Abstract
The ubiquitin ligases RNF8 and RNF168 orchestrate DNA damage signalling through the ubiquitylation of histone H2A and the recruitment of downstream repair factors. Here, we demonstrate that RNF8, but not RNF168 or the canonical H2A ubiquitin ligase RNF2, mediates extensive chromatin decondensation. Our data show that CHD4, the catalytic subunit of the NuRD complex, interacts with RNF8 and is essential for RNF8-mediated chromatin unfolding. The chromatin remodelling activity of CHD4 promotes efficient ubiquitin conjugation and assembly of RNF168 and BRCA1 at DNA double-strand breaks. Interestingly, RNF8-mediated recruitment of CHD4 and subsequent chromatin remodelling were independent of the ubiquitin-ligase activity of RNF8, but involved a non-canonical interaction with the forkhead-associated (FHA) domain. Our study reveals a new mechanism of chromatin remodelling-assisted ubiquitylation, which involves the cooperation between CHD4 and RNF8 to create a local chromatin environment that is permissive to the assembly of checkpoint and repair machineries at DNA lesions.
Keywords: chromatin, DNA damage response, DSB repair, histone ubiquitin ligase, RNF8
Introduction
DNA lesions trigger a signalling cascade that leads to the activation of cell-cycle checkpoints and DNA repair (Dinant et al, 2009; Huen and Chen, 2010). DNA damage-induced ubiquitylation of the core histone H2A plays an important role in DNA damage signalling in response to ultraviolet light-induced DNA lesions (Bergink et al, 2006) and DNA double-strand breaks (DSBs) (Mailand et al, 2007). The ubiquitin ligase RNF8 is recruited to sites of DNA damage through phospho-specific interactions with MDC1, where it, in concert with the ubiquitin ligase RNF168, mediates ubiquitylation of histone H2A and likely other unidentified substrates (Huen et al, 2007; Mailand et al, 2007; Doil et al, 2009; Marteijn et al, 2009; Stewart et al, 2009). These non-proteolytic ubiquitin conjugates at DNA lesions serve as binding sites for the recruitment of a number of downstream factors, including BRCA1, p53-binding protein 1 (53BP1) and RAD18 (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Doil et al, 2009; Huang et al, 2009; Marteijn et al, 2009; Stewart et al, 2009; Acs et al, 2011). Thus, the DNA damage-induced ubiquitylation of histone H2A by the concerted action of RNF8 and RNF168 plays a central role in DNA damage signalling and repair.
While RNF8 and RNF168 mediate H2A ubiquitylation in the context of the DNA damage response (DDR), the ubiquitin ligase RNF2, which is part of the Polycomb repressive complex 1, is the canonical ubiquitin ligase of histone H2A (Wang et al, 2004). It is well established that the RNF2-mediated ubiquitylation of H2A is linked to transcriptional repression and a tightly compacted chromatin state (Francis et al, 2004; Simon and Kingston, 2009). Whereas RNF8-mediated H2A ubiquitylation has also been linked to transcriptional silencing at DSBs (Chou et al, 2010; Miller et al, 2010; Shanbhag et al, 2010), consistent with the repressive nature of this histone modification, how RNF8 establishes a chromatin environment that is accessible to the binding of downstream DDR proteins such as BRCA1 remains enigmatic. To address this issue, we assessed the impact of the ubiquitin ligases RNF8 and RNF168 on chromatin structure. We found that RNF8, but not RNF168 or RNF2, promotes extensive decondensation of higher-order chromatin structure. RNF8-mediated chromatin unfolding is dependent on the recruitment and the ATPase activity of the chromodomain helicase DNA-binding protein 4 (CHD4), which is an SNF2-like chromatin remodelling factor that is required for an efficient DDR (Larsen et al, 2010; Polo et al, 2010; Smeenk et al, 2010). Once recruited to DSBs, the chromatin remodelling activity of CHD4 promotes efficient ubiquitin conjugation by RNF8 and subsequent assembly of the downstream repair factors RNF168 and BRCA1. Interestingly, our data show that both recruitment of CHD4 and chromatin decondensation are independent of the ubiquitin-ligase activity of RNF8, unveiling the involvement of a new non-catalytic function of RNF8 in DNA damage signalling. We propose that RNF8-induced higher-order chromatin decondensation and histone H2A ubiquitylation act synergistically in facilitating the DDR.
