Abstract
The nuclear import receptor Kap114 carries transcription factors and other cargos across nuclear pores into the nucleus. Here we show that yeast Kap114 is modified by SUMO (small ubiquitin-related modifier) and that sumoylation is required for Kap114-mediated nuclear import. Among the four known SUMO-specific E3 ligases in yeast, Mms21 is the preferred E3 enzyme responsible for the covalent attachment of SUMO to the Kap114 protein. Kap114 is sumoylated on lysine residue 909, which is part of a ΨKxD/E sumoylation consensus motif. Kap114 containing a lysine-to-arginine point mutation at position 909 mislocalizes to the nucleus and is defective in promoting nuclear import. Similarly, mutants defective in sumoylation or desumoylation specifically accumulate Kap114 in the nucleus and are blocked in import of Kap114 cargos. Ran–GTP is not sufficient to disassemble Kap114/cargo complexes, which necessitates additional cargo release mechanisms in the nucleus. Remarkably, sumoylation of Kap114 greatly stimulates cargo dissociation in vitro. We propose that sumoylation occurs at the site of Kap114 cargo function and that SUMO is a cargo release factor involved in intranuclear targeting.
Keywords: Kap114, nuclear pore complex, nucleocytoplasmic transport, Ran GTPase, SUMO
Introduction
Proteins are selectively transported through the central channel of the nuclear pore complex (NPC) in and out of the nucleus. The majority of macromolecules is actively translocated by specific transport factors belonging to the importin-β superfamily, which includes importins and exportins. The small GTPase Ran (Gsp1 in Saccharomyces cerevisiae), which is predominantly bound to GTP in the nucleus and to GDP in the cytoplasm, determines the direction of nucleocytoplasmic transport. The interaction with nuclear Ran–GTP has opposite consequences for importins and exportins. Binding of Ran–GTP to importin/cargo complexes dissociates cargos, while binding of Ran–GTP to exportins is required for cargo association (for reviews see Fried and Kutay (2003), Pemberton and Paschal (2005) and Cook et al (2007)). In mammalian cells, 20 members of the importin-β superfamily have been identified, whereas the yeast S. cerevisiae contains 14 β-receptors (Quan et al, 2008), which separate into ten importins and four exportins. Typical characteristics of these transport factors are around 40 continuous α-helical structures building so-called HEAT repeats. The N terminus forms the Ran/Gsp1 binding domain, while the cargo interacts with a large concave surface of a superhelix provided by the C-terminal domain (Fried and Kutay, 2003; Pemberton and Paschal, 2005; Cook et al, 2007). Most of the β-receptors usually interact directly with their transport substrate. Several transport substrates for the non-essential yeast importin Kap114 have been identified. It directly binds to its import cargos in a Gsp1–GTP-sensitive manner and the nuclear import of its cargos is affected in kap114 knockout cells. Kap114 mediates the nuclear import of the TATA-binding protein Tbp1 (Spt15; Morehouse et al (1999); Pemberton et al (1999)) and of the transcription factor IIB Sua7 (Hodges et al, 2005). Together with other β-receptors, Kap114 is the importin for the histones H2A and H2B (Mosammaparast et al, 2001; Greiner et al, 2004) and for the ribosomal assembly factor Rpf1 (Caesar et al, 2006). Kap114 is also involved in the nuclear import of the nucleosome assembly factor Nap1 (Mosammaparast et al, 2002).
Sumoylation of proteins is a reversible process, in which SUMO (small ubiquitin-related modifier) is covalently and posttranslationally attached to target proteins. Four SUMO genes are present in mammalian cells, whereas there is only one essential gene, SMT3, in yeast. In contrast to ubiquitination, sumoylation of proteins does not induce their degradation but contributes to the regulation of various cellular mechanisms like transcription, DNA repair, chromosomal stability, and cell cycle progression (reviewed in Hay, (2005); Geiss-Friedlander and Melchior (2007); Wilkinson and Henley (2010)). Similar to ubiquitination, three steps are necessary to attach Smt3 to its targets. After Smt3 precursor processing by the specific protease Ulp1 (Li and Hochstrasser, 2000), the E1 heterodimeric enzyme Aos1/Uba2 activates Smt3 in an ATP-dependent manner, forming a high energetic thioester bond between Uba2 and Smt3 (Dohmen et al, 1995; Johnson et al, 1997). Smt3 is then conjugated to the E2 enzyme Ubc9 via a thioester bridge (Johnson and Blobel, 1997). Finally, Smt3 is covalently bound to the target protein with the assistance of E3 ligases, which determine the target specificity. Four E3 enzymes have been identified in yeast, Siz1 (Takahashi et al, 2001), Nfi1 (Siz2; Johnson and Gupta, 2001), Mms21 (Zhao and Blobel, 2005), and Cst9 (Zip3) (Cheng et al, 2006), of which only Mms21 is essential for viability. In the target protein, a lysine residue is sumoylated that is usually part of the consensus sequence ΨKxD/E, where Ψ is a large hydrophobic residue. The Smt3-specific proteases Ulp1 and Ulp2 deconjugate Smt3 from their targets. Therefore, sumoylation is a reversible process.
In this study we show that sumoylation of yeast Kap114 is required for its function as an importin. Kap114 is SUMO-modified on lysine residue 909 in vivo and in vitro. A replacement of lysine residue 909 by an arginine residue leads to a nuclear accumulation of Kap114 and the simultaneous cytoplasmic mislocalization of the Kap114 import cargos like Tbp1 and Sua7. Similarly, the nuclear import of Kap114-specific transport substrates is inhibited in mutants affected in the sumoylation cycle. We show that sumoylation of Kap114 facilitates the dissociation of the Kap114/cargo complexes mediated by Gsp1–GTP, indicating that sumoylation plays a role in intranuclear targeting.
Results
Kap114 is sumoylated in vitro and in vivo
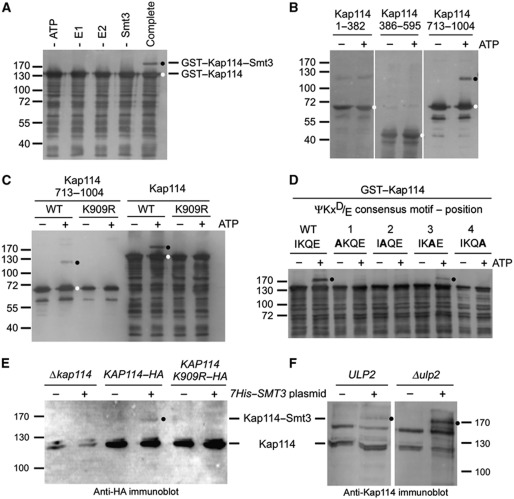
To identify modifications by Smt3 potentially influencing nucleocytoplasmic trafficking, we screened all β-receptors of the yeast S. cerevisiae as GST fusion proteins using an in vitro sumoylation assay with recombinant proteins purified from E. coli lysates. The receptors were incubated with the Aos1/Uba2 E1 heterodimer, the E2 enzyme Ubc9, Smt3, and ATP. A SUMO modification is indicated by a band shift to higher molecular weight in SDS gels. Such a band shift was readily detected for the importin Kap114 even in the absence of an E3 ligase (Figure 1A). The formation of the high molecular weight product was sumoylation-specific as the omission of single components of the sumoylation reaction did not result in modified Kap114. Furthermore, Kap114–Smt3 was immunodetectable with anti-GST or anti-Kap114 antibodies (Figure 1A).
Figure 1.
