Abstract
Epigenetic mechanisms such as DNA hypermethylation and modifications of histone amino acids are known to play an important role in the control of gene expression both in normal human development and tumorigenesis. Hypermethylation of CpG islands within promoter regions of tumor suppressor genes is associated with transcriptional inactivation and represents, in addition to genetic aberrations, an important mechanism of gene silencing in the pathogenesis of human cancer. Inter-α-trypsine inhibitors (ITIs) are a family of serine protease inhibitors consisting of one light chain (bikunin) and two heavy chains (ITI heavy chains, ITIHs). ITIHs stabilize the extracellular matrix (ECM) by interacting with hyaluronic acid, which is a major ECM component. Hypermethylation in the upstream region of the promoter-associated CpG island of ITIH5, the most recently described member of the ITIH family, has been previously detected in breast cancer and was associated with an adverse outcome. In this study, we determined the DNA methylation status of the promoter region near the transcription start site of the ITIH5 tumor suppressor gene in leukemia cell lines and primary samples from patients with acute myeloid leukemia (AML) as well as the potential use of demethylating agents to restore a demethylated state of the promoter. Aberrant ITIH5 promoter hypermethylation occurred in 15 of 104 (14.4%) diagnostic AML samples. There were no statistically significant correlations between the ITIH5 methylation status and clinical prognostic parameters. Our results indicate that aberrant ITIH5 promoter hypermethylation is a novel epigenetic event in AML.
Keywords: DNA hypermethylation, Epigenetics, Acute myeloid leukemia, Inter-α-trypsine inhibitor, Tumor suppressor gene
Introduction
The pathogenesis of human cancer is characterized by a multistep process involving multiple molecular alterations leading to dysregulation of a variety of signaling pathways (Hanahan and Weinberg 2011). Acute myeloid leukemia (AML) is a clonal disorder of hematopoietic stem cells characterized by disrupted maturation and uncontrolled proliferation of immature progenitor cells and subsequent suppression of normal hematopoiesis (Löwenberg et al. 1999). Normal hematopoietic stem cells (HSC) are defined by their ability to exert self-renewal and multilineage differentiation and maturation. In contrast, leukemic stem cells appear to be transformed HSCs with a loss of control of both proliferation and maturation through accumulating genetic and epigenetic aberrations (Huntly and Gilliland 2005). Important genetic findings in AML comprise chromosomal translocations involving different transcription factors and activating point mutations in multiple signal transduction pathways (Tenen 2003). Furthermore, the ineffective hematopoiesis in AML is thought to be the result of dysregulation between the blasts and the surrounding hematopoietic microenvironment consisting of a variety of cells, including bone marrow stromal cells and their products, especially cytokines and extracellular matrix (ECM) molecules (Mayani 1996). This bone marrow microenvironment supports and regulates the proliferation and differentiation of hematopoietic cells (Bhatia et al. 1993). However, the role of the microenvironment itself in AML has not yet been well characterized.
DNA methylation of CpG islands associated with gene promoter regions is the most extensively studied epigenetic mechanism, which is not only crucial for the regulation of gene expression during normal mammalian development but also contributes to silencing of cancer-related genes in tumorigenesis (Herman and Baylin 2003; Esteller 2008). Recent advances in the rapidly evolving field of cancer epigenetics have shown extensive reprogramming of every component of the epigenetic machinery in cancer including DNA methylation, histone modifications, nucleosome positioning, and non-coding RNAs, specifically microRNA expression (Sharma et al. 2010). Additionally, epigenetic changes play an important role as an alternative mechanism of transcriptional inactivation of cancer-related genes in hematopoietic malignancies. These aberrations may thus contribute to enhanced proliferation and self-renewal, differentiation arrest as well as impaired apoptosis of leukemic blasts (Galm et al. 2006). Patterns of DNA methylation are non-random and tumor type specific, and this could also be shown in AML (Figueroa et al. 2010).