Results
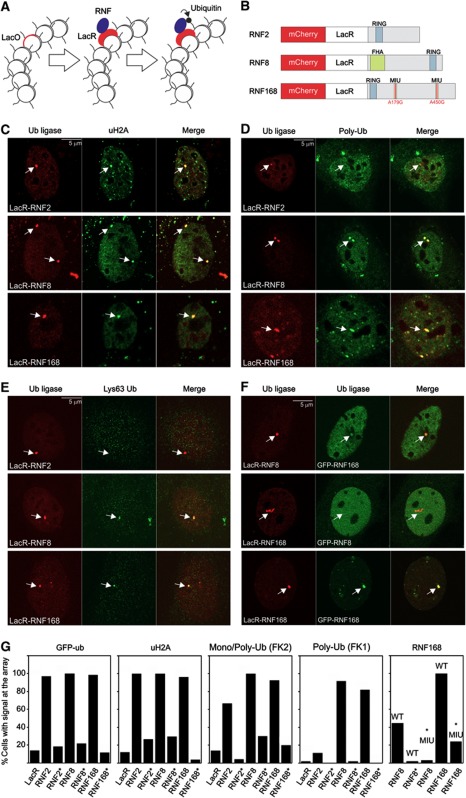
Functional tethering of RNF8, RNF168 and RNF2 to chromatin induces local H2A ubiquitylation
In order to assess the impact of H2A ubiquitin ligases on chromatin compaction, we utilized an in vivo targeting system for tethering proteins-of-interest to chromatin (Figure 1A). This system is based on the expression of a protein-of-interest fused to the Escherichia coli lactose repressor protein (LacR) in cell lines that contain genomic insertions of multiple copies of the bacterial lactose operator (LacO) sequence (Robinett et al, 1996; Tumbar et al, 1999; Soutoglou and Misteli, 2008). To target and visualize RNF8, RNF168 and RNF2, we fused these ubiquitin ligases to LacR and the fluorescent protein mCherry (Figure 1B). We found that the mCherry-LacR-RNF8 localized to laser-inflicted DNA DSBs in U2OS cells confirming the functionality of RNF8 in this fusion (Supplementary Figure S1A). It is noteworthy that also the RNF168 fusion was recruited to DSBs even though the two motifs interacting with ubiquitin (MIU) in RNF168 fusion protein had been mutated in order to prevent the mCherry-LacR-RNF168 fusion from binding to chromatin-associated ubiquitin conjugates (Supplementary Figure S1B). A likely explanation may be the recent identification of a third ubiquitin-binding domain in RNF168 that is sufficient for the localization of this ubiquitin ligase to DSBs (Pinato et al, 2011). Importantly, these experiments show that the mCherry-LacR-tagged RNF8 and RNF168 proteins efficiently localized to laser-inflicted DSBs (Huen et al, 2007; Mailand et al, 2007; Doil et al, 2009; Marteijn et al, 2009; Stewart et al, 2009).
Figure 1.
Tethering histone ubiquitin ligases triggers localized ubiquitylation of histone H2A. (A) Schematic representation of the targeting approach. The white circles represent nucleosomes some of which contain LacO repeats. (B) Schematic representation of the mCherry-LacR-RNF fusion proteins. Mutations in the MIU domains of RNF168 are indicated in red. (C–E) Tethering of ubiquitin ligases (red) in NIH2/4 cells and analysis of (C) uH2A, (D) poly-ubiquitin conjugates (FK1) and (E) Lys63-linked poly-ubiquitin signal at the array (green). (F) Cross-recruitment of ubiquitin ligases. (G) Quantification of the percentage of cells with positive signals at the array after tethering the indicated fusion proteins. Catalytic inactive ubiquitin ligases are indicated with an asterisk. Note that in the RNF168 panel, we have tethered either LacR-RNF8, LacR-RNF8*RING or LacR-RNF168 as indicated on the x-axis, while we monitored the recruitment of GFP-RNF168WT or GFP-RNF168*MIU as indicated above the bars. Values represent the mean of two independent experiments (n=50 cells). The scale bars are 5 μm.
Expression of mCherry-LacR-RNF8, RNF168 or RNF2 in NIH2/4 mouse cells, harbouring 256 repeats of the LacO sequence stably integrated in chromosome 3 (Soutoglou et al, 2007), resulted in clear localization of the fusion proteins to the LacO array (Figure 1C). Tethering of RNF8, RNF168 or RNF2 resulted in an increase in ubiquitylated H2A (uH2A) at the array (Figure 1C and G) as well as elevated levels of green fluorescent protein (GFP)-tagged ubiquitin (Figure 1G; Supplementary Figure S1C), showing that the immobilization of these ubiquitin ligases to chromatin indeed induced H2A ubiquitylation. Importantly, when catalytically inactive ubiquitin ligases harbouring inactivating amino-acid substitutions in their RING domain (C90S for RNF2, C403S for RNF8 or C16S for RNF168) were tethered, the accumulation of uH2A or GFP-ubiquitin at the array was reduced to background levels that were observed when tethering mCherry-LacR (Figure 1G).
It has been well documented that the DDR ubiquitin ligases RNF8 and RNF168 generate Lys63-linked poly-ubiquitin chains (Doil et al, 2009; Shao et al, 2009; Stewart et al, 2009), whereas RNF2 mediates mono-ubiquitylation of H2A (Wang et al, 2004). To investigate the ubiquitylation status in our experimental system, we stained cells with ubiquitin-specific antibodies. Consistent with the GFP-ubiquitin accumulation and uH2A staining, we observed that tethering of each of the three ubiquitin-ligase fusions resulted in an increase in endogenous ubiquitin conjugates at the array as revealed by staining with an antibody that recognizes both mono- and poly-ubiquitin conjugates (FK2; Figure 1G; Supplementary Figure S1D). Staining with an antibody specific for poly-ubiquitin conjugates (FK1) confirmed that tethering of RNF8 and RNF168, but not RNF2, resulted in poly-ubiquitylation at the targeted locus (Figure 1D and G). In contrast to RNF2, both RNF8 and RNF168 generated Lys63-linked poly-ubiquitin chains at the array (Figure 1E). Tethering of catalytically inactive ubiquitin ligases reduced the formation of ubiquitin conjugates at the array to background levels, underscoring that the local ubiquitylation is dependent on the enzymatic activity of these ubiquitin ligases (Figure 1G).