Kap114 is sumoylated in vitro and in vivo on lysine residue 909. (A) Kap114 is sumoylated in vitro in the absence of an E3 ligase. Purified GST–Kap114 was incubated with 2 μg Aos1/Uba2 (E1), 0.75 μg Ubc9 (E2), 7.5 μg Smt3 (SUMO), and 5 mM ATP (complete mixture), as described in Materials and methods, at 30°C for 90 min. In control reactions, ATP, E1, E2, or Smt3 was omitted as indicated. (B) Kap114 is sumoylated at its C terminus. GST fusion proteins with Kap1141–382, Kap114386–595, and Kap114713–1004 were incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP at 30°C for 90 min. (C) Sumoylation of Kap114 occurs on lysine residue 909. GST fused to Kap114713–1004 or full-length Kap114 either unmutated (WT) or carrying the point mutation K909R was incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP. After sumoylation, GST fusion proteins were purified by pulldown assays using glutathione sepharose (A–C). (D) Sumoylation occurs at the ΨKxD/E consensus motif between residues 908 and 911 of Kap114. GST fusion proteins with full-length WT Kap114 or the point mutants I908A, K909A, Q910A, and E911A were incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP at 30°C for 90 min. (E) Kap114 is sumoylated in vivo. Yeast cells deleted for KAP114 transformed with plasmids pRS426–KAP114–HA, pRS426–KAP114 K909R–HA, and YEp181–CUP–HIS7–SMT3 as indicated were grown in liquid cultures in the presence of copper sulphate for synthesis of His-tagged Smt3. (F) Sumoylation of Kap114 is increased in Δulp2 mutants. WT cells or ulp2 knockout cells transformed with plasmids pRS426–KAP114–HA and YEp181–CUP–HIS7–SMT3 were grown in liquid cultures in the presence of copper sulphate for synthesis of His-tagged Smt3. The cells were collected and resuspended in buffer containing 6 M guanidine–HCl. After glass bead lysis, cleared lysates were incubated with Ni-NTA agarose. Bound proteins were washed with buffer containing 8 M urea and eluted with SDS buffer (E, F). All proteins were separated by SDS–PAGE and detected by immunoblotting analysis using Kap114-specific affinity-purified antibodies (A–D, F) or anti-HA antibodies (E) and peroxidase-coupled secondary antibodies. Molecular weight markers are shown in kDa. Unmodified proteins are marked with a white dot, sumoylation products are labelled with a black dot.
In order to identify the sumoylation site, we divided Kap114, which comprises 1004 amino acid residues, into three non-overlapping fragments covering residues 1–382, 386–595, and 713–1004, respectively. Only the fragment containing Kap114 residues 713–1004 was modified by Smt3, indicating that Kap114 is sumoylated near its C terminus (Figure 1B). During sumoylation the carboxyl group of the C-terminal Smt3 glycine residue is covalently linked via an isopeptide bond to a lysine side chain of the target. The sequence around the sumoylation site typically fits to the consensus ΨKxD/E. Computer programs such as the SUMOplot software (Abgent, San Diego, CA) predicted the motif 908IKQE911 within the Kap114 sequence as a high probability sumoylation site. Therefore, a point mutation was introduced by site-directed mutagenesis to change the lysine residue 909 of Kap114 into an arginine residue. We observed that neither the Kap114 713–1004 fragment nor full-length Kap114 containing this K909R mutation was SUMO-modified, while the unmutated proteins were sumoylated (Figure 1C). This shows that the lysine residue 909 of Kap114 can be modified by Smt3. To investigate whether the Kap114 sequence 908IKQE911 represents a ΨKxD/E sumoylation consensus motif, we performed an alanine scan of the region encompassing the putative motif (Figure 1D). We found that the conserved residues at positions 1, 2, and 4 of the sequence are essential for sumoylation in vitro. The Q910 residue at position 3 is not strictly required for sumoylation, but contributes to efficient modification of Kap114 by Smt3. In accordance with earlier reports (Rodriguez et al, 2001; Sampson et al, 2001), this verifies that the 908IKQE911 sequence of Kap114 represents a typical ΨKxE sumoylation consensus motif.
We next investigated whether Kap114 is sumoylated in vivo. Yeast cells deleted for the KAP114 gene were transformed with plasmids encoding Kap114 fused to a C-terminal haemagglutinin (HA) tag. The cells synthesizing Kap114–HA or Kap114–HA containing the K909R mutation were additionally transformed with a plasmid coding for 7His-tagged Smt3. To prevent desumoylation by the Smt3-specific deconjugases, SUMO conjugates were purified from cell lysates by nickel pulldown assays under denaturing conditions. Figure 1E shows that sumoylated Kap114–HA was detected in nickel eluates by western blotting using anti-HA antibodies. While some unmodified Kap114–HA bound unspecifically to Ni-NTA beads, Smt3-conjugated Kap114–HA was observed only in KAP114–HA cells expressing 7His–SMT3. The appearance of Smt3-modified Kap114 was undetectable in the absence of 7His–Smt3 and in Δkap114 cells. Therefore, the sumoylation was specific. Furthermore, no sumoylated Kap114 was found in KAP114–HA K909R cells. This shows that the lysine residue at position 909 of Kap114 is sumoylated in vivo. We observed only a low efficiency of Kap114 sumoylation, which is usually reported also for many other SUMO targets and is partly due to the extremely rapid deconjugation of sumoylation products during cell lysis even though denaturing conditions are used. Because sumoylation is very dynamic, the SUMO modification has a short half-life based on constant cycles of deconjugation and conjugation. This explains the commonly observed very low steady-state levels of substrate modification (Johnson, 2004; Hay, 2005). For this reason, we aimed to specifically block SUMO deconjugation by examining the Δulp2 mutant. We found that the degree of sumoylation indeed dramatically increased in these cells (Figure 1F).
The localization of Kap114 is regulated by sumoylation
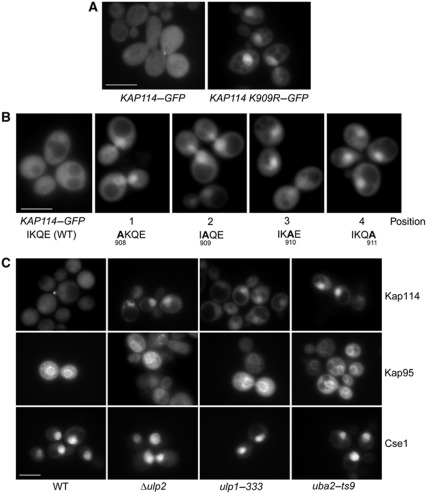
To analyse whether sumoylation of Kap114 affects its cellular localization, we introduced the K909R mutation into a plasmid encoding Kap114–GFP. KAP114 knockout cells containing this plasmid were compared to cells carrying non-mutated KAP114–GFP by direct fluorescence microscopy. In accordance with earlier observations (Pemberton et al, 1999), Kap114–GFP was equally distributed in the nucleoplasm and the cytoplasm (Figure 2A). In contrast, Kap114 K909R–GFP is located primarily in the nucleus, indicating that sumoylation of Kap114 affects its cellular distribution. As Kap114 is a protein that shuttles between the cytoplasm and the nucleoplasm, the nuclear accumulation is probably caused by either nuclear retention or by an inhibition of export to the cytoplasm.
Figure 2.
Sumoylation of Kap114 is required for its proper cellular localization. (A) Kap114 K909R accumulates in the nucleus. Δkap114 cells containing pRS316–KAP114–GFP or pRS316–KAP114 K909R–GFP were cultured in liquid medium at 30°C. (B) The mutagenesis of the ΨKxD/E sumoylation consensus motif between residues 908 and 911 affects the cellular distribution of Kap114. Cells deleted for KAP114, transformed with pRS316–KAP114–GFP plasmids, and carrying the unmutated allele or the indicated KAP114 mutations were grown in liquid medium. (C) WT cells, Δulp2, ulp1–333, or uba2–ts9 mutants containing the plasmids pRS316–KAP114–GFP, pRS316–GFP–KAP95, or pRS316–GFP–CSE1 were cultured in liquid media at 30°C (WT and Δulp2) or 25°C (ulp1–333 and uba2–ts9). Logarithmically growing cells were analysed by direct fluorescence microscopy using a Zeiss Axioscope microscope at 1000-fold magnification to determine the localization of the GFP fusion proteins. Scale bar, 5 μm.
We then introduced the above described alanine point mutations into Kap114–GFP (Figure 2B). The mutagenesis at positions 1, 2, and 4 of the 908IKQE911 sequence led to nuclear accumulation of Kap114 similar to the K909R mutant. The Q910A (position 3) mutant showed an intermediate phenotype, most cells accumulated Kap114 in the nucleus but many cells exhibited a weaker mislocalization. This indicates that, like it was observed in vitro, the Q residue at position 3 is of minor importance for sumoylation of Kap114. These observations together confirm that the Kap114 908IKQE911 peptide forms a ΨKxD/E sumoylation consensus motif.