Inter-α-trypsine inhibitors (ITIs) are a family of plasma protease inhibitors consisting of one light chain (bikunin) and a variable set of two homologous heavy chains (ITIH1-5) which are linked by chondroitin sulfate, a glycosaminoglycan. ITIHs stabilize the ECM by covalently binding to hyaluronic acid, which is a major component of the ECM, forming so-called cable-like structures (Enghild et al. 1991; Chen et al. 1994; Bost et al. 1998). ITIs can be found at high concentrations in human plasma as well as in other compartments especially in the connective tissue (Zhuo et al. 2004). ITIs have been found to have inhibitory effects on tumor progression and metastasis in vitro owing to the protease inhibiting function of bikunin and the stabilization of the ECM by the ITIHs (Kobayashi et al. 1995; Bourguignon et al. 1999). ITIH5 is a novel member of the ITIH family and the only ITIH gene with a CpG-rich promoter region. Aberrant DNA hypermethylation associated with transcriptional silencing of the putative tumor suppressor gene ITIH5 has previously been found in breast cancer and reportedly has a negative prognostic impact in this disease (Himmelfarb et al. 2004; Veeck et al. 2008). In this study, using a candidate gene approach, we determined the methylation status of the promoter-associated CpG island of ITIH5 in leukemia cell lines and primary AML patient samples.
Materials and methods
Human tissue samples
After informed consent was given, bone marrow (BM) and peripheral blood (PB) specimens (72 BM and 32 PB) were obtained at the time of diagnosis during routine clinical assessment of 104 patients with AML, who presented at the University Hospital Aachen, Germany, between 1995 and 2008. Cytogenetic data were available in 90 of 104 patients. PB samples from healthy volunteers served as controls. Mononuclear cells from BM and PB were separated by density gradient centrifugation prior to further analysis. The clinical characteristics of the patient cohort are summarized in Table 1.
Table 1.
Characteristics of the patient cohort
| Total number of patients, n | 104 |
|---|---|
| Age (years, median and range) | 60.4 (21.0–89.1) |
| Gender, n | |
| Male | 50 |
| Female | 54 |
| Source of material, n | |
| BM | 72 |
| PB | 32 |
| FAB subtype, n | |
| M0 | 5 |
| M1 | 34 |
| M2 | 14 |
| M4 | 27 |
| M4eo | 11 |
| M5 | 12 |
| M7 | 1 |
| Karyotype, n | |
| Favorable | 16 |
| Intermediate | 60 |
| Unfavorable | 14 |
| No data | 14 |
| Laboratory parameters (median and range) | |
| WBC (109/l) | 21.4 (0.9–354) |
| Hemoglobin (g/l) | 91 (41–142) |
| Platelet count (109/l) | 60.5 (3–680) |
| LDH (U/l) | 496 (139–3761) |
FAB French–American–British, WBC white blood cell, LDH lactate dehydrogenase
Cell culture and drug treatment
We obtained the AML cell lines KG1a, HL-60, and GDM-1 as well as the Hodgkin’s lymphoma cell lines L-428, L-540, and L-1236 from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Cells were routinely cultured in RPMI 1640 (Invitrogen, Karlsruhe, Germany) supplemented with 10–20% fetal calf serum (Biochrom AG, Berlin, Germany). For demethylation studies, the AML cell lines HL-60 and KG1a were incubated with or without a final concentration of 1.0 μM 5-Aza-2′-deoxycytidine (DAC; Sigma, St. Louis, MO, USA) for 96 h.
Sodium bisulfite treatment and methylation-specific polymerase chain reaction
Genomic DNA was isolated from cell lines and primary tissues using standard methods. A purification of leukemic blasts prior to further analysis was not performed owing to the high sensitivity of the methylation-specific polymerase chain reaction (MSP) technique (Herman et al. 1996). Approximately 1 μg DNA was sodium bisulfite-modified and subjected to MSP with primers specifically recognizing the unmethylated or the methylated sequence of the ITIH5 gene, respectively. Although MSP primers and reaction conditions for the ITIH5 gene have been published previously (Veeck et al. 2008), we decided to design new primers annealing closer to the transcription start site (Fig. 1). The primer-specific reaction temperature for MSP was 63°C. The primer sequences used in this study are shown in Table 2.
Fig. 1.
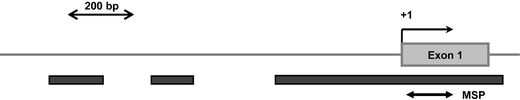
Schematic representation of the human ITIH5 promoter region. Dark gray bars indicate location of the three promoter-associated CpG islands. The bold double-headed arrow shows the region amplified by MSP
Table 2.
MSP primer sequences specific for the ITIH5 promoter region
| Primer | Primer sequence | Amplicon length (bp) |
|---|---|---|
| ITIH5 U sense | G TTG GAG TTT TGG GTG TTG TAA AGT | 141 |
| ITIH5 U antisense | CCC AAC TCT ACA CCT CTT CTT ACA | |
| ITIH5 M sense | TTG GAG TTT TGG GCG TTG TAA AGC | 139 |
| ITIH5 M antisense | CCA ACT CTA CGC CTC TTC CTA CG |
Statistical analysis
Overall survival curves were plotted according to the method of Kaplan and Meier and compared using the log-rank test. Survival was calculated from the date of diagnosis until the patients’ death or last visit. Correlations between variables were analyzed using the Fisher’s exact two-sided test. A p value <0.05 was considered to be statistically significant. All calculations were performed using the SAS statistical software version 9.1.3 (SAS Institute Inc., Cary, NC, USA).
Results
MSP analysis revealed that the ITIH5 promoter region was fully hypermethylated in the AML cell lines HL-60, KG1a, and GDM-1 as well as in the Hodgkin’s lymphoma cell lines L-428, L-540, and L-1236 (Fig. 2). In the cell lines HL-60 and KG1a, treatment with the demethylating agent DAC for 96 h at a 1.0 μM concentration induced a partial promoter demethylation (Fig. 3). We then analyzed the ITIH5 methylation status in mononuclear cells obtained from 104 patients with newly diagnosed AML. The frequency of aberrant methylation among the primary patient samples was 14.4% (15 of 104; Fig. 4).
Fig. 2.
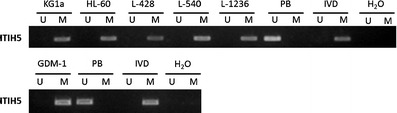
Representative MSP analyses of the ITIH5 gene in cell lines (AML: KG1a, HL-60, GDM-1; Hodgkin’s lymphoma: L-428, L-540, L-1236). DNA from peripheral blood cells of healthy donors, in vitro methylated DNA, and water served as controls. Lane U amplified product with primers recognizing the unmethylated ITIH5 sequence, lane M amplified product recognizing the methylated ITIH5 sequence
Fig. 3.
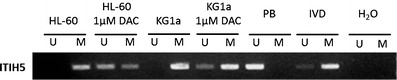
Representative MSP analyses of the ITIH5 gene in AML cell lines HL-60 and KG1a after treatment with 1 μM DAC for 96 h. DNA from peripheral blood cells of healthy donors, in vitro methylated DNA, and water served as controls. Lane U amplified product with primers recognizing the unmethylated ITIH5 sequence, lane M amplified product recognizing the methylated ITIH5 sequence
Fig. 4.