Having established that tethering of the ubiquitin ligases RNF8, RNF168 and RNF2 mediated localized H2A ubiquitylation, we subsequently analysed the interdependency of their recruitment. Importantly, immobilized RNF2 did not recruit GFP–RNF8 (Supplementary Figure S1F) and vice versa (data not shown), ruling out that histone ubiquitylation is caused by cross-recruitment of these proteins. Previous studies have shown that RNF168 acts downstream of RNF8, as it is recruited to DSBs by binding to ubiquitin conjugates catalysed by RNF8 (Doil et al, 2009; Stewart et al, 2009). Accordingly, we observed that mCherry-LacR-RNF8 recruited GFP-RNF168, which was dependent on the ubiquitin-ligase activity of RNF8 and the ubiquitin-binding domains of RNF168 (Figure 1F and G). In line with the model that RNF168 acts downstream of RNF8, we found that mCherry-LacR-RNF168 did not recruit GFP-tagged RNF8 (Figure 1F; Supplementary Figure S1F). Consistent with the recently reported homo-dimerization of RNF8 (Bekker-Jensen et al, 2010), we found that tethering of mCherry-LacR-RNF8 resulted in recruitment of GFP-RNF8 (Supplementary Figure S1E and F). RNF8 recruitment required a functional RING domain in the tethered LacR-RNF8 (Supplementary Figure S1E and F), suggesting that RNF8 dimerization depends on the RING domain, similar to what has been reported for several other RING ubiquitin ligases (Brzovic et al, 2001; Huang et al, 2011). Furthermore, mCherry-LacR-RNF168 tethering resulted in robust accumulation of GFP-RNF168 in an MIU-dependent manner (Figure 1F and G), indicating that chromatin-bound RNF168 initiates a positive feedback loop. These findings show that prolonged binding of RNF8 in the absence of DNA lesions triggers the recruitment of RNF168, which, in turn, initiates a positive feedback loop resulting in additional accumulation of RNF168.
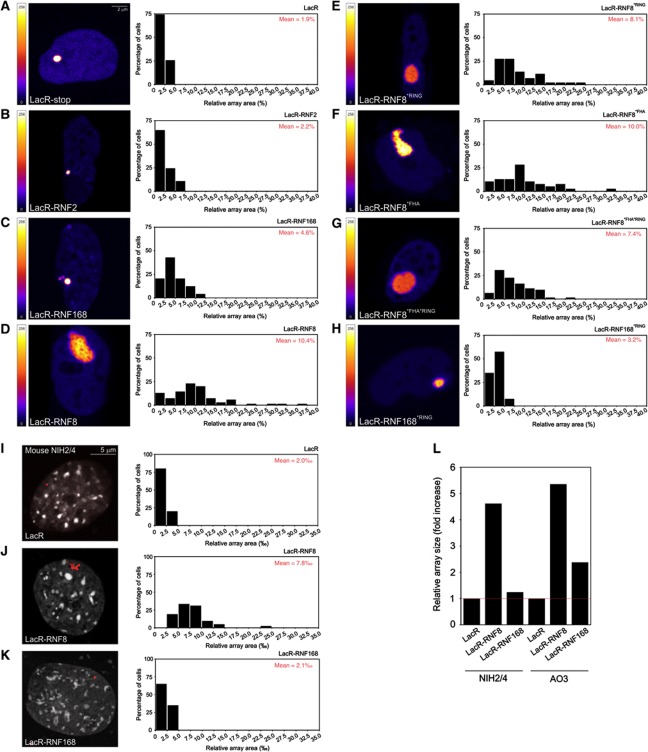
RNF8 tethering induces alterations in higher-order chromatin structure
To assess the impact of H2A ubiquitin ligases on higher-order chromatin structure, we utilized an assay to directly monitor and visualize changes in chromatin compaction (Tumbar et al, 1999; Verschure et al, 2005). For this purpose, we used cells that harbour a 90-Mbp heterochromatic region consisting of LacO repeats interspersed with the dihydrofolate reductase gene and co-amplified genomic DNA (Robinett et al, 1996; Tumbar et al, 1999). mCherry-LacR-tagged ubiquitin ligases were targeted to the compacted chromatin region harbouring the LacO repeats to study their ability to promote changes in chromatin structure. We captured 8-bit greyscale confocal images of cells expressing mCherry-LacR-tagged ubiquitin ligases, which were converted to coloured images using a look-up table based on the pixel intensities in the original image ranging from 0 (black) to 256 (white). While mCherry-LacR (Figure 2A), mCherry-LacR-RNF2 (Figure 2B) and mCherry-LacR-RNF168 (Figure 2C) did not cause changes in chromatin compaction, we observed extensive chromatin decondensation by mCherry-LacR-RNF8 (Figure 2D). Quantitative analysis of the size of the LacO array in living cells showed that the array occupied ∼2% of the nuclear area when tethering mCherry-LacR or mCherry-LacR-RNF2 (Figure 2A and B). Immobilizing RNF168 or RNF168*RING resulted in a slight increase in the array size (5 and 3%, respectively; Figure 2C and H). In contrast, the array unfolded extensively to 10% of the nuclear area upon tethering RNF8 (Figure 2D), which corresponds to an ∼5-fold increase in the surface area occupied by the LacO array.