To analyse whether the SUMO modification pathway has an impact on the localization of Kap114, we used the mutants ulp1–333, Δulp2, and uba2–ts9. These strains are expected to have defects in either sumoylation (uba2–ts9) or desumoylation (ulp1–333, Δulp2; Schwienhorst et al (2000), Bylebyl et al (2003), Li and Hochstrasser (2003)). Additionally, ulp1 mutants are defective in the conversion of the Smt3 precursor to the mature protein (Li and Hochstrasser, 2000). Figure 2C shows that Kap114–GFP accumulated in the nucleus of these mutants. The localization pattern is comparable to that of Kap114 K909R–GFP. The temperature-sensitive uba2–ts9 and ulp1–333 mutants showed this mislocalization phenotype even at the permissive temperature. The nuclear accumulation of Kap114–GFP in these strains was somewhat more pronounced after a 1-h shift to the non-permissive temperature of 37°C (not shown). The localization of the importins Kap120 (not shown) and Kap95 (importin-β), as well as the localization of the exportin Cse1 in the examined mutants was determined as controls (Figure 2C). In contrast to Kap114, the localization of these β-receptors was the same in SUMO pathway mutants and in wild-type (WT) cells. Thus, Kap114 specifically mislocalizes to the nucleus in mutants defective in sumoylation and deconjugation. Considering the predominantly nuclear localization of the SUMO modification pathway proteins (Li and Hochstrasser, 2000; Huh et al, 2003; Makhnevych et al, 2007; Sydorskyy et al, 2010), our results altogether indicate that continuous sumoylation and desumoylation of Kap114 in the nucleoplasm is required for its proper cellular distribution.
The nuclear import of Kap114 cargos is specifically inhibited in sumoylation mutants
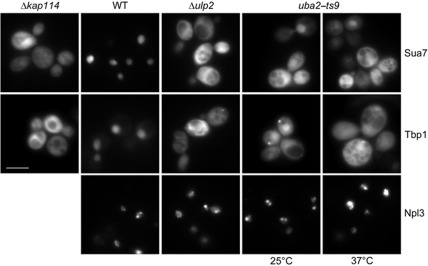
Next we investigated whether the nuclear accumulation of Kap114 in sumoylation pathway mutants affects the nuclear uptake of Kap114-specific transport substrates. The essential proteins Sua7 and Tbp1 are both involved in transcriptional processes and are completely nuclear in WT cells. To increase the size of these proteins above the diffusion limit of the nuclear pore (∼50 kDa), we fused them to a GST–GFP moiety. In accordance with earlier observations (Morehouse et al, 1999; Pemberton et al, 1999; Hodges et al, 2005), the Sua7 and Tbp1 GFP fusion proteins mislocalize to the cytoplasm in kap114 null cells (Figure 3). However, both proteins were not excluded from the nucleus suggesting that other importins can also mediate the import to some degree. In Δulp2 mutants, Sua7 and Tbp1 accumulate strongly in the cytoplasm indicating a severe import defect. The nuclear uptake of the reporter proteins was similarly defective in uba2–ts9 cells already at the permissive temperature. After a shift to the non-permissive temperature, nuclear import was inhibited stronger (Figure 3). On the contrary, the nuclear uptake of the mRNA-binding protein Npl3, which is transported through the NPC by the importin Mtr10 (Pemberton et al, 1997; Senger et al, 1998), was not affected in Δulp2 and uba2–ts9 cells indicating that the investigated mutants do not exhibit a general import defect. This suggests that the sumoylation of Kap114 specifically regulates the nuclear import of its cargos.
Figure 3.
Sua7 and Tbp1 accumulate in the cytoplasm of sumoylation mutants. Δkap114 cells, WT cells as well as Δulp2 and uba2–ts9 mutants were transformed with plasmids YCpGAL–GST–GFP–SUA7, YCpGAL–GST–GFP–TBP1, or YEpGAL–GFP–NPL3. The strains were cultured in liquid media containing 2% raffinose at 25°C (uba2–ts9 only) or at 30°C. For induction of the GFP fusion protein synthesis, the cells were complemented with 2% galactose and incubated further for 2 h. After 1 h of galactose induction, the uba2–ts9 culture was split and one-half was shifted to 37°C for 1 h. The cellular distribution of the GFP fusion proteins was analysed by direct fluorescence microscopy. Scale bar, 5 μm.
If mislocalization of Sua7 and Tbp1 in ulp2 and uba2 mutants is Kap114-specific, overexpression of KAP114 is expected to rescue this effect. To test this we transformed single copy (CEN) or multicopy (2 μ) KAP114 plasmids into Δulp2 and uba2–ts9 cells. We determined by immunoblot analysis that the amount of Kap114 was ∼2.5-fold higher in cells containing the KAP114 CEN plasmid, whereas it was ∼4-fold higher when the KAP114 2μ plasmid had been transformed (Supplementary Figure 1B). As anticipated, Sua7 and Tbp1 were completely nuclear in Δkap114 cells transformed with the KAP114 plasmids (Supplementary Figure 1A). The localization defect of Sua7 and Tbp1 in ulp2 and uba2 mutants was rescued by KAP114 overexpression as well. A substantial recovery was mediated by the single copy KAP114 plasmid with a predominant nuclear localization of Sua7 and Tbp1, whereas these proteins were completely nuclear in cells harbouring the multicopy plasmid (Supplementary Figure 1A). The dose-dependent rescue indicates that Kap114 is limiting in the cytoplasm of sumoylation mutants.
Lysine residue 909 of Kap114 is required for efficient nuclear import
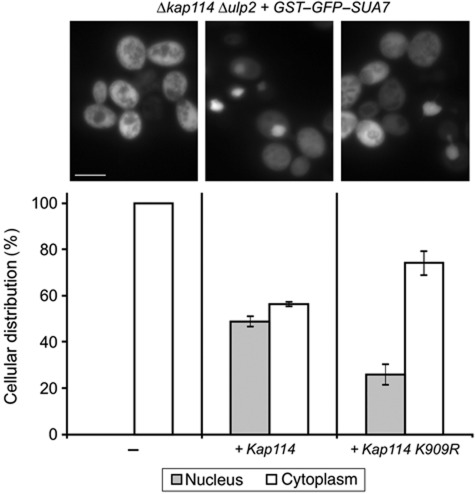
To directly test whether sumoylation of Kap114 is required for the function of Kap114 as an importin, we constructed a Δkap114 Δulp2 double mutant. These cells were transformed with single copy plasmids either containing WT KAP114 or the unsumoylatable lysine-to-arginine substitution mutant KAP114 K909R. The nuclear import of Sua7 viewed by fluorescence microscopy was inhibited in 100% of Δkap114 Δulp2 cells (Figure 4, left panel). Import was partially restored in Δkap114 Δulp2 mutants containing the KAP114 plasmid. Overall, 45% of the cells showed a nuclear localization of Sua7 (Figure 4, middle panel). The higher import rate compared to Δulp2 cells (see Figure 3) is explained by the ∼50% higher expression level of plasmid-encoded Kap114 versus genomically encoded Kap114 (Supplementary Figure 1B). Remarkably, the Δkap114 Δulp2 KAP114 K909R mutants exhibited a significantly lower import rate with only 25% of the cells accumulating Sua7 within the nucleus (Figure 4, right panel), indicating that sumoylation of lysine residue at position 909 in Kap114 is required for efficient nuclear import. A very similar phenotype was observed in Δkap114 ULP2 mutants (data not shown). To test with isolated proteins whether Kap114 K909R has a defect in Gsp1–GTP binding, we bound 6His-tagged Kap114 and Kap114 K909R to immobilized Gsp1–GTP and found no difference in their binding efficiency (Supplementary Figure 2). Thus, we can conclude that purified Kap114 K909R is functional with respect to interaction with the Ran GTPase.
Figure 4.
