Abstract
Emerging evidence points to a role for long non-coding RNAs in the modulation of epigenetic states and transcription in human cells. New insights, using various forms of small non-coding RNAs, suggest that a mechanism of action is operative in human cells, which utilizes non-coding RNAs to direct epigenetic marks to homology containing loci resulting ultimately in the epigenetic-based modulation of gene transcription. Importantly, insights into this mechanism of action have allowed for certain target sequences, which are either actively involved in RNA mediated epigenetic regulation or targets for non-coding RNA based epigenetic regulation, to be selected. As such, it is now feasible to utilize small antisense RNAs to either epigenetically silence a gene expression or remove epigenetic silencing of endogenous non-coding RNAs and essentially turn on a gene expression. Knowledge of this emerging RNA-based epigenetic regulatory network and our ability to cognitively control gene expression has deep implications in the development of an entirely new area of pharmacopeia.
Keywords: Non-coding RNA, Epigenetic, Transcription, Pseudogene, Tumor suppressor gene, Antisense RNA
Introduction
An enigma in modern-day molecular biology has been determining what exactly the role is for the massive amount of non-protein-coding transcripts found in the human cell. Non-coding transcripts have been estimated to account for ∼70–98% of the genome (reviewed in (Taft et al. 2007; Morris 2009a)). These non-coding RNAs (ncRNAs) emanate from non-coding DNA, regions in the genome that have been dubbed the “genomic dark matter”; as their functional relevance is questionable. If the non-coding regions of the human genome are not thought to be involved in protein coding then what, if anything, are these non-coding RNAs (ncRNAs) doing? And why would the cell transcribe vast tracks of DNA to never produce a protein?
RNA-mediated transcriptional gene silencing
Evidence addressing the central question of what the role is for ncRNAs in human cells has emerged from studies carried out with RNA interference (RNAi). RNAi is a process whereby small double-stranded interfering RNA (siRNAs) molecules functionally target and direct the degradation of a homology containing mRNA (reviewed in (Hannon 2002; Hannon and Rossi 2004)). This form of RNA-based regulation was termed; post-transcriptional gene silencing (PTGS). In PTGS, siRNAs are targeted to messenger RNA, recruiting the native cellular DICER machinery to degrade the transcript and provide gene suppression. While PTGS is a potent mechanism for the targeted degradation of targeted mRNAs, its gene suppressive effect is transient, lasting only until new transcripts are generated or the siRNAs are degraded. While several studies have been carried out using RNAi to suppress gene expression in a PTGS-based manner, one underappreciated study has created quite a wave of insight. In this study carried out in human cells, siRNAs were designed to target a gene promoter instead of the mRNA/coding region. The result of this siRNA targeting of a gene promoter was the suppression of gene expression at the transcriptional level, upstream of the well-documented PTGS pathway (Morris et al. 2004). Interestingly, this mode of RNA-based transcriptional suppression appeared to rely on an epigenetic component, as Trichostatin A and 5′ Aza-cytadine suppressed the obsiRNA-mediated transcriptional gene silencing (TGS). Notably, the observed siRNA-mediated TGS was similar to previous work carried out in plants and yeast (Matzke et al. 2004).
During the course of investigation as to how siRNAs were capable of directing TGS in human cells, one striking observation was generated. Weinberg and colleagues (2006) discovered, quite serendipitously, that only the antisense strand of the siRNA was required for TGS in human cells. Other studies have further validated this requirement (Turner et al. 2009), suggesting that the endogenous mechanism in human cells whereby ncRNAs regulate gene transcription may involve forms of RNA beyond double-stranded RNAs. Later work by several groups have collectively led to several insights suggesting a model for how small RNAs can function in controlling gene transcription in human cells (reviewed in (Morris 2009b))(Fig. 1). In this model, the RNA guides an epigenetic remodeling complex to the targeted promoter loci which contains homology to the RNA guide, ultimately resulting in the epigenetic remodeling of the target site to a transcriptionally silent state (Fig. 1). While much has been gleamed from these collective bodies of work, it had remained unknown whether or not human cells generated endogenous RNAs that might be regulating gene transcription in this manner.