Figure 2.
Higher-order chromatin alterations by tethering histone ubiquitin ligases in AO3 cells. (A–H) AO3 cells containing a 90-Mbp heterochromatic array were transfected with (A) mCherry-LacR, (B) mCherry-LacR-RNF2, (C) mCherry-LacR-RNF168, (D) mCherry-LacR-RNF8, (E) mCherry-LacR-RNF8*RING, (F) mCherry-LacR-RNF8*FHA, (G) mCherry-LacR-RNF8*FHA*RING and (H) mCherry-LacR-RNF168*RING fusion protein. Images show confocal slices of the nuclear distribution of the mCherry-LacR fusion proteins. The LUT for the colour coding is shown left of all panels. Cells were co-transfected with GFP-NLS (not shown) to measure the surface area of the nucleus. Quantitative measurements on the relative size of the array (area of the array/area of the nucleus) on a large numbers of cells (∼100 cells for each condition) are shown next the images as histograms. The mean size of the array (in % of nuclear area) for each condition is shown in red in the histograms. The scale bar is 2 μm. NIH2/4 cells containing a 256 × LacO array (10 kbp) were transfected with (I) mCherry-LacR, (J) mCherry-LacR-RNF8 or (K) mCherry-LacR-RNF168 (red). An overlay of the mCherry-LacR signal and counterstaining with Hoechst 33258 (white) is shown. Quantitative measurements on the relative size of the array (in ‰ of nuclear area) are shown next to the images as histograms. Values represent the mean of ∼50 cells from two independent experiments. The scale bar is 5 μm. Note that the array in NIH2/4 cells (∼10 kbp) is about 9000 times smaller than the array in AO3 cells (∼90 Mbp). (L) The relative increase in the array size induced by RNF8 in NIH2/4 and AO3 cells, about five-fold, is very similar between these two cell types. Thus, RNF8 unfolds chromatin structure ∼5-fold irrespective of cell type.
To validate the results obtained in AO3 hamster cells, we analysed the impact of RNF8 and RNF168 on chromatin structure in murine NIH2/4 cells, which harbour a significantly smaller array of only 256 repeats (∼10kbp) of the LacO sequence (Soutoglou et al, 2007). For this purpose, we captured high-resolution images of the array in NIH2/4 cells expressing LacR (Figure 2I), mCherry-LacR-RNF8 (Figure 2J) or mCherry-LacR-RNF168 (Figure 2K). Quantitative analysis of high-resolution images revealed that tethering of RNF8 also resulted in decondensation of the LacO array in NIH2/4 (Figure 2J). Like in AO3 cells, chromatin unfolding in NIH2/4 was specific for RNF8 as tethering of RNF168 did not cause decondensation (Figure 2K). Notably, the relative RNF8-induced unfolding of the LacO array was remarkably similar in NIH2/4 and AO3 cells (Figure 2L). Finally, we analysed whether an alternative way of tethering RNF8 to chromatin would also trigger chromatin unfolding. For this purpose, we fused RNF8 to the reverse tetracycline repressor protein (rTetR; Gossen et al, 1995) and expressed the GFP-rTetR-RNF8 fusion in human U2OS 2-6-3 cells, which harbour ∼200 copies of a TetO-containing cassette integrated in the genome (Janicki et al, 2004). Quantitative analysis showed a significant increase in the array size in cells expressing GFP-rTetR-RNF8 as compared with cells that expressed GFP-rTetR (Supplementary Figure S2). Together, these data show that immobilization of RNF8 on chromatin, irrespective of using a LacR- or TetR-based tethering approach, is sufficient to trigger chromatin unfolding and that this phenomenon is conserved as it was observed in murine, Chinese hamster and human cell lines.