Rescue of Sua7 mislocalization by WT and mutant KAP114. The Δkap114 Δulp2 double mutant carrying the plasmid YCpGAL–GST–GFP–SUA7 was transformed with empty vector, pRS315–KAP114, or pRS315–KAP114 K909R. The cells were cultured in liquid medium containing raffinose, and synthesis of the GFP–Sua7 fusion protein was induced by the addition of 2% galactose. After 2 h, the strains were analysed by direct fluorescence microscopy. The number of cells showing a predominantly nuclear versus a cytoplasmic localization of the GST–GFP–Sua7 fusion protein was quantified. Data shown in the bar graphs represent the average values±standard deviations of three independent experiments. Scale bar, 5 μm.
Mms21 is the preferred E3 ligase for Kap114
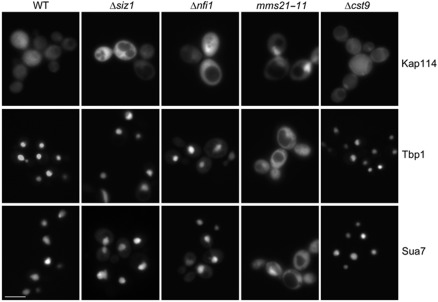
Four E3 ligases have been identified in S. cerevisiae. To determine which protein is the corresponding E3 ligase for Kap114 in vivo, we analysed the localization of Kap114 in E3 mutants (Figure 5). Whereas Kap114 was located throughout the cell in WT and Δcst9 cells (but was excluded from the vacuoles), it was primarily nuclear in mms21-11 mutants. In Δsiz1 and Δnfi1 cells, Kap114 showed a predominantly even cellular distribution. However, some cells possessed a weak nuclear signal. We then assayed for the nuclear import of Sua7 and Tbp1 in these mutants (Figure 5). The Kap114 cargo molecules were entirely nuclear in WT and Δcst9 cells. However, in mms21-11 cells they were strongly mislocalized to the cytoplasm. This import defect was again rescued by overexpression of KAP114 (Supplementary Figure 1A), which suggests that Kap114 is limiting for import in the cytoplasm of mms21-11 mutants. Sua7 and Tbp1 were mostly nuclear in Δsiz1 and Δnfi1 mutants, but many cells additionally exhibited a faint cytoplasmic signal indicating a very weak import defect. We conclude that Cst9 is not required for proper localization of Kap114 and its import substrates (IS). However, Mms21 is necessary for both the correct cellular localization of Kap114 and for efficient nuclear uptake of Tbp1 and Sua7. This suggests that Mms21 constitutes the Kap114-specific E3 ligase. Owing to the weak mislocalization phenotype in Δsiz1 and Δnfi1 mutants, the Siz1 and Nfi1 enzymes might also contribute to Kap114 SUMO conjugation to a lower extent. This is supported by a remarkable correlation between the mislocalization of Kap114 and its transport cargos.
Figure 5.
Kap114, Tbp1, and Sua7 mislocalize in the E3 mutant mms21-11. WT, Δsiz1, Δnfi1, mms21-11, or Δcst9 cells containing the plasmids pRS316–KAP114–GFP, YCpGAL–GST–GFP–TBP1, or YCpGAL–GST–GFP–SUA7 were cultured at 30°C in liquid media. Strains expressing KAP114–GFP were grown in medium containing 2% glucose, whereas cells transformed with plasmids coding for GST–GFP fusions with Tbp1 and Sua7 were cultured in medium containing 2% raffinose and were then grown in 2% galactose for 2 h. The localization of the GFP fusion proteins was examined by direct fluorescence microscopy. Scale bar, 5 μm.
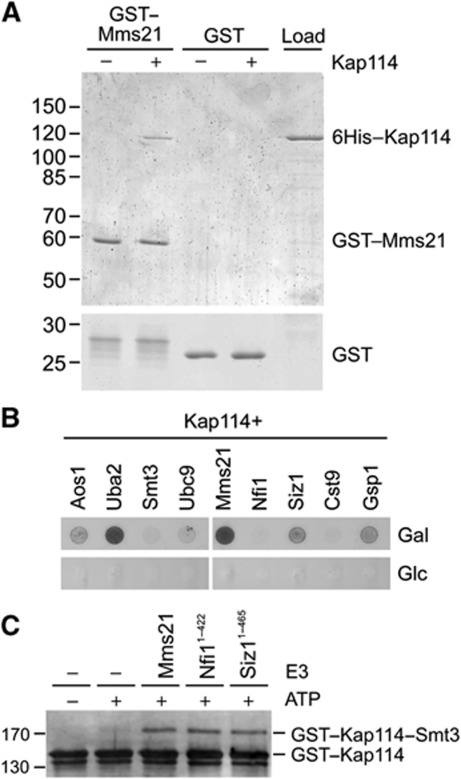
To ascertain whether Mms21 directly binds to Kap114, we performed pulldown experiments using bacterially purified GST–Mms21 and 6His-tagged Kap114 (Figure 6A). GST–Mms21 was immobilized on glutathione sepharose and incubated with recombinant Kap114. After washing, the bound material was eluted and separated by SDS–PAGE. Whereas Kap114 did not bind to GST alone, it bound to GST–Mms21, which was readily detectable in Coomassie Blue-stained gels. We therefore conclude that Mms21 and Kap114 directly interact in the absence of other cellular components. If Kap114 would act as an importin for Mms21, the binding to Mms21 would expected to be Gsp1-sensitive. However, Mms21 was not displaced from Kap114 by Gsp1–GTP (Supplementary Figure 3). Thus, the Kap114/Mms21 binding evidently represents not an importin/cargo interaction but rather a SUMO target/E3 interaction. In pulldown assays, we did not observe a direct binding of Kap114 to Siz1-N or Nfi1-N (not shown).
Figure 6.
Kap114 interacts with the E3 ligase Mms21. (A) Kap114 directly binds to Mms21. Purified GST–Mms21 or GST as the control were immobilized on glutathione sepharose and incubated with 6His-tagged Kap114 at 4°C for 1 h. After washing, bound proteins were eluted, separated by SDS–PAGE and stained with Coomassie Brilliant Blue. In the right lane, 25% of the Kap114 input is shown. Molecular weight markers are given in kDa. (B) Kap114 interacts with sumoylation components in two-hybrid assays. A LexA operon–LEU2 reporter yeast strain was transformed with a HIS3 bait plasmid encoding a fusion protein between the LexA DNA–binding domain and Kap114 as well as with TRP1 prey plasmids coding for fusions of a DNA activation domain to the proteins indicated. The synthesis of the activation domain hybrids is controlled by the GAL1 promoter that is inducible by galactose. The cells were grown on media lacking histidine, tryptophan, and leucine in the presence of either galactose (top) or glucose (bottom). The spots in each row originate from the same plate. (C) Mms21, Nfi1, and Siz1 stimulate sumoylation of Kap114 in vitro. GST–Kap114 was sumoylated as described in Figure 1A, except that all reactions contained only 0.19 μg Ubc9 (177 nM final concentration) and that the E3 derivatives GST–Mms21, Nfi11–422, or Siz11–465 were added as indicated. The samples were analysed by SDS–PAGE and western blotting using anti-Kap114 antibodies. Figure source data can be found with the Supplementary data.
To test whether Mms21 and Kap114 also interact in vivo, we performed two-hybrid interaction assays using a LexA-based system. Kap114 fused to a LexA DNA–binding domain was co-synthesized in cells containing a LEU2 reporter gene together with several activation domain fusions. The synthesis of correctly sized fusion proteins in these cells was confirmed by western blotting (data not shown). Besides the E3 ligases, our analysis also included other sumoylation factors and Gsp1 as a positive control (Figure 6B). As expected, Kap114 interacted with the Gsp1 GTPase. Because the activation domain fusions were expressed under the control of the galactose-inducible GAL1 promoter, a specific interaction is indicated by growth on leucine-deficient agar plates containing galactose as the carbon source and the failure to grow on plates containing glucose. Whereas the co-synthesis of Kap114/Smt3 or Kap114/Ubc9 hybrid proteins led only to very weak growth on galactose plates, Kap114 clearly interacted with the E1 components Aos1 and Uba2. This observation provides independent evidence for an interaction of Kap114 with the sumoylation machinery. Among the E3 ligases, we observed no interaction between Kap114 and Nfi1 or Cst9, respectively. However, the simultaneous expression of the Kap114/Mms21 and Kap114/Siz1 hybrids allowed growth on galactose-containing plates, indicating that these factors interact under in vivo conditions.