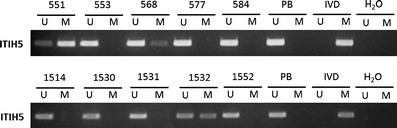
Representative MSP analyses of the ITIH5 gene in primary AML patient samples. DNA from peripheral blood cells of healthy donors, in vitro methylated DNA, and water served as controls. Lane U amplified product with primers recognizing the unmethylated ITIH5 sequence, lane M amplified product recognizing the methylated ITIH5 sequence
Among clinical prognostic parameters, we found no significant correlation between hypermethylation of ITIH5 and cytogenetics (Table 3), elevated serum levels of lactate dehydrogenase, French–American–British subtype, gender, age, peripheral blood cell counts, and overall survival (OS; p = 0.08; Fig. 5). Notably, median WBC was elevated in patients with ITIH5 hypermethylation compared to those without, though the difference was not statistically significant (73.3 × 109/l vs. 49.9 × 109/l, p = 0.24).
Table 3.
Association between the ITIH5 methylation status and cytogenetic findings in the patient cohort
| Cytogenetic risk | ITIH5 hypermethylation (%) |
|---|---|
| Total number of patients (n = 104) | 14.4% (15/104) |
| Favorable risk (n = 16) | 18.8% (3/16) |
| inv(16) (n = 11) | 27.3% (3/11) |
| t(8;21) (n = 5) | 0% |
| Intermediate risk (n = 60) | 11.7% (7/60) |
| Unfavorable risk (n = 14) | 7.1% (1/14) |
| Karyotype n.a. (n = 14) | 28.6% (4/14) |
n.a. not available
Fig. 5.
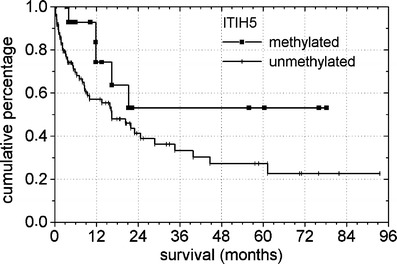
Survival curves of patients with primary AML according to the ITIH5 methylation status at diagnosis (p = 0.08). In a total of 104 patients analyzed, 15 showed aberrant ITIH5 promoter hypermethylation
Discussion
Recent advances in the field of epigenetics have shown that human cancer cells harbor global epigenetic abnormalities in addition to numerous genetic alterations (Sharma et al. 2010). Among these epigenetic aberrations, DNA hypermethylation is the one which has been most extensively studied. With regard to our findings, we conclude that promoter hypermethylation of ITIH5 is a novel epigenetic event in AML that may contribute to leukemogenesis by interfering with the interaction between the ECM and the leukemic clone. It has been shown that in the BM of AML patients, the ECM is enriched in hyaluronic acid, although the impact of this alteration still remains poorly defined (Sundström et al. 2005). Since there was a trend toward an association between increased WBC and ITIH5 promoter hypermethylation, we hypothesize that epigenetic dysregulation of ITIH5 may result in an impaired interaction between the leukemic clone and its’ surrounding ECM in the BM. This may potentially lead to either increased peripheral WBC or a higher turnover of leukemic cells. Additionally, the OS curve implies a slight advantage for patients who carry a hypermethylated ITIH5 promoter region, though this difference was not statistically significant. However, since little is known about the interaction between leukemic blasts and their surrounding ECM, further studies are required to better understand the functional consequences and the impact of a dysregulation of the blast–matrix interaction on the pathogenesis and the course of AML as well as the function of ITIH5 in this setting. Additionally, our cell line data indicate that epigenetic dysregulation of ITIH5 may also play a role in the pathogenesis of Hodgkin’s lymphoma.
Epigenetic silencing of cancer-related genes has been shown to be reversible. Consequently, epigenetic alterations are potential targets of great interest for a molecular targeted therapy in human malignancies (Gilbert et al. 2004). In this study, we have shown that treatment of the AML cell lines KG1a and HL-60 with the demethylating agent DAC resulted in partial demethylation of the ITIH5 promoter region thus allowing the restoration of a potentially silenced gene expression. The use of the demethylating agents 5-azacytidine and DAC has already been approved for the treatment of myelodysplastic syndromes (Kantarjian et al. 2006). The partial reversal of the ITIH5 promoter hypermethylation may contribute to the beneficial effect of an epigenetically targeted therapy in AML. Further studies are warranted to assess the impact of ITIH5 promoter hypermethylation on ITIH5 expression levels.
Since our patient cohort only consisted of 104 patient samples and was heterogeneous for age, risk factors, and treatment regimens, no definitive conclusion can be retained from these data regarding the prognostic impact of ITIH5 promoter hypermethylation. Therefore, the analysis of a larger number of well-characterized patient samples may be useful to further clarify the possible role of aberrant ITIH5 methylation as a biomarker in AML. Consequently, additional studies are necessary to elucidate the functional role of epigenetic dysregulation of ITIH5 in leukemogenesis.
Acknowledgments
We thank Claudia Schubert, Melanie Hoffmann, Lucia Vankann, and Peter Glatte for the expert technical assistance.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Bhatia R, McGlave PB, Dewald GW, Blazar BR, Verfaillie CM. Abnormal function of the bone marrow microenvironment in chronic myelogenous leukemia: role of malignant stromal macrophages. Blood. 1993;85:3636–3645. [PubMed] [Google Scholar]
- Bost F, Diarra-Mehrpour M, Martin JP. Inter-alpha-trypsin inhibitor proteoglycan family—a group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem. 1998;252:339–346. doi: 10.1046/j.1432-1327.1998.2520339.x. [DOI] [PubMed] [Google Scholar]
- Bourguignon J, Borghi H, Sesboüé R, Diarra-Mehrpour M, Bernaudin JF, Métayer J, Martin JP, Thiberville L. Immunohistochemical distribution of inter-alpha-trypsin inhibitor chains in normal and malignant human lung tissue. J Histochem Cytochem. 1999;47:1625–1632. doi: 10.1177/002215549904701214. [DOI] [PubMed] [Google Scholar]
- Chen L, Mao SJ, McLean LR, Powers RW, Larsen WJ. Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J Biol Chem. 1994;269:28282–28287. [PubMed] [Google Scholar]
- Enghild JJ, Salvesen G, Hefta SA, Thøgersen IB, Rutherfurd S, Pizzo SV. Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-alpha-inhibitor. J Biol Chem. 1991;266:747–751. [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Gilbert J, Gore SD, Herman JG, Carducci MA. The clinical application of targeting cancer through histone acetylation and hypomethylation. Clin Cancer Res. 2004;10:4589–4596. doi: 10.1158/1078-0432.CCR-03-0297. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb M, Klopocki E, Grube S, Staub E, Klaman I, Hinzmann B, Kristiansen G, Rosenthal, Dürst M, Dahl E. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004;204:69–77. doi: 10.1016/j.canlet.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Huntly BJP, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, Bennet JM, Albitar M, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Gotoh J, Hirashima Y, Fujie M, Sugino D, Terao T. Inhibitory effect of a conjugate between human urokinase and urinary trypsin inhibitor on tumor cell invasion in vitro. J Biol Chem. 1995;270:8361–8366. doi: 10.1074/jbc.270.14.8361. [DOI] [PubMed] [Google Scholar]
- Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- Mayani H. Composition and function of the hemopoietic microenvironment in human myeloid leukemia. Leukemia. 1996;10:1041–1047. [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström G, Dahl IM, Hultdin M, Lundström B, Wahlin A, Engström-Laurent A. Bone marrow hyaluronan distribution in patients with acute myeloid leukemia. Med Oncol. 2005;22:71–78. doi: 10.1385/MO:22:1:071. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Veeck J, Chorovicer M, Naami A, Breuer E, Zafrakas M, Bektas N, Dürst M, Kristiansen G, Wild PJ, Hartmann A, Knuechel R, Dahl E. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene. 2008;27:865–876. doi: 10.1038/sj.onc.1210669. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Hascall VC, Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein–glycosaminoglycan–protein complex. J Biol Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]