We then asked whether chromatin decondensation by RNF8 requires its ubiquitin-ligase activity or phospho-specific interactions through the RING domain and forkhead-associated (FHA) domain, respectively (Mailand et al, 2007). For this purpose, we utilized RNF8 mutants harbouring a single amino-acid substitution that inactivates the catalytic activity of the RING finger (C403S) or phospho-specific binding of the FHA domain (R42A). Surprisingly, we found that tethered RNF8*RING still mediated chromatin unfolding in AO3 cells, suggesting that this phenomenon is independent of its ubiquitin-ligase activity (Figure 2E). Notably, we did not detect any quantitative differences in the extent of decondensation induced by tethered wild-type (WT) RNF8 compared with its mutant RNF8*RING counterpart, underscoring the RING- and dimerization-independent nature of this phenomenon. Like in NIH2/4 cells (Supplementary Figure S1E and F), we found that tethering of LacR-RNF8 in AO3 cells resulted in robust accumulation of GFP-RNF8 to the unfolded array (Supplementary Figure S3). However, tethering of LacR-RNF8*RING, although still resulting in extensive chromatin unfolding, failed to recruit GFP-RNF8 to the array (Supplementary Figure S3). To validate the inactivation of the FHA domain of RNF8 in the tethering system, we monitored the interaction between immobilized RNF8 and MDC1, which is known to require a functional FHA domain (Huen et al, 2007; Mailand et al, 2007). While tethering of WT RNF8 resulted in robust accumulation of GFP-MDC1, mCherry-LacR-RNF8*FHA immobilization failed to recruit MDC1 to the array in NIH2/4 cells (Supplementary Figure S1G and H). Notably, tethering RNF8*FHA to the array in AO3 cells still triggered extensive chromatin decondensation (Figure 2F), similar to WT RNF8 (Figure 2D). As we found that RNF8 dimerizes through its RING domain (Supplementary Figures S1E and F and S3), the possibility remained that mCherry-LacR-RNF8*FHA mediated chromatin unfolding by dimerizing with endogenous RNF8. However, the double-mutant RNF8*FHA/*RING, which is impaired in phospho-specific binding as well as RNF8-RNF8 dimerization, also triggered robust chromatin decondensation, conclusively showing that phospho-specific interactions with the FHA domain are not required to unfold chromatin (Figure 2G). Together, our results provide direct evidence for RNF8-mediated decondensation of higher-order chromatin structure independently of the canonical phospho-specific interaction through the FHA domain or the catalytic activity of the RING finger domain.
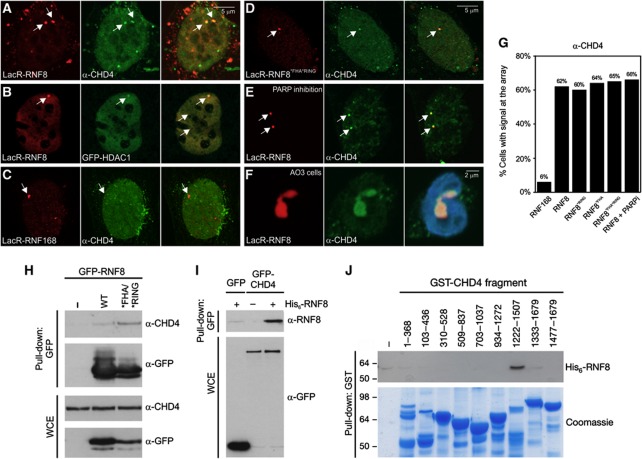
The SNF2-like chromatin remodeler CHD4 is recruited upon RNF8 tethering
Our findings show that prolonged binding of RNF8 promotes large-scale changes in chromatin structure, which prompted us to test whether RNF8 recruits a chromatin remodelling enzyme to the array, which, in turn, modulates chromatin structure. To address this possibility, we tethered mCherry-LacR-RNF8 to the array in NIH2/4 cells and subsequently analysed the localization of a collection of fluorescent protein-tagged chromatin remodelling enzymes that have previously been linked to DNA repair: BRG1 (Park et al, 2006; Lee et al, 2010), ACF1 (Ura et al, 2001; Luijsterburg et al, 2009; Lan et al, 2010), ALC1 (Ahel et al, 2009; Gottschalk et al, 2009) and CHD4, which is the ATPase subunit of the NuRD (Nucleosome Remodelling and histone Deacetylation) complex (Xue et al, 1998), that has recently been implicated in the DDR (Pegoraro et al, 2009; Larsen et al, 2010; Polo et al, 2010; Smeenk et al, 2010; Urquhart et al, 2011). While BRG1, ACF1 and ALC1 did not co-localize with tethered RNF8 (Supplementary Figure S4), recruitment of the SNF2-like chromatin remodeller CHD4 was observed upon immobilization of mCherry-LacR-RNF8 (Figure 3A and G). Notably, GFP-tagged HDAC1, another subunit of the NuRD complex (Xue et al, 1998), also localized to the array upon immobilization of RNF8 (Figure 3B). Recruitment of CHD4 was found to be specific for RNF8 and was not observed when tethering RNF168 (Figure 3C and G), in line with the notion that RNF8, but not RNF168, mediates higher-order chromatin unfolding (Figure 2C and D). Moreover, recruitment of CHD4, like chromatin decondensation by RNF8 (Figure 2E–G), did not require phospho-specific interactions through the FHA or the catalytic activity of the RING domain of RNF8 (Figure 3D and G). Although CHD4 has been suggested to bind to chromatin in a poly(ADP-ribose) (PAR)-dependent manner (Polo et al, 2010), the interaction between RNF8 and CHD4 at the array was independent of this modification (Figure 3E and G). In agreement with our findings in NIH2/4 cells, staining of AO3 cells, containing the 90-Mbp LacO array, with CHD4 antibodies showed that the RNF8-induced unfolded chromatin structures are decorated with CHD4 (Figure 3F).
Figure 3.
RNF8 recruits chromatin remodelling ATPase CHD4. Tethering of mCherry-LacR-RNF8 (red) to the LacO array in NIH2/4 cells triggers recruitment of (A) endogenous CHD4 (green), and (B) GFP-HDAC1 (green), while (C) mCherry-LacR-RNF168 (red) immobilization does not trigger endogenous CHD4 recruitment (green) to the array. Tethering of (D) mCherry-LacR-RNF8*FHA/*RING and (E) mCherry-LacR-RNF8 following treatment with PARP inhibitors also results in endogenous CHD4 recruitment (green). (F) mCherry-LacR-RNF8-mediated unfolded chromatin structures (red) in AO3 cells are decorated with endogenous CHD4 (green). The merge also shows staining with Hoechst 33258 (2 μg/ml). (G) Quantification of the percentage of cells with positive signal (CHD4 staining) at the array after tethering the indicated mCherry-LacR fusion proteins. Values represent the mean of two independent experiments (n=50 cells). (H) GFP-RNF8WT or GFP-RNF8*FHA/*RING was immunoprecipitated from U2OS cells followed by immunoblotting with CHD4 and GFP antibodies. WCE; whole-cell lysates. (I) HEK293T cells were transfected with plasmids encoding GFP or GFP-CHD4 for 24 h, and cells were lysed under denaturing conditions and subjected to GFP immunoprecipitation (IP). The beads were then washed extensively in denaturing buffer, re-equilibrated in native buffer and incubated or not with 100 ng bacterially purified, recombinant His6-RNF8 as indicated. After thorough washing, the bound complexes were resolved on SDS–PAGE and immunoblotted with RNF8 and GFP antibodies. (J) A series of overlapping GST-tagged CHD4 fragments were immobilized on GSH Sepharose and incubated with purified, recombinant His6-RNF8. Bound complexes were analysed by immunoblotting with RNF8 antibody. Figure source data can be found with the Supplementary data.
RNF8 physically interacts with CHD4
To analyse whether RNF8 and CHD4 interact biochemically, we performed co-immunoprecipitation experiments and found that endogenous CHD4 was indeed present in RNF8 immunoprecipitates from cells expressing either epitope-tagged WT RNF8 or RNF8*FHA/*RING (Figure 3H). Notably, CHD4 associated more avidly with the double FHA/RING mutant compared with WT RNF8. To test if RNF8 binds directly to CHD4, we immobilized GFP-CHD4 under mildly denaturing conditions to disrupt protein–protein interactions on beads, which were subsequently incubated with recombinant RNF8. Western blotting showed that recombinant RNF8 bound to the immobilized GFP-CHD4, but not to GFP alone (Figure 3I), suggesting that a direct interaction between CHD4 and RNF8 may be responsible for RNF8-dependent chromatin changes. We subsequently used GST fusion fragments spanning the coding region of CHD4 (Urquhart et al, 2011). The CHD4 fragments were bound to glutathione agarose beads and tested for their ability to bind recombinant RNF8. Notably, a CHD4 region spanning amino acids 1222–1507, but not the other CHD4 fragments, bound to recombinant RNF8 (Figure 3J). Together, these findings suggest that a region on CHD4 between amino acids 1222 and 1507 mediates the interaction with RNF8.
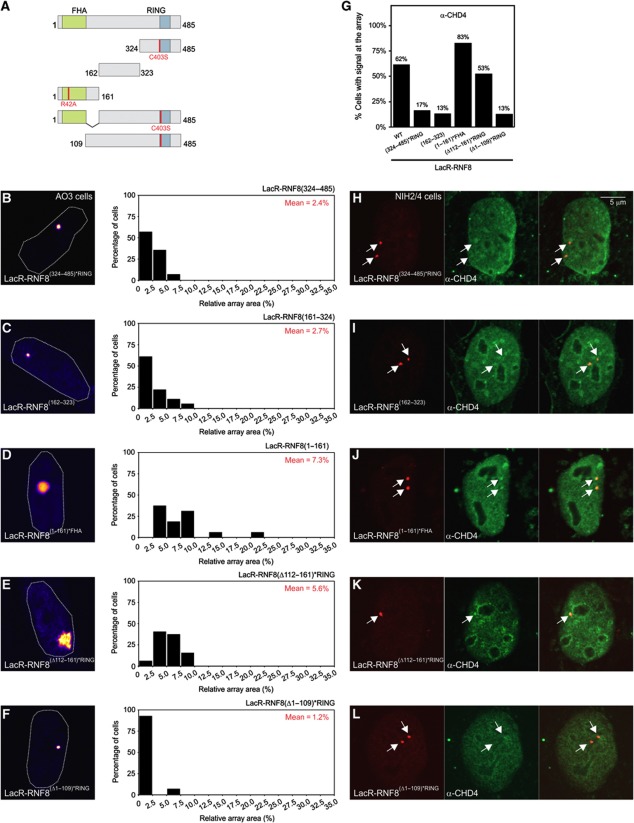
RNF8 interacts with CHD4 through its FHA domain
To map which region of RNF8 is necessary to mediate chromatin decondensation, we generated RNF8 deletion mutants in which the FHA and RING domains were inactivated (Figure 4A) to prevent phospho-specific interactions or dimerization with WT RNF8. Tethering of mCherry-LacR-RNF8(324−485)*RING (Figure 4B) or mCherry-LacR-RNF8(162–323) (Figure 4C) did not result in chromatin decondensation, while the N-terminal fragment mCherry-LacR-RNF8(1−161)*FHA mediated efficient chromatin decondensation (Figure 4D). Consistent with a role for CHD4 in RNF8-mediated chromatin unfolding, RNF8(1−161)*FHA also still recruited CHD4 (Figure 4G and J), while the other deletion mutants failed to do so (Figure 4G–I). The N-terminal RNF8 fragment that is sufficient to unfold chromatin and recruit CHD4 encompasses the conserved FHA domain (residues 17–111) as well as an α-helical extension (residues 130–140) that is located away from the phosphopeptide-interacting surface (Huen et al, 2007). Tethering of deletion mutant RNF8(Δ112−161)*RING still resulted in chromatin unfolding (Figure 4E) and CHD4 recruitment (Figure 4G and K), suggesting that the α-helical extension is not involved in chromatin decondensation. Conversely, deletion of the FHA domain (RNF8(Δ1−109)*RING) completely abolished chromatin unfolding (Figure 4F) and CHD4 recruitment (Figure 4G and L). In summary, RNF8 mediates chromatin unfolding and the recruitment of the chromatin remodeller CHD4 independently of canonical phospho-specific binding through the FHA domain or the catalytic activity of the RING finger domain. Interestingly, the FHA domain is essential for chromatin unfolding and CHD4 recruitment, suggesting that modulation of chromatin structure involves a new phospho-independent function of the FHA domain of RNF8.