We next investigated whether Mms21 can mediate sumoylation of Kap114 in vitro. We employed the sumoylation assay containing Smt3, E1, and E2 described in Figure 1, but reduced the concentration of the E2 enzyme Ubc9 to amounts that did no longer allow sumoylation of Kap114. We hypothesized that under these conditions the sumoylation reaction becomes E3-dependent. Besides the above described full-length Mms21 fusion protein, we aimed to utilize recombinant Nfi1 (726 residues) and Siz1 (904 residues). Because we were unable to purify these enzymes as full-length proteins in an active form, we constructed the C-terminal deletions Nfi11–422 and Siz11–465, both containing the conserved and catalytically active Siz/PIAS–RING domain (Reindle et al, 2006; Yunus and Lima, 2009), as fusion proteins carrying a 6His tag. Figure 6C shows that all three purified E3 constructs were equally active with respect to sumoylation of Kap114 in vitro.
Sumoylation of Kap114 stimulates the dissociation of the Kap114/cargo complexes
To test whether sumoylation affects the Kap114-mediated transport mechanism, we analysed the interaction with Kap114 binding partners. For this purpose, we performed pulldown experiments using in vitro sumoylation reactions, which contain both sumoylated and unsumoylated Kap114. The mixture was incubated with immobilized GST–Sua7, GST–Tbp1, and GST–Gsp1–GTP (Supplementary Figure 4). In these assays, we observed no significant differences in the binding behaviour of Kap114 versus Kap114–Smt3 towards Sua7 or Gsp1–GTP, indicating that the binding of Kap114 to these factors is not detectably changed by sumoylation. In contrast, Kap114–Smt3 bound almost 1.5 times stronger to Tbp1 compared with unmodified Kap114.
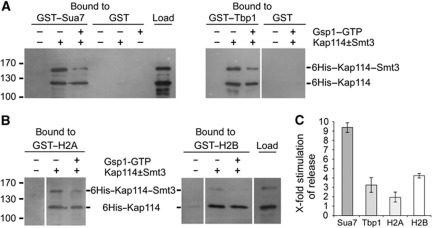
We next investigated whether sumoylation of Kap114 is involved in the Gsp1-mediated cargo release in the nucleus. It was reported that Gsp1–GTP alone is not sufficient to efficiently release the transport cargos Tbp1 (Pemberton et al, 1999), Rpf1 (Caesar et al, 2006), as well as the histones H2A and H2B (Greiner et al, 2004) from Kap114 in vitro. Therefore, other nuclear factors are necessary to assist in cargo release. To examine whether Smt3 is a potential cargo release factor, we bound the mixture of sumoylated and unmodified Kap114 to immobilized GST–Sua7 or GST–Tbp1. During a subsequent incubation of Kap114/cargo complexes for 1 h with buffer alone, they remained completely stable (not shown). Remarkably, when the import complexes were incubated in the presence of excess amounts of Gsp1–GTP, Kap114–Smt3 was considerably more efficiently released than unmodified Kap114 (Figure 7A). The quantification in Figure 7C shows that ∼3.2 times more Kap114–Smt3 was released from Tbp1 compared with unmodified Kap114. The release efficiency of Kap114–Smt3 from Sua7 was even ∼9.3 times higher compared with unsumoylated Kap114. Similarly, as shown before (Greiner et al, 2004), the H2 histones formed stable complexes with Kap114 and Kap114–Smt3. Again, the Gsp1-mediated cargo release was considerably stimulated by the SUMO modification (Figure 7B). Approximately two-fold more Kap114–Smt3 was released from H2A compared with unmodified Kap114 in the presence of Gsp1–GTP, whereas the average release from H2B increased by a factor of 4.3 (Figure 7C). The preferential release of sumoylated Kap114 from its cargos indicates that import complex dissociation is regulated by sumoylation of Kap114 and that Smt3 functions as a cargo release factor.
Figure 7.
Sumoylation of Kap114 stimulates the Gsp1–GTP-mediated dissociation of Sua7, Tbp1, and the H2 histones from Kap114. (A, B) Purified 6His–Kap114 was sumoylated in vitro as described in Figure 1A. Subsequently, the mixture (25% of the load is shown) was incubated at 4°C for 1 h with GST–Sua7, GST–Tbp1, and GST (A) or GST–H2A and GST–H2B (B) immobilized on glutathione sepharose. After three washing steps, the reactions were further incubated with buffer alone or with excess amounts of 6His–Gsp1 Q71L–GTP at 4°C for 1 h. After washing, bound proteins were eluted with SDS sample buffer, separated by SDS–PAGE, and detected by western blotting using anti-Kap114 antibodies. Molecular weight markers are shown in kDa. The lanes in each gel block originate from the same gel. (C) The stimulation of Gsp1–GTP-mediated cargo release by sumoylation of Kap114 was quantified. Data are given as x-fold stimulation of release (average values±s.d., n=3) of Sua7, Tbp1, H2A, and H2B from sumoylated versus unmodified Kap114 in the presence of Gsp1–GTP. The ratio of bound unmodified Kap114 after incubation with buffer alone and unmodified Kap114 after incubation with Gsp1–GTP was divided by the ratio of bound sumoylated Kap114 after incubation with buffer alone and sumoylated Kap114 after incubation with Gsp1–GTP. This value defined the factor of x-fold stimulation of cargo release plotted on the y axis. Figure source data can be found with the Supplementary data.
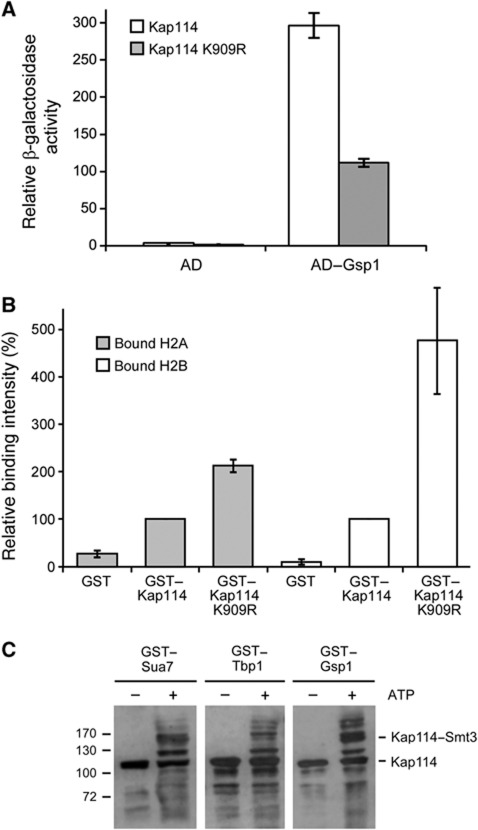
We finally performed experiments which aimed to test the idea that SUMO acts as a cargo release factor for Kap114. Smt3 apparently facilitates the interaction with Gsp1 and enhances the dissociation efficiency. This is supported by our finding that unsumoylatable Kap114 interacts about three times weaker with Gsp1 than WT Kap114 in vivo using a two-hybrid assay (Figure 8A). A prediction for unsumoylatable Kap114 is that the interaction with cargo is stabilized within the nucleus. To test this, we expressed plasmid-encoded GST–KAP114 and GST–KAP114 K909R in yeast cells. After lysis, the synthesis of the fusion proteins was verified (not shown) and they were affinity-purified using glutathione sepharose beads. As predicted, the amounts of H2 histones bound to Kap114 increased when the unsumoylatable mutant had been synthesized (Figure 8B). About 2.1 times more H2A bound to Kap114 K909R as compared with non-mutated Kap114, whereas roughly 4.8 times more H2B bound to unsumoylatable Kap114 in comparison with Kap114.
Figure 8.
The K909R mutation affects in vivo interactions of Kap114. (A) Quantification of the two-hybrid interaction between Gsp1 and Kap114 versus Kap114 K909R. LexA operon–LEU2 reporter cells containing plasmids encoding the activation domain (AD) alone (empty vector) or fused to Gsp1 and plasmids encoding the DNA-binding domain fused to Kap114 or Kap114 K909R were grown in liquid cultures. The synthesis of the AD fusions was induced by addition of 2% galactose for 2 h. Then relative β-galactosidase activity units were determined in liquid assays. Data represent the average value of three experiments. (B) The binding of H2A and H2B histones to Kap114 K909R is increased in comparison with non-mutated Kap114. Yeast cells deleted for KAP114 were transformed with plasmids YEpGAL–GST, YEpGAL–GST–KAP114, or YEpGAL–GST–KAP114 K909R and cultured in liquid media containing 2% raffinose at 30°C. For induction of the GST fusion protein synthesis, the cells were complemented with 2% galactose and incubated further for 3 h. The cells were collected, and the cleared lysates were used in pulldown assays with glutathione sepharose to purify proteins bound to the GST fusions, which were then analysed by SDS–PAGE and immunoblotting with antibodies against H2A and H2B. Signal intensities were determined by densitometry, the value for H2A and H2B bound to GST–Kap114 was set as 100% and compared to the amount of bound histones to GST–Kap114 K909 as well as the GST control (average value of three experiments). (C) Kap114 bound to cargo or Gsp1 can be sumoylated in vitro. A quantity of 10 μg of 6His-tagged Kap114 was bound to 6 μg of GST–Sua7, GST–Tbp1, or GST–Gsp1–GTP, which were immobilized before on glutathione sepharose. After washing three times with TEFP buffer, sumoylation components were added as described in Figure 1A, and the reactions were incubated with or without 5 mM ATP at 30°C for 90 min. After washing, elution with SDS sample buffer, and SDS–PAGE, Kap114 was detected by immunoblotting using Kap114-specific antibodies. Molecular weight markers are in kDa.
Another prediction from our model is that sumoylation can occur on cargo-bound and perhaps also on Ran-bound Kap114. To this end, we prebound Kap114 to immobilized Sua7, Tbp1, or Gsp1–GTP and then performed an in vitro sumoylation reaction. Figure 8C shows that all three states of Kap114 are sumoylatable. The cargo proteins as well as Gsp1 were not sumoylated under these conditions (not shown).
Discussion
In this study we show that the importin Kap114 is sumoylated and that sumoylation regulates its function. Our results demonstrate for the first time that the SUMO conjugation system specifically controls nuclear trafficking by modifying an importin β-like transport receptor. Several lines of evidence establish that Kap114 is a genuine SUMO target: Kap114 can be sumoylated in vivo and in vitro; Kap114 interacts with SUMO conjugation factors by two-hybrid analysis and directly binds to Mms21; the Kap114 sequence 908IKQE911 constitutes a typical ΨKxD/E sumoylation motif; Smt3 conjugation of the Kap114 K909R mutant is defective in vivo and in vitro; Kap114 and its transport cargos mislocalize in sumoylation mutants; and finally, the K909R mutant mislocalizes and is disturbed in its function.
We show that two different Lys909 point mutants, K909R and K909A, are inactive. Together with the alanine scan results, it seems unlikely that the investigated individual mutations cause conformational changes that inhibit sumoylation of another residue. It was found that the typical sumoylation motif is usually not part of a helix (Pichler et al, 2005). According to the secondary structure prediction programme YASPIN (Lin et al, 2005), the region around residue 909 of Kap114 (predicted to be a sumoylation motif with a probability of 94%) is non-helical, whereas the two other high probability SUMO sites at residues 261 (score 94%) and 494 (score 91%) are both situated within α-helical regions. This favours the idea that SUMO consensus sites are modified only when present in unstructured regions. We assume that the sumoylation site is located between the next to last and the last HEAT repeat of Kap114. It has to be noted that we cannot exclude that the Kap114 K909R mutant has defects in addition to a block of sumoylation.
It has been reported that sumoylation affects the import of classical NLS proteins, which is mediated by importin-α and β (Stade et al, 2002). The results of that study showed that nuclear accumulation of importin-α is accompanied by cytoplasmic mislocalization of classical NLS cargos in sumoylation mutants. However, as the SUMO target has not yet been identified, it is unclear whether importin-α or another factor is modified by sumoylation. We can exclude a general regulation of Ran-dependent nucleocytoplasmic traffic by SUMO modification. This can be concluded from the localization of importin-β (Kap95), Kap104-mediated import, mRNA export (Stade et al, 2002), the localization of the exportin Cse1 or the importin Kap120, and from Mtr10-mediated import (this study), which are all not affected in SUMO modification pathway mutants.
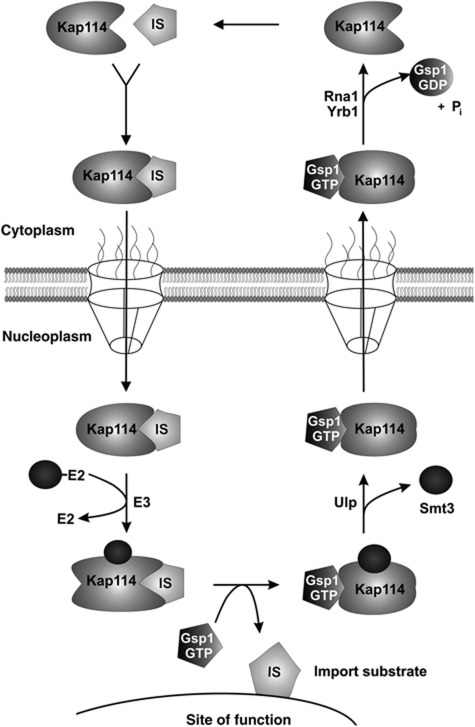
Our data are in agreement with a model for the role of sumoylation of Kap114 illustrated in Figure 9. After import, the Kap114/cargo complex cannot be effectively disassembled by Gsp1–GTP and migrates to the site of function of the cargo. Sumoylation of Kap114 is a prerequisite of efficient cargo dissociation and takes place in the immediate neighbourhood of the site of function (see below). After cargo release, Kap114 is desumoylated on its way back to the cytoplasm. Smt3 deconjugation is mediated by the mostly nuclear Ulp2 isopeptidase and/or by Ulp1, which is located at the nuclear side of the NPC (Panse et al, 2003; Geiss-Friedlander and Melchior, 2007). In the cytoplasm, dissociation of the Gsp1 GTPase and Kap114 is triggered by GTP hydrolysis catalysed by the GTPase activation protein Rna1 (Fried and Kutay, 2003; Pemberton and Paschal, 2005). Interestingly, the disassembly of the Kap114/Gsp1–GTP export complex requires in addition to Rna1 the cytoplasmic release factor Yrb1 (Schlenstedt et al, 1995, 1997; Bischoff and Görlich). Yrb1 can be viewed as the cytoplasmic counterpart to the nuclear release factor SUMO. After release of the free form of Kap114, it can bind a cargo molecule to enter another transport cycle. Both sumoylation and desumoylation of Kap114 occur within the nucleus, which is consistent with our observation that defects in both processes result in the same phenotype. A defect in sumoylation will lead to inactivation of Kap114. The import receptor is unable to release its cargo and accumulates in the nuclear interior. It is unclear why a defect in desumoylation also results in nuclear accumulation of Kap114. Presumably, the Smt3-conjugated form of Kap114 adopts a conformation incompatible with export. Defects in sumoylation and desumoylation are both accompanied by a depletion of cytoplasmic Kap114, and consequently, an inhibition of import.
Figure 9.
Model of the regulation of Kap114 by sumoylation. After binding of the IS to Kap114 in the cytoplasm, the Kap114/IS complex passes the NPC. In the nucleoplasm, the Kap114/IS complex migrates to the vicinity of the site of cargo function where Kap114 is sumoylated. The release of the IS is assisted by Smt3 and requires binding of Gsp1–GTP to Kap114. Before export, Kap114 is deconjugated from Smt3 by Ulp isopeptidases. In the cytoplasm, Kap114 dissociates from Gsp1 after GTP hydrolysis, which is mediated by Rna1 (GTPase-activating protein) and stimulated by Yrb1 (yeast RanBP1).
The general cargo release factor Gsp1–GTP, although present at high concentrations in the nucleoplasm, is insufficient for the disassembly of cargo/Kap114 complexes. This was shown before for several cargos, which are all nucleic acid binding proteins (Pemberton et al, 1999; Greiner et al, 2004; Caesar et al, 2006). We show here that the transcription factor Sua7 bound to Kap114 also constitutes a Gsp1–GTP resistant cargo/receptor complex. Although structural data about Kap114 are not available, it is clear that the sumoylation site of Kap114 is situated close to the substrate binding region, whereas the Gsp1-binding domain is located at the N terminus (Hodges et al, 2005; Cook et al, 2007). The modification of Kap114 with Smt3 apparently neither results in a diminished affinity for import cargos nor an inhibition of binding to Gsp1–GTP. It can be predicted that in the cargo-bound conformation of Kap114 the N-terminal Gsp1-binding domain is not or only partially accessible for Gsp1–GTP. This could be attributed to a direct shielding caused by the binding of the C-terminal and N-terminal domains. The covalent attachment of Smt3 to the C terminus of Kap114 would lead to a conformational change that is compatible with binding to Gsp1–GTP.
It has been reported that interaction partners of the cargos at the site of function are able to stimulate cargo dissociation from Kap114. This might be accomplished by competition for binding sites or by binding to alternate sites resulting perhaps in allosteric effects (Pemberton et al, 1999; Greiner et al, 2004). Interestingly, it is known also for other importins that intranuclear targets are necessary to assist in cargo release (Senger et al, 1998; Caesar et al, 2006). Many of these cargos seem to be nucleic acid binding factors. It is necessary especially for cargos containing exposed basic domains that a premature disassembly of the importin/cargo complex must be prevented to avoid aggregation of the cargo (Jäkel et al, 2002). It is indeed known for free histones that they are toxic to cells (Sutton et al, 2001), which suggests that histone importins act as nuclear chaperones. Regulation by sumoylation represents an additional tool to achieve the goal of coordinated intranuclear targeting. Moreover, sumoylation offers the possibility to rapidly respond to cellular signals affecting Kap114-mediated nucleocytoplasmic communication. Because unsumoylated Kap114 is non-functional as an import receptor, it is a potential target for shutting down an entire import pathway by modulating its sumoylation status.
It was shown that the E1 and E2 enzymes are sufficient for sumoylation in vitro in the presence of ATP, while an E3 ligase increases the specificity of the reaction (Johnson and Blobel, 1997). We found that three different E3 ligases are able to promote sumoylation of Kap114 in vitro. However, the analysis of mutants in vivo revealed that Mms21 constitutes the Kap114-specific E3 enzyme. In mms21 mutants, Kap114 accumulates in the nucleus and import of Kap114 cargos is defective. Our findings that Siz1 and Nfi1 can also stimulate sumoylation of Kap114 in vitro and that the respective mutants show weak Kap114 transport defects favour the idea that these two E3 enzymes also contribute to Kap114 sumoylation to a limited extent. Interestingly, Mms21 was found in a multiprotein complex associated with DNA repair and chromosomal organization (Zhao and Blobel, 2005). Thus, it seems that Mms21 is especially concentrated at chromosomal regions, which supports our view that sumoylation of Kap114 occurs at the site of function, which represents DNA for the histones, the TATA-binding protein, and the transcription factor Sua7.
Materials and methods
Yeast strains and plasmids
All strains and plasmids used in this study are listed in Supplementary Tables I and II. The genomic integration of GFP and the HA tag to create C-terminal Kap114 fusions as well as the integration of knockout cassettes to generate KAP114::TRP1, ULP2::HIS3, and CST9::HIS3 strains were performed as described (Longtine et al, 1998). The correct integration into each strain was verified by several control PCRs and by immunoblotting analysis. The genomic DNA of KAP114–GFP and KAP114–HA strains served as PCR templates for the construction of yeast expression plasmids. The respective genes or coding sequences were amplified by PCR from genomic DNA using Pwo polymerase (Roche) as well as appropriate primers and cloned into pQE vectors (Qiagen), pGEX plasmids (GE Healthcare), pRS yeast vectors (Sikorski and Hieter, 1989; Christianson et al, 1992), pEG–Bam (pGS897), pJG–Bam (pGS898; Caesar et al, 2006), pJG4–5 (pGS476; OriGene), or YCpGAL–GST–GFP (pGS846; Solsbacher et al, 2000). The K909R point mutation in KAP114 was generated using the QuikChange site-directed mutagenesis kit (Stratagene). All plasmids were sequenced to confirm that they did contain the correct sequence. Oligonucleotide sequences and plasmid construction details are available upon request.
Detection of in vivo sumoylation
To identify sumoylated proteins in living cells, a denaturing nickel pulldown assay was performed. Yeast cells were transformed with plasmids encoding C-terminally HA-tagged Kap114 and with YEp181–CUP–HIS7–SMT3 coding for 7His-tagged Smt3. The strains were cultured in selective liquid medium containing 2% glucose and 100 μM CuSO4 at 30°C until the OD600 reached values between 0.5 and 1. Subsequently, 50 OD units of cells were resuspended in 1 ml buffer A (6 M guanidine–HCl, 100 mM NaH2PO4, 10 mM Tris–HCl, 10 mM β-mercaptoethanol, 5 mM imidazol, pH 8) with N-ethyl maleimide (NEM, final concentration 2.5 μg/ml) and protease inhibitors (pepstatin A, leupeptin, antipain, chymostatin, each 200 μg/ml). Then 250 μl glass beads (0.5 mm, Sigma) were added and the cells were lysed using a Precellys 24 homogenizer (Peqlab) at 6.500 r.p.m. for 150 s with cooling intervals after 30 s. After centrifugation for 20 min at 19.000 g, the supernatant was incubated overnight at 4°C with 40 μl nickel-nitrilotriacetic acid (NTA) agarose (Qiagen) beads equilibrated with buffer A. After washing three times with 1 ml buffer B (8 M urea, 100 mM NaH2PO4, 10 mM Tris–HCl, 3.5 mM β-mercaptoethanol, 0.1% Triton X-100, pH 6.3), bound proteins were eluted with 30 μl HU buffer (8 M urea, 200 mM Tris–HCl, 1 mM EDTA, 5% SDS, 0.1% bromophenol blue, 97 mM DTT, pH 6.8) at 60°C for 10 min. Proteins were separated by SDS–PAGE and detected by immunoblotting using anti-HA antibodies (Covance) plus peroxidase-coupled anti-mouse secondary antibodies (Sigma) or affinity-purified anti-Kap114 antibodies plus anti-rabbit secondary antibodies (Sigma). The signals were visualized with the enhanced chemiluminescence detection system ECL+ (GE Healthcare). Control western blots using anti-HA and anti-Smt3 antibodies were performed to demonstrate that HA-tagged Kap114 was soluble and that the majority of Smt3 bound to nickel agarose.
Protein purification
The synthesis of recombinant proteins in E. coli and the preparation of cell lysates was performed as described (Schlenstedt et al, 1997). The purification of GST fusion proteins was carried out by affinity chromatography on glutathione sepharose (GE Healthcare) and was followed by buffer exchange to PBSKMT buffer (150 mM NaCl, 25 mM sodium phosphate, 3 mM KCl, 1 mM MgCl2, 0.1% Tween, pH 7.3). Fusion proteins to the 6-histidine tag were purified on Ni-NTA agarose from bacterial extracts. The separation of 6His-tagged Gsp1 Q71L–GTP was described before (Maurer et al, 2001). Aos1 and Uba2 were co-synthesized and co-purified (Johnson and Gupta, 2001). Proteins used in the in vitro sumoylation assay were stored in TEFP buffer (100 mM NaCl, 50 mM Bis-Tris, 10 mM MgCl2, pH 6.5). Recombinant WT Smt3 or the triple point mutant Smt3 K11,15,19R protein correspond to the processed SUMO form ending with double glycine residues at their C terminus.
For purification of GST fusion proteins from yeast, cell lysates were prepared as described (Schlenstedt et al, 1997) and were incubated with glutathione sepharose at 4°C for 1 h. The beads were washed three times with PBSKMT buffer and bound proteins were eluted with SDS sample buffer. The bound histone proteins were detected by immunoblotting using anti-H2A and anti-H2B antibodies (Santa Cruz Biotechnology).
In vitro sumoylation assays
To analyse sumoylation in vitro, 3 μg of target protein were mixed with 2 μg Aos1/Uba2 (E1; 300 nM final concentration), 0.75 μg Ubc9 (E2; 700 nM final concentration), 7.5 μg Smt3 K11,15,19R (which was somewhat more efficient than Smt3; 10 mM final concentration), and 0.1 mM DTT in a final volume of 60 μl TEFP buffer in the presence or absence of 5 mM ATP (GE Healthcare) and incubated at 30°C for 90 min. To examine E3-dependent sumoylation, 2 μg GST–Mms21, 6His–Nfi11–422, or 6His–Siz11–465 were added to the mixture. GST-tagged sumoylated proteins were optionally separated from the sumoylation enzymes by incubating the mixture with 30 μl of glutathione sepharose beads in 500 μl of PBSKMT buffer at 4°C for 1 h. After washing three times, 5 μl of 1 M DTT and 17 μl of three-fold SDS sample buffer (without DTT) were added to the beads in order to elute the proteins by heating at 65°C for 20 min. Proteins were separated by SDS–PAGE and detected by immunoblotting analysis using affinity-purified polyclonal anti-Kap114 antibodies.
Two-hybrid interaction analysis
Two-hybrid assays were carried out using the DupLEX-A system (OriGene Technologies, Rockville, MD) as described (Caesar et al, 2006). The strain EGY48 holding a LexA operon–LEU2 reporter was transformed with the bait plasmid pEG–Bam–KAP114 encoding the LexA DNA–binding domain fused to Kap114 and with various pJG4–5-derived prey vectors containing GAL1 promoter–driven B42 activation domain fusions to sumoylation factors. The synthesis of all hybrid proteins in the transformants was confirmed by immunoblotting with anti-LexA antibodies (Santa Cruz Biotechnology) and anti-HA antibodies. The cells were spotted onto agar plates with synthetic complete leucine-deficient media containing 2% galactose or 2% glucose and incubated at 30°C for 2 days. Values for β-galactosidase activity were calculated from liquid assays using o-nitrophenyl-β-D-galactopyranosid as described (Miller, 1972).
Protein interaction and Gsp1–GTP dissociation assays
Purified GST fusion proteins (12 μg) were immobilized on 30 μl glutathione sepharose beads in PBSKMT buffer at 4°C for 1 h. After three washing steps with 1 ml buffer, potential binding partners were added and the reactions were further incubated in a volume of 500 μl at 4°C for 1 h. After washing, proteins were eluted with 30 μl 2 × SDS sample buffer by incubation at 95°C for 5 min. For Gsp1–GTP dissociation assays, immobilized GST–Tbp1 and GST–Sua7 were incubated with sumoylation reactions containing sumoylated and unsumoylated Kap114. After washing, a five-fold molar excess of 6His–Gsp1 Q71L–GTP was added to induce dissociation, and the reactions were further incubated at 4°C for 1 h, which was followed by washing and elution with SDS sample buffer.
Supplementary Material
Acknowledgments
We are grateful for generous gifts of strains and plasmids to Erica Johnson (Philadelphia), Katrin Stade (Berlin), Pamela Silver (Boston), Helle Ulrich, Adelina Davies (London), Jürgen Dohmen (Cologne), and Xiaolan Zhao (New York City). We thank Karsten Mayr and Silke Guthörl for technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft and by the Homburger Forschungsförderung (HOMFOR).
Author contributions: UR and GS designed the experiments and wrote the manuscript. UR performed most of the experiments and analysed the results. MS was involved in Kap114 in vitro sumoylation. YC established the in vitro sumoylation assay and purified recombinant proteins. SC generated the Kap114 Ala point mutants and performed experiments shown in Figures 1D, 2B, and , and Supplementary Figure S3.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bischoff FR, Görlich D (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. . F EBS Lett 419: 249–254 [DOI] [PubMed] [Google Scholar]
- Bylebyl GR, Belichenko I, Johnson ES (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278: 44113–44120 [DOI] [PubMed] [Google Scholar]
- Caesar S, Greiner M, Schlenstedt G (2006) Kap120 functions as a nuclear import receptor for ribosome assembly factor Rpf1 in yeast. Mol Cell Biol 26: 3170–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-H, Lo Y-H, Liang S-S, Ti S-C, Lin F-M, Yeh C-H, Huang H-Y, Wang T-F (2006) SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev 20: 2067–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multi-functional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E (2007) Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671 [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrov H, Kolarov J, Goffeau A, Varshavsky A (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem 270: 18099–18109 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Greiner M, Caesar S, Schlenstedt G (2004) The histones H2A/H2B and H3/H4 are imported into the yeast nucleus by different mechanisms. Eur J Cell Biol 83: 511–520 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Hodges JL, Leslie JH, Mosammaparast N, Guo Y, Shabanowitz J, Hunt DF, Pemberton LF (2005) Nuclear import of TFIIB is mediated by Kap114p, a karyopherin with multiple cargo-binding domains. Mol Biol Cell 16: 3200–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Jäkel S, Mingot J-M, Schwarzmaier P, Hartmann E, Görlich D (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 21: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272: 26799–26802 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J 16: 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-J, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-J, Hochstrasser M (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 160: 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Simossis VA, Taylor WR, Heringa J (2005) A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21: 152–159 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Makhnevych T, Ptak C, Lusk CP, Aitchison JD, Wozniak RW (2007) The role of karyopherins in the regulated sumoylation of septins. J Cell Biol 177: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K, Schlenstedt G (2001) The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p). Mol Biol Cell 12: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1972) InExperiments in Molecular Genetics pp73–75Cold Spring Harbor, New York: Cold Spring Harbor Laboratory, [Google Scholar]
- Morehouse H, Buratowski RM, Silver PA, Buratowski S (1999) The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc Natl Acad Sci USA 96: 12542–12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Ewart CS, Pemberton LF (2002) A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J 21: 6527–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF (2001) Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol 153: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Küster B, Gerstberger T, Hurt E (2003) Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol 5: 21–27 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM (2005) Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol 145: 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum SR, Blobel G (1997) A distinct parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol 139: 1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK (2005) SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol 12: 264–269 [DOI] [PubMed] [Google Scholar]
- Quan Y, Ji Z-L, Wang X, Tartakoff AM, Tao T (2008) Evolutionary and transcriptional analysis of karyopherin β superfamily proteins. Mol Cell Proteomics 7: 1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119: 4749–4757 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276: 12654–12659 [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem 276: 21664–21669 [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff FR (1997) Yrb4p, a yeast Ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO J 16: 6237–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA (1995) Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J 14: 5367–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst I, Johnson ES, Dohmen RJ (2000) SUMO conjugation and deconjugation. Mol Gen Genet 263: 771–786 [DOI] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J 17: 2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J, Maurer P, Vogel F, Schlenstedt G (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol Cell Biol 20: 8468–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Vogel F, Schwienhorst I, Meusser B, Volkwein C, Nentwig B, Dohmen RJ, Sommer T (2002) A lack of SUMO conjugation affects cNLS-dependent nuclear protein import in yeast. J Biol Chem 277: 49554–49561 [DOI] [PubMed] [Google Scholar]
- Sutton A, Bucaria J, Osley MA, Sternglanz R (2001) Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydorskyy Y, Srikumar T, Jeram SM, Wheaton S, Vizeacoumar FJ, Makhnevych T, Chong YT, Gingras A-C, Raught B (2010) A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol Cell Biol 30: 4452–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Kahyo T, Toh-e A, Yasuda H, Kikuchi Y (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem 276: 48973–48977 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol Cell 35: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.