Fig. 1.
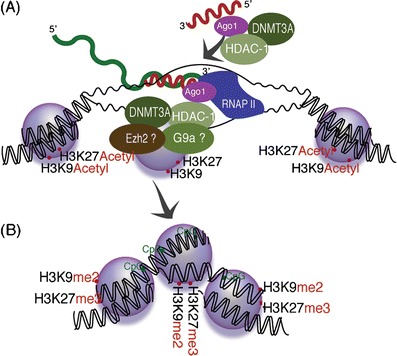
Small antisense RNA-directed transcriptional gene silencing—a de novo derived small antisense RNAs, designed to target a promoter of choice, can interact with promoter-associated RNAs at the transcribed promoter (Hawkins et al. 2009; Han et al. 2007) and facilitate the recruitment of epigenetic remodeling complexes (Weinberg et al. 2006; Turner et al. 2009; Hawkins et al. 2009; Han et al. 2007; Kim et al. 2006; Suzuki et al. 2005; Suzuki et al. 2008). b The result of small antisense RNA targeting is the epigenetic remodeling and stable silencing of transcription from the targeted promoter
RNAi-mediated suppression of the endogenous non-coding RNA suppressor results in activation of gene expression in human cells
Despite earlier observations indicating that small ncRNAs, derived de novo could affect gene transcription, little was known as to whether or not there were active ncRNAs modulating gene transcription in human cells. Some studies provided evidence that micro-RNAs (miRNAs) could function as endogenous regulators of gene transcription (Klase et al. 2007; Omoto and Fujii 2005; Omoto et al. 2004; Tan et al. 2009; Kim et al. 2008). Hints to a deeper layer of RNA-based transcriptional regulation arose from one study which set out to target de novo derived siRNAs to the p21 and E-cadherin promoters, specifically targeting A:T-rich regions in these promoters. Notably, A:T-rich regions of the genome are generally found in the coding bodies of genes, while gene promoters tend to exhibit CpG islands and palindromic sequences with high GC content, often times used for transcription factor or protein binding (Alberts et al. 1994). In this study, it was determined that these p21 and E-cadherin promoter targeted siRNAs were able to activate gene expression, though activation of actual transcription was not demonstrated by nuclear run-on analysis or RNA polymerase occupancy at the respective promoters (Li et al. 2006; Place et al. 2008). In these studies, it was also determined that Argonaute 2 (Ago-2) was required for the observed activation, suggesting an RNAi-like effect was responsible for the observed activation (Li et al. 2006; Place et al. 2008). This observation is noteworthy as siRNAs shown to modulate TGS have generally been observed to require the action of Argonaute 1 (Ago-1), DNA methyltransferase (3a DNMT3a), and histone deacetylase 1 (HDAC-1) (reviewed in detail in (Morris 2009a; Morris 2009b))(Fig. 1). Nonetheless, the answer to the question as to how siRNAs targeted to A:T-rich regions in the p21 and E-cadherin promoters were utilizing Ago-2 to activate gene expression remained unknown.
During a foray into the mechanism of siRNA-mediated activation of p21 and E-cadherin, it was determined mechanistically that the A:T-rich siRNAs were actually targeting RNAi to an endogenous long antisense ncRNA (Morris et al. 2008). This observation suggested that possibly long antisense ncRNAs might be functioning as endogenous suppressors of p21 and E-cadherin, possibly regulating their epigenetic states and transcription. Notably, other tumor suppressor genes also appeared to be under this newly discovered regulatory mechanism of long antisense non-coding RNA (Yu et al. 2008). A model has begun to emerge based on several different bodies of work suggesting that long antisense ncRNAs are active regulators of gene transcription in human cells (Fig. 2). Mechanistically, this model suggests that the siRNA targeting and degradation of the gene-specific long antisense ncRNAs affects the ability of these RNAs to interact with and guide epigenetic silencing complexes to their particular targeted loci in either gene bodies or promoters of the sense/mRNA loci for the gene. The result of this targeting is the activation of gene expression (Fig. 2)(Morris et al. 2008; Yu et al. 2008; Camblong et al. 2007; Cho et al. 2005; Ebralidze et al. 2008; Katayama et al. 2005; Knee and Murphy 1997; Li et al. 2009; Tufarelli et al. 2003; Wahlestedt 2006).
Fig. 2.
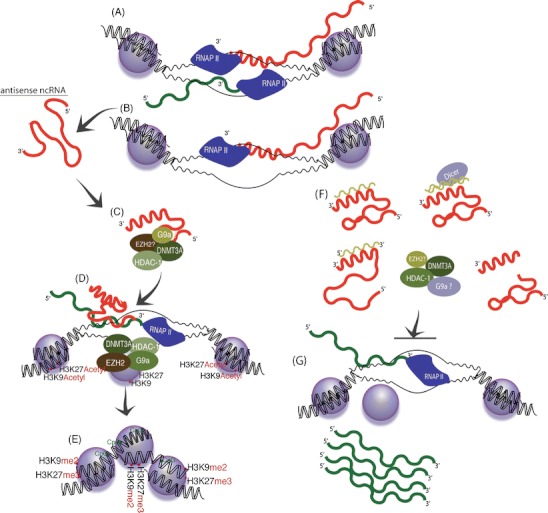
Endogenous antisense non-coding RNA mediated transcriptional silencing and activation. Long antisense non-coding RNAs emanating from either a bidirectionally transcribed, b alternative alleles, or pseudogenes, (Morris et al. 2008; Hawkins and Morris 2010) can fold into secondary structures (c) and interact with various proteins as well as target these proteins to homology containing loci in the genome (d). The antisense RNA protein complexes can then target the corresponding homology containing loci in the genome where epigenetic remodeling of the local chromatin via directed histone and DNA methylation can occur (e). When the antisense non-coding RNAs are targeted by either single-stranded oligonucleotides or siRNAs, the secondary structure is changed or interactions with the epigenetic remodeling protein complexes are blocked (f). The result is a loss of antisense non-coding RNA-directed epigenetic regulation of gene expression and subsequent activation of gene transcription (g)
Some antisense ncRNAs can function in trans to epigenetically regulate gene expression. In a recent study, an Oct-4 regulatory long-antisense ncRNA emanating from Oct-4 pseudogene 5 (Hawkins and Morris 2010) was found to be involved in epigenetically regulating Oct-4. Notably, this particular antisense ncRNA is not poly-adenylated, contains Oct-4 intronic sequences, and was found to specifically overlap segments of the Oct-4 promoter. The suppression of this Oct-4 antisense ncRNA resulted in a loss of H3K27 and H3K9 methylation at the Oct-4 promoter and activation of Oct-4 transcription (Hawkins and Morris 2010). The suppression of Ezh2 and G9a also had an impact on Oct-4 transcription; the loss of these factors resulted in increased transcription of Oct-4, suggesting that Oct-4 might be under epigenetic regulation via the action of this Oct-4 targeted long antisense ncRNA emanating from Oct-4 pseudogene 5 (Hawkins and Morris 2010).
Taken together, these data, along with those from studies carried out on p21 and p15 (Morris et al. 2008; Yu et al. 2008; Hawkins and Morris 2010), indicated that siRNA-directed gene activation is in essence “RNAi-mediated suppression of the endogenous ncRNA epigenetic suppressor”, which results in a loss of endogenous epigenetic brakes being instilled in the particular gene by the action of the long antisense ncRNA (Fig. 2 and reviewed in (Morris 2009a; Morris 2009b)). Others have also found ncRNAs acting in the modulation of epigenetic states of imprinted genes (Tsai et al. 2010). These new findings strongly implicate long antisense ncRNAs as epigenetic regulators of gene expression in human cells. Such observations offer significant insights into a major question in biology: How do epigenetic changes get instilled at particular loci?
While the insights gained from these bodies of work indicate that a vastly more complex regulatory network is operative in human cells they also suggest that some form of regulation can be instilled in a gene-specific manner, i.e., we can turn a gene on or off transcriptionally. Evidence with small RNA-targeted TGS has demonstrated that stable long-term silencing can be established after a relatively short duration of small RNA targeting to a gene promoter (Turner et al. 2009; Hawkins et al. 2009). This form of long-term silencing required Histone Deacetylase 1 (HDAC1), DNA methyltransferase 3A (DNMT3a), and epigenetic changes such as histone 3 lysine 9 di-methylation as well as histone 3 lysine 27 tri-methylation at the targeted site for the initiation of silencing. Conversely, the long-term maintenance of silencing required the action of DNA methyltransferase 1 (DNMT1) and DNMT3a as well as DNA methylation at the targeted promoter (Turner et al. 2009; Hawkins et al. 2009). The impact of a set of molecules that can govern this form of control cannot be overstated as long-lasting effects can, by extension, have an impact on the evolution of the cell. Interestingly, to date, the majority of genes exhibiting long antisense ncRNAs appear to be tumor suppressors (reviewed (Morris and Vogt 2010)). These observations might speak to a role for dysregulated long antisense ncRNA-based networks in the evolution of epigenetic silencing and cancer (Morris and Vogt 2010)?
Conclusion
The observations that antisense ncRNAs are involved in regulating gene expression in differentiated cells (reviewed in (Morris 2009b)) adds to the growing complexity of known long ncRNAs involved in cellular regulation. Examples include the Xist antisense ncRNA TSIX, which is involved in dosage compensation and X-inactivation in undifferentiated primordial cells (reviewed (Lee 2009)). Other examples of ncRNAs involved in epigenetic regulation can be found in the imprinted genes (reviewed in (Latos and Barlow 2009)). These long ncRNAs also appear to function by targeting the recruitment of different epigenetic regulatory complexes to their intended targets. Clearly, the emerging evidence suggests that in human cells many more genes than previously envisioned, beyond X-inactivation and imprinted genes might actually be regulated by long ncRNAs. In fact, ncRNAs might be actively switching on and off genes in an orchestral regulation that governs the fidelity of the cell and functions in cellular adaption. Knowledge of this mechanism can no doubt prove useful in future therapeutics aimed at controlling gene expression in a transcriptional manner. Future therapies may utilize small RNAs or various chemically modified oligonucleotides to epigenetically modulate a gene transcription or to target endogenous ncRNAs actively involved in regulating particular genes expression. While we may be clearly on the verge of a new therapeutic platform that allows for the targeted control of gene transcription via controlled epigenetic targeting, one issue will continue to remain: delivery.
Acknowledgments
This project is funded by NIH R01 HL083473 and NIH R01 AI084406 to KVM.
References
- Alberts BBD, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 3. New York: Garland Publishing; 1994. [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, Jorda MA, Petkova V, Rosenbauer F, Huang G, Dayaram T, Klupp J, O’Brien KB, Will B, Hoogenkamp M, Borden KL, Bonifer C, Tenen DG. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element . Genes Dev. 2008;22:2085–2092. doi: 10.1101/gad.1654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee R, Murphy PR. Regulation of gene expression by natural antisense RNA transcripts. Neurochem Int. 1997;31:379–392. doi: 10.1016/S0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- Latos PA, Barlow DP. Regulation of imprinted expression by macro non-coding RNAs. RNA Biol. 2009;6:100–106. doi: 10.4161/rna.6.2.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2009;115:133–139. doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Morris KV. Non-coding RNAs, epigenetic memory, and the passage of information to progeny. RNA Biol. 2009;6:242–247. doi: 10.4161/rna.6.3.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Vogt PK. Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle. 2010;9:2542–2545. doi: 10.4161/cc.9.13.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86:751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. Journal of RNAi and Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–23363. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Turner AM, Cruz J, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol Ther. 2009;17:360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]