Figure 4.
The FHA domain of RNF8 regulates chromatin decondensation and CHD4 recruitment in a phospho-independent manner. (A) Schematic representation of the RNF8 deletion mutants used in (A–J). The mutations to inactive the FHA and RING domain are indicated in red. (B–F) Chromatin decondensation in AO3 cells upon tethering the indicated RNF8 deletion mutants to the array. (G) Quantification of the percentage of cells with positive signal (CHD4 staining) at the array after tethering the indicated RNF8 fusion proteins. Values represent the mean of two independent experiments (n=30 cells). (H–L) Recruitment of endogenous CHD4 in NIH2/4 cells induced by tethering of the indicated RNF8 deletion mutants.
The ATPase activity of CHD4 is required for RNF8-mediated decondensation
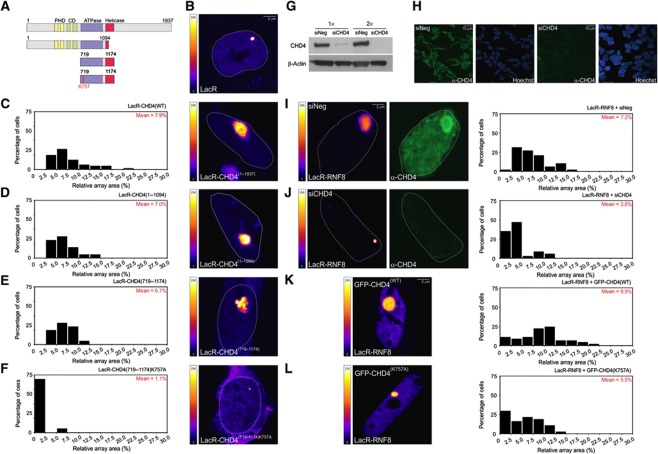
The finding that CHD4 is recruited by RNF8 raised the question whether the chromatin remodeller CHD4 is responsible for RNF8-mediated chromatin unfolding. At first glance, it might be surprising that the NuRD ATPase CHD4, which is implicated in transcriptional repression (Xue et al, 1998; Denslow and Wade, 2007), a phenomenon often linked to chromatin condensation, may be involved in chromatin unfolding. To assess the impact of CHD4 on chromatin structure, we constructed an mCherry-LacR-CHD4 fusion protein (Figure 5A). Tethering of CHD4, but not LacR alone (Figure 5B and C) triggered extensive chromatin decondensation to a similar extent as mCherry-LacR-RNF8 (Figure 5C compare with Figure 2D), suggesting that CHD4 may be responsible for RNF8-induced chromatin remodelling. CHD4 contains a conserved ATPase/helicase domain that is also found in other SNF2 family members (Woodage et al, 1997; Hall and Georgel, 2007). Tethering an N-terminal fragment of CHD4(1−1094), which contains the intact ATPase domain, but lacks the conserved helicase motifs (V and VI; Figure 5A), also efficiently triggered chromatin decondensation (Figure 5D), indicating that the putative helicase function of CHD4 may not be required for the observed chromatin decondensation. We subsequently tethered a region spanning the conserved ATPase/helicase domain of CHD4(719−1174) (Figure 5A), which unfolded the array similar to full-length CHD4 (Figure 5E). Interestingly, tethering the CHD4 ATPase/helicase domain in which the ATPase activity was inactivated by an amino-acid substitution (K757A; Figure 5A) failed to mediate chromatin unfolding, showing that CHD4-mediated chromatin decondensation strictly requires the ATPase activity of CHD4 (Figure 5F). To directly address whether CHD4 is involved in RNF8-mediated chromatin changes, we reduced CHD4 levels by RNA interference and measured how this affected RNF8-mediated chromatin unfolding. Western blot analysis (Figure 5G) and immunofluorescence microscopy (Figure 5H) confirmed that siRNAs directed against CHD4 efficiently lowered the CHD4 protein levels in AO3 cells. We subsequently quantified the size of the array upon tethering mCherry-LacR-RNF8 in cells transfected with control or CHD4 siRNAs. Our analysis showed that siRNA-mediated knockdown of CHD4 markedly impaired chromatin decondensation by RNF8 (Figure 5I and J). To test if RNF8-mediated chromatin decondensation requires the ATPase activity of CHD4, we expressed the dominant-negative GFP-CHD4K757A to interfere with the ATPase function of endogenous CHD4 (Smeenk et al, 2010). In contrast to expression of GFP-CHD4WT, expression of GFP-CHD4K757A markedly reduced chromatin decondensation upon RNF8 tethering (Figure 5K and L). These results demonstrate that RNF8-mediated alterations in higher-order chromatin structure are dependent on the ATPase activity of the chromatin remodeller CHD4.
Figure 5.
RNF8-dependent chromatin decondensation depends on the ATPase activity of CHD4. (A) Schematic representation of the CHD4 fusion proteins. (B–F) Chromatin decondensation in AO3 cells upon tethering the indicated CHD4 fusion proteins or LacR to the array. (G) Western blot analysis of AO3 cells transfected with control siRNA or CHD4 siRNA. (H) Immunofluorescence microscopy on cells transfected with control siRNA or CHD4 siRNA. Cells were pre-extracted, fixed and stained with antibodies against CHD4. (I–L) Chromatin decondensation upon tethering mCherry-LacR-RNF8 to the array in AO3 transfected with (I) siNeg, (J) siCHD4, (K) GFP-CHD4WT or (L) dominant-negative GFP-CHD4K757A. Quantitative measurements on the relative size of the array (in % of nuclear area) are shown next to the images as histograms. The values represent the mean of ∼50 cells from two independent experiments. The mean size of the array (in % of nuclear area) for each condition is shown in red in the histograms. The LUT for the colour coding is shown left of the panel. Figure source data can be found with the Supplementary data.
RNF8 recruits CHD4 to DNA damage independently of its ubiquitin-ligase activity
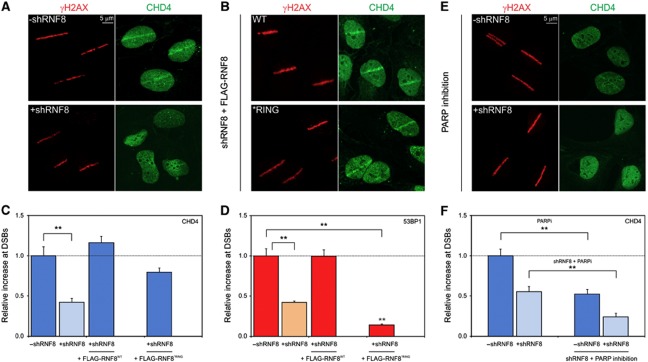
Having established that RNF8 recruits CHD4 in the targeting system, resulting in chromatin decondensation, we next examined whether CHD4 is also recruited to sites of DNA damage by RNF8 in the physiological context of the DDR. DSBs were inflicted by micro-irradiation of cells that were sensitized with 5′-bromo-2-deoxyuridine (BrdU) (Lukas et al, 2003; Luijsterburg et al, 2009). To test if RNF8 is involved in CHD4 recruitment to DSBs, we used a U2OS cell line stably expressing a doxycycline-inducible shRNA that targets RNF8 (Mailand et al, 2007). Endogenous CHD4 was recruited to laser-generated DSBs marked by phosphorylated histone H2AX (γH2AX; Figure 6A), in line with several recent reports (Chou et al, 2010; Larsen et al, 2010; Polo et al, 2010; Smeenk et al, 2010). Importantly, we found that depletion of RNF8 significantly reduced the recruitment of CHD4 to laser-generated DSBs (Figure 6A and C), consistent with an important role of RNF8 in CHD4 recruitment. As previously described (Mailand et al, 2007), RNF8 depletion also significantly reduced 53BP1 accumulation at DSBs (Figure 6D). We subsequently expressed WT RNF8 (RNF8WT) or mutant RNF8*RING that were rendered insensitive to the RNF8-specific shRNA (Mailand et al, 2007). Ectopic expression of WT RNF8 following knockdown of endogenous RNF8 restored the recruitment of CHD4 (Figure 6B and C) and 53BP1 (Figure 6D) to laser-generated DSBs. Introducing ectopic RNF8*RING also restored the recruitment of endogenous CHD4 (Figure 6B and C), in contrast to 53BP1 (Figure 6D), which is consistent with the RING-independent recruitment of CHD4 to tethered RNF8 (Figure 3D and G) as well as the RING-independent nature of the CHD4-RNF8 interaction (Figures 3H and 4J). It should be noted that the levels of CHD4 at DSBs were slightly lower in cells expressing RNF8*RING as compared with WT RNF8-expressing cells and, although this difference was not statistically significant, the possibility remains that RING-dependent RