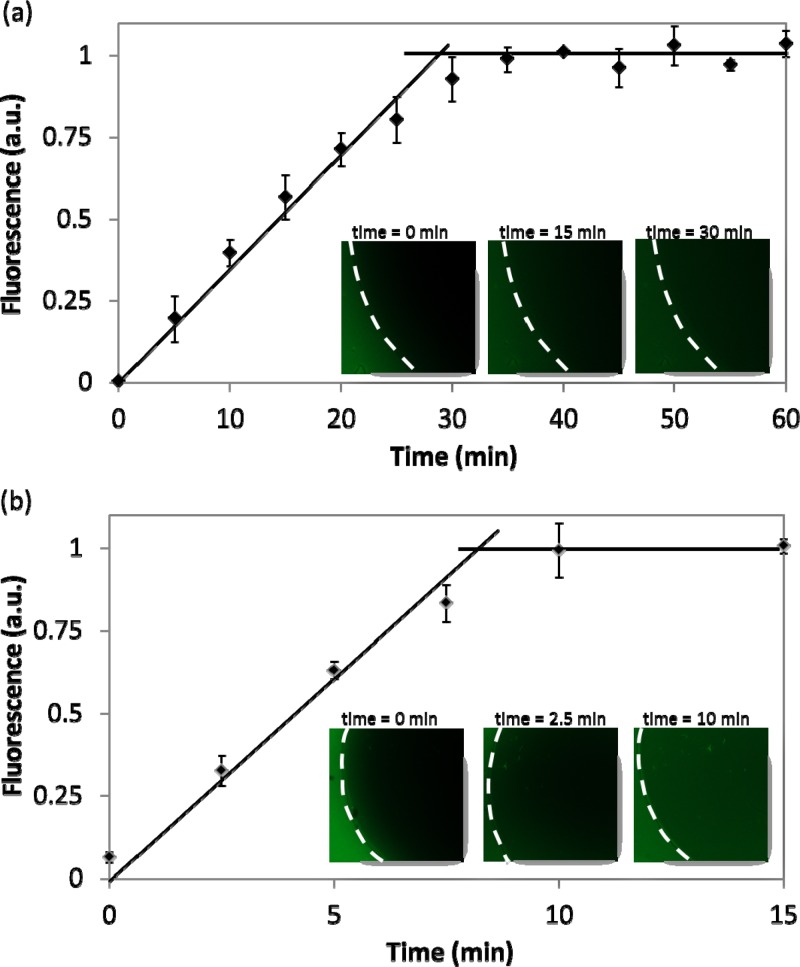
A third question about the techniques described here was the nature and rate of analyte exchange between gel disc and droplet during droplet movement. To address this question, droplets containing fluorescein were merged with gels and were incubated either passively (i.e., with no further droplet actuation) or actively (i.e., with continuous actuation through the gel) as described in the Methods and Materials section. The fluorescence intensities measured in the gel discs were plotted as a function of time. As shown in Figure 3, the trends for solution exchange with the gel disc have a similar profile for both active and passive cases; the fluorescence intensity increases linearly until it saturates and is constant thereafter. The inflection point between the linear increase and the saturation state represents the time required for fluorescein molecules to completely fill the gel. For passive exchange this process required 28.8 ± 1.5 min, while active exchange was ∼3 × faster, at 8.7 ± 0.6 min. From this, we estimate the diffusion coefficient for the passive case to be 0.58 × 10−9 m2/s and the apparent rate for active transport to be 1.9 × 10−9 m2/s. The measured diffusion coefficient for fluorescein in the gel disc is very similar to the known diffusion coefficient for fluorescein in water, 0.49 × 10−9 m2/s.62 This is expected, as the 4% agarose used here has large pores (∼243 nm for 4 wt. % agarose63) which should not impede the movement of small molecules such as fluorescein. The faster exchange observed for the active case suggests that the electromechanical droplet actuation facilitates convection into the gel, but more work is needed to probe this phenomenon.
Figure 3.
Transport assay: Droplets containing fluorescein were merged with gel discs and allowed to incubate either passively (a) or actively (b) and fluorescence in the gel was plotted as a function of time. The insets are representative fluorescent images of gels at different times (the dashed white lines indicate the edge of the discs). In both the passive and active cases, the fluorescence increases linearly inside the gel until saturation. The inflection point between the linear increase and saturation regions represents the time required for fluorescein to permeate through the whole gel. This time is ∼3 × less for active transport relative to passive transport. The experiments were replicated 3 times on three different gels and error bars are ±1 S.D.
The observations of gel disc behaviour in digital microfluidic systems reported above are interesting, and they suggest a host of future studies into the effects (both for 2D transport, as shown in Figure 3, and for 3D transport, not evaluated here) of gel size, droplet volume, pore size, molecule size, and actuation velocity. But the observations recorded here demonstrate proof of principle for the combination of digital microfluidics with hydrogels and were sufficient to inspire exploration of whether this combination is useful for applications involving immobilized enzymes and cells, described below.
Application #1: Enzyme microreactor
As a first demonstration of an application for digital microfluidics and hydrogel discs, we explored the implementation of enzyme microreactors. Enzyme microreactors are often used to perform small-scale enzymatic digestions of small amounts of rare protein samples or for multiplexed screening of many protein samples. For such applications, is advantageous to immobilize the enzyme onto a substrate in a bio-compatible niche to preserve functionality.25 Unfortunately, most gel-based enzyme reactors are addressable only by pipetting into multiwell plates. In laboratories lacking robotic dispensers and aspirators, which forms a practical limit to the extent of multiplexing that is possible. This limitation motivates the development of methods such as digital microfluidics (which are readily automated61, 64 and useful for multiplexing51) for implementing enzyme microreactors.
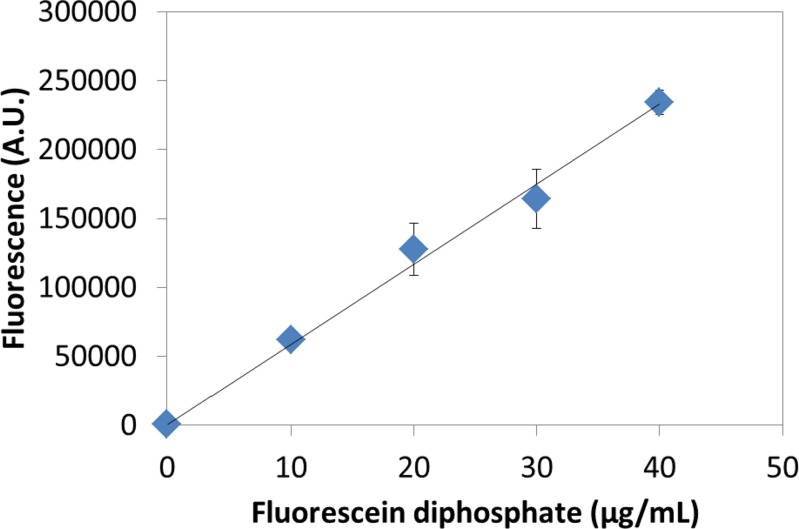
Here, we report the first hydrogel enzyme microreactor system addressed by digital microfluidics. In this system, alkaline phosphatase molecules were immobilized to agarose gel discs by oxidizing alcohol groups on the gel to aldehydes, and then coupling the aldehyde groups to protein amine groups. Droplets containing fluorescein diphosphate (which is non-fluorescent) were dispensed and merged onto the gels to allow for the enzyme to cleave off the phosphate groups, generating the fluorescent product, fluorescein. After incubating, the droplets were dispensed and their fluorescence was measured. Figure 4 shows the fluorescence of the dispensed droplets as a function of fluorescein diphosphate concentration. As expected, the fluorescence increased linearly with substrate concentration.
Figure 4.
Enzyme microreactor: Gel discs were formed with covalently linked alkaline phosphatase. Droplets containing fluorescein diphosphate were merged onto the discs and after incubation, the fluorescence of dispensed droplets was reported as a function of the concentration of fluorescein diphosphate. The experiments were replicated 3 times and error bars are ±1 S.D. A least squares line of best fit through the data has an R2 = 0.9929.
The digital microfluidic enzymatic microreactor described here has a number of differences relative to those formed in conventional enclosed microchannel systems.25, 28, 31 For example, microchannel systems are well suited for very low (∼nl) sample volumes. This property makes microchannel enzyme microreactors useful for analytical applications, but such systems are often not easily adapted for use for preparative applications. In contrast, the digital microfluidic system described here is appropriate for larger (∼μl) sample volumes, with straightforward means of recovery of products. In addition, in the digital microfluidic system, the reagents and products are isolated in an immobile droplet (surrounded by air), such that products cannot diffuse away. We propose that the two types of systems are complementary, with application dictating the appropriate format. Further, we propose that future systems engineered for multiplexed analysis of different conditions51 will be a welcome advance for labs that are currently limited to manual pipetting and well plates.
Application #2: 3D cell culture
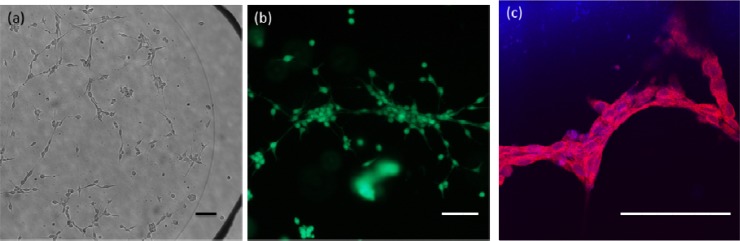
As a second demonstration of an application for digital microfluidics and hydrogel discs, we explored the use of this system for 3D cell culture. NIH-3T3 fibroblasts were seeded in a solution of ultra-low gelling temperature agarose, which was then gelled into discs and incorporated into DMF devices. The ability of the cells to form 3D networks was found to be highly dependent on agarose concentration, which is consistent with the previous reports.65 Gel discs with agarose concentrations greater than 1 wt. % resulted in cells with (undesirable) spherical morphologies, and discs with agarose densities lower than 0.45 wt. % had morphologies that were similar to those found in conventional two-dimensional systems (not shown). It is likely that high-concentration agarose gels tightly constrain the cells, while low-concentration agarose solutions failed to fully transition into gels. Regardless, as demonstrated in Figure 5, 0.58 wt. % agarose gel was a useful concentration for forming 3D networks of cells.
Figure 5.
Three-dimensional cell culture: NIH-3T3 cells were cultured and imaged in 0.58 wt. % agarose gel discs on DMF devices. Cells were imaged through transparent windows on DMF devices using bright field (a), fluorescence (b) with calcein AM dye and confocal fluorescence (c) with DAPI (shown in red) and phalloidin (shown in blue) microscopies. The scale bar in each image is 25 μm.
As shown in Figure 5a, fibroblasts cultured in the digital microfluidic/hydrogel disc format exhibited stellate bipolar morphologies consistent with culture in high-modulus extracellular matrix66 and formed 3D networks of interconnected cells. This morphology is different than what is observed in cells grown in traditional 2D systems (such cells have flat and unconnected morphologies, not shown). As shown in Figure 5b, cells embedded in gel discs remained viable as determined by staining with calcein AM, and high cell viability was observed for up to 7 days, after which experiments were discontinued. Cells were also fixed and stained for nuclei and F-actin in situ using digital microfluidic actuation. This process (described in detail in the Methods and Materials section) underscores the robustness of the new methods reported here—gels were sequentially exposed to seven sets of droplets (each as depicted in Figure 2) to rinse, fix, rinse, dehydrate, rinse, stain, and rinse the cells, respectively.
As shown in Figure 5c, cells embedded in 0.58 wt. % agarose discs self-organized into tightly packed networks with cell-cell interaction (clustering). The dense packing of cells is a key feature of the engineering of tissues and is known to affect cell phenotype by creating molecular gradients and affecting matrix stiffness.67 In addition to the tight packing of cells, the shift from cell-substrate interactions typical of 2D cell culture to cell-cell interactions in 3D is likely to change cell behaviour as a result of increased cellular communication.68 Given the widespread interest in the development of 3D cell culture systems as a better model of in vivo phenotypes69 (relative to traditional 2D culture systems), we propose that the techniques reported here, in which 3D scaffolds can be independently and sequentially addressed with reagents in an automated, miniaturized format, will be useful for numerous applications in tissue engineering.
CONCLUSION
We have demonstrated that cylindrical hydrogel discs can be incorporated in digital microfluidic devices, and that droplets of reagents can be systematically addressed to them. The gel discs can be functionalized with proteins to act as enzymatic microreactors, as demonstrated in the action of alkaline phosphatase on fluorescein diphosphate. The gel discs can also be used for 3D cell culture on DMF devices, as demonstrated by the behaviour of NIH-3T3 cells grown in such systems, which formed tightly packed networks rife with cell-cell connections. We propose that combining hydrogels with DMF will be useful for many applications in the future.
ACKNOWLEDGMENTS
We thank the Natural Sciences and Engineering Research Council and the Canadian Cancer Society for financial support. We thank Ilya Gourevich at the Nano Imaging Facility at the Department of Chemistry at the University of Toronto for assistance obtaining confocal images. E.K. and A.R.W. thank the Canada Research Chair (CRC) program for CRCs.
References
- Osada Y., Ping Gong J., and Tanaka Y., J. Macromol. Sci. 44, 87 (2004). 10.1081/MC-120027935 [DOI] [Google Scholar]
- Tumarkin E. and Kumacheva E., Chem. Soc. Rev. 8, 2161 (2009). 10.1039/b809915b [DOI] [PubMed] [Google Scholar]
- Eddington D. T. and Beebe D. J., Adv. Drug Delivery Rev. 56(2), 199 (2004). 10.1016/j.addr.2003.08.013 [DOI] [PubMed] [Google Scholar]
- Lee S., Eddington D. T., Kim Y., Kim W., and Beebe D. J., J. MEMS 12(6), 848 (2003). 10.1109/JMEMS.2003.820292 [DOI] [Google Scholar]
- Rivest C., Morrison D. W. G., Ni B., Rubin J., Yadav V., Mahdavi A., Karp J. M., and Khademhosseini A., J. Mech. Mater. Struct. 2(6), 1103 (2007). 10.2140/jomms [DOI] [Google Scholar]
- Blanco R. M., Calvete J. J., and Guisan J. M., Enzyme Microb. Technol. 11, 353 (1989). 10.1016/0141-0229(89)90019-7 [DOI] [Google Scholar]
- Huang X., Yin Y., Tang Y., Bai X., Zhang Z., Xu J., Liu J., and Shen J., Soft Matter 5, 1905 (2009). 10.1039/b816888a [DOI] [Google Scholar]
- Retama J. R., Lopez-Ruiz B., and Lopez-Cabarcos E., Biomaterials 24(17), 2965 (2003). 10.1016/S0142-9612(03)00095-4 [DOI] [PubMed] [Google Scholar]
- Courtenay V. D., Selby P. J., Smith I. E., Mills J., and Peckham M. J., Br. J. Cancer 38(1), 77 (1978). 10.1038/bjc.1978.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger S., Copeland M. F., Downes E. M., Braasch A. T., Tuson H. H., Eun Y.-J., and Weibel D. B., J. Am. Chem. Soc. 133, 5966 (2011). 10.1021/ja111131f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe B. D., Brown M. A., UIyer R. K., Radisic M., and Murthy S. K., Lab Chip 9, 1507 (2009). 10.1039/b823523f [DOI] [PubMed] [Google Scholar]
- Hatch A., Hansmann G., and Murthy S. K., Langmuir 27, 4257 (2011). 10.1021/la105016a [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan A. P., Bruzewicz D. A., Glavan A., Butte M., and Whitesides G. M., PLoS ONE 3(5), e2258 (2008). 10.1371/journal.pone.0002258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun Y.-J., Utada A. S., Copeland M. F., Takeuchi S., and Weibel D. B., ACS Chem. Biol. 6, 260 (2011). 10.1021/cb100336p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T. G. and Hoffman A. S., J. Appl. Polym. Sci. 46(4), 659 (2003). 10.1002/app.1992.070460413 [DOI] [Google Scholar]
- Brondsted H. and Kopecek J., pH-Sensitive Hydrogels (ACS, Salt Lake City, UT, 1992). [Google Scholar]
- Hovgaard L. and Brondsted H., J. Controlled Release 36(1–2), 159 (1995). 10.1016/0168-3659(95)00049-E [DOI] [Google Scholar]
- Boissiere M., Meadows P. J., Bayner R., Helary C., Livage J., and Coradin T., J. Mater. Chem. 16, 1178 (2006). 10.1039/b515797h [DOI] [Google Scholar]
- Islam M. T., Rodriguez-Hornedo N., Ciotti S., and Ackermann C., Pharm. Res. 21, 1192 (2004). 10.1023/B:PHAM.0000033006.11619.07 [DOI] [PubMed] [Google Scholar]
- Lopez V. C., Hadgraft J., and Snowden M. J., Int. J. Pharm. 292, 137 (2005). 10.1016/j.ijpharm.2004.11.040 [DOI] [PubMed] [Google Scholar]
- Popanda O., Ebbeler R., Twardella D., Helmbold I., Gotzes F., Schmezer P., Thielmann H. W., von Fournier D., Haase W., Sautter-Bihl M. L., Wenz F., Bartsch H., and Chang-Claude J., Int. J. Radiat. Oncol., Biol., Phys. 55, 1216 (2003). 10.1016/S0360-3016(02)04415-2 [DOI] [PubMed] [Google Scholar]
- de Vicente J., Stokes J. R., and Spikes H. A., Food Hydrocolloids 20, 483 (2006). 10.1016/j.foodhyd.2005.04.005 [DOI] [Google Scholar]
- Khan A. A., Khan H. M., and Delince H., Food Control 16, 141 (2005). 10.1016/j.foodcont.2004.01.009 [DOI] [Google Scholar]
- Villarini M., Scassellati-Sforzolini G., Moretti M., and Pasquini R., Cell Biol. Toxicol. 16, 285 (2000). 10.1023/A:1026794213308 [DOI] [PubMed] [Google Scholar]
- Koh W.-G. and Pishko M., Sens. Actuators B 106(1), 335 (2005). 10.1016/j.snb.2004.08.025 [DOI] [Google Scholar]
- Koh W.-G. and Pishko M., Anal. Bioanal. Chem. 385, 1389 (2006). 10.1007/s00216-006-0571-6 [DOI] [PubMed] [Google Scholar]
- Srinivasan A., Bach H., Sherman D. H., and Dordick J. S., Biotechnol. Bioeng. 88, 528 (2004). 10.1002/(ISSN)1097-0290 [DOI] [PubMed] [Google Scholar]
- Wong A., Perez-Castillejos R., Love J. C., and Whitesides G. M., Biomaterials 29(12), 1853 (2008). 10.1016/j.biomaterials.2007.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajar C. M., Chen X., Harris J. W., Suresh V., Hughes C. C. W., Jeon N. L., Putnam A. J., and Georges S. C., Biophys. J. 94(5), 1930 (2008). 10.1529/biophysj.107.120774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu B., Yang P., and Girault H. H., Anal. Bioanal. Chem. 390, 227 (2008). 10.1007/s00216-007-1664-6 [DOI] [PubMed] [Google Scholar]
- Sundararaghavan H. G., Monteiro G. A., Firestein B. L., and Shreiber D. I., Biotechnol. Bioeng. 102(2), 632 (2008). 10.1002/bit.v102:2 [DOI] [PubMed] [Google Scholar]
- Kuckling D., Harmon M. E., and Frank C. W., Macromolecules 35, 6377 (2002). 10.1021/ma0203041 [DOI] [Google Scholar]
- Sundararaghavan H. G., Masand S. N., and Shreiber D. I., J. Neurotrauma 28, 2377 (2011). 10.1089/neu.2010.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni K. P., Ramarathinam S. H., Friend J., Yeo L., Purcell A. W., and Perlmutter P., Lab Chip 10, 1518 (2010). 10.1039/c001501f [DOI] [PubMed] [Google Scholar]
- Lee J., Moon H., Fowler J., Schoellhammer T., and Kim C. J., Sens. Actuators A 95, 259 (2002). 10.1016/S0924-4247(01)00734-8 [DOI] [Google Scholar]
- Pollack M. G., Fair R. B., and Shenderov A. D., Appl. Phys. Lett. 77, 1725 (2000). 10.1063/1.1308534 [DOI] [Google Scholar]
- Wheeler A. R., Science 322, 539 (2008). 10.1126/science.1165719 [DOI] [PubMed] [Google Scholar]
- Chatterjee D., Shepherd H., and Garrell R. L., Lab Chip 9, 1219 (2009). 10.1039/b901375j [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Torii T., and Higuchi T., Lab Chip 2, 19 (2002). 10.1039/b108739h [DOI] [PubMed] [Google Scholar]
- Srinivasan V., Pamula V. K., and Fair R. B., Lab Chip 4(4), 310 (2004). 10.1039/b403341h [DOI] [PubMed] [Google Scholar]
- Miller E. M. and Wheeler A. R., Anal. Chem. 80(5), 1614 (2008). 10.1021/ac702269d [DOI] [PubMed] [Google Scholar]
- Shah G. J., Ohta A. T., Chiou E. P. Y., Wu M. C., and Kim C. J., Lab Chip 9, 1732 (2009). 10.1039/b821508a [DOI] [PubMed] [Google Scholar]
- Fan S. K., Huang P. W., Wang T. T., and Peng Y. H., Lab Chip 8, 1325 (2008). 10.1039/b803204a [DOI] [PubMed] [Google Scholar]
- Barbulovic-Nad I., Au S., and Wheeler A. R., Lab Chip 10, 1536 (2010). 10.1039/c002147d [DOI] [PubMed] [Google Scholar]
- Barbulovic-Nad I., Yang H., Park P. S., and Wheeler A. R., Lab Chip 8, 519 (2008). 10.1039/b717759c [DOI] [PubMed] [Google Scholar]
- Au S. H., Shih S. C. C., and Wheeler A. R., Biomed. Microdevices 13, 41 (2011). 10.1007/s10544-010-9469-3 [DOI] [PubMed] [Google Scholar]
- Witters D., Vergauwe N., Vermeir S., Ceyssens F., Liekens S., Puers R., and Lammertyn J., Lab Chip 11, 2790 (2011). 10.1039/c1lc20340a [DOI] [PubMed] [Google Scholar]
- Vergauwe N., Witters D., Ceyssens F., Vermeir S., Verbruggen B., Puers R., and Lammertyn J., J. Micromech. Microeng. 21(5), 054026 (2011). 10.1088/0960-1317/21/5/054026 [DOI] [Google Scholar]
- Srigunapalan S., Eydelnant I. A., Simmons C. A., and Wheeler A. R., Lab Chip 12, 369 (2012). 10.1039/c1lc20844f [DOI] [PubMed] [Google Scholar]
- Eydelnant I. A., Uddayasankar U., Li B., Liao M. W., and Wheeler A. R., Lab Chip 12, 750 (2012). 10.1039/c2lc21004e [DOI] [PubMed] [Google Scholar]
- Bogojevic D., Chamberlain M. D., Barbulovic-Nad I., and Wheeler A. R., Lab Chip 12, 627 (2012). 10.1039/c2lc20893h [DOI] [PubMed] [Google Scholar]
- Fair R. B., Khlystov A., Tailer T. D., Ivanov V., Evans R. D., Griffin P. B., Srinivasan V., Pamula V. K., Pollack M. G., and Zhou J., IEEE Des. Test Comput. 24(1), 10 (2007). 10.1109/MDT.2007.8 [DOI] [Google Scholar]
- Jebrail M. J., Ng A. H. C., Rai V., Hili R., Yudin A. K., and Wheeler A. R., Angew. Chem., Int. Ed. 49, 8625 (2010). 10.1002/anie.201001604 [DOI] [PubMed] [Google Scholar]
- Miller E. M. and Wheeler A. R., Anal. Chem. 80, 1614 (2008). 10.1021/ac702269d [DOI] [PubMed] [Google Scholar]
- Mateo C., Abian O., Fernandez-Lafuente R., and Guisan J. M., Enzyme Microb. Technol. 26(7), 509 (2000). 10.1016/S0141-0229(99)00188-X [DOI] [PubMed] [Google Scholar]
- Mateo C., Grazu V., Palomo J. M., Lopez-Gallego F., Fernandez-Lafuente R., and Guisan J. M., Nat. Protoc. 2, 1022 (2007). 10.1038/nprot.2007.133 [DOI] [PubMed] [Google Scholar]
- Luk V. N., Fiddes L. K., Luk V. M., Kumacheva E., and Wheeler A. R., “Digital microfluidic-hydrogel microreactors for proteomics,” Proteomics (in press). 10.1002/pmic.201100608 [DOI] [PubMed]
- Luk V. N., Mo G. C., and Wheeler A. R., Langmuir 24, 6382 (2008). 10.1021/la7039509 [DOI] [PubMed] [Google Scholar]
- Au S. H., Kumar P., and Wheeler A. R., Langmuir 27, 8586 (2011). 10.1021/la201185c [DOI] [PubMed] [Google Scholar]
- Yang H., Mudrik J. M., Jebrail M. J., and Wheeler A. R., Anal. Chem. 83, 3824 (2011). 10.1021/ac2002388 [DOI] [PubMed] [Google Scholar]
- Gong J. and Kim C. J., Lab Chip 8, 898 (2008). 10.1039/b717417a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S. A., Pitts B., and Stewart P. S., Antimicrob. Agents Chemother. 49(2), 728 (2005). 10.1128/AAC.49.2.728-732.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernodet N., Maaloum M., and Tinland B., Electrophoresis 18, 55 (1997). 10.1002/(ISSN)1522-2683 [DOI] [PubMed] [Google Scholar]
- Shih S. C. C., Fobel R., Kumar P., and Wheeler A. R., Lab Chip 11, 535 (2011). 10.1039/c0lc00223b [DOI] [PubMed] [Google Scholar]
- Kumachev A., Greener J., Tumarkin E., Eiser E., Zandstra P. W., and Kumacheva E., Biomaterials 32(6), 1477 (2011). 10.1016/j.biomaterials.2010.10.033 [DOI] [PubMed] [Google Scholar]
- Grinnell F., Trends Cell Biol. 13(5), 264 (2003). 10.1016/S0962-8924(03)00057-6 [DOI] [PubMed] [Google Scholar]
- Griffith L. G. and Swartz M. A., Nat. Rev. Mol. Cell Biol. 7, 211 (2006). 10.1038/nrm1858 [DOI] [PubMed] [Google Scholar]
- Folch A. and Toner M., Annu. Rev. Biomed. Eng. 2, 227 (2000). 10.1146/annurev.bioeng.2.1.227 [DOI] [PubMed] [Google Scholar]
- Barralet J. E., Wang L., Lawson M., Triffitt J. T., Cooper P. R., and Shelton R. M., J. Mater. Sci.: Mater. Med. 16, 515 (2005). 10.1007/s10856-005-0526-z [DOI] [PubMed] [Google Scholar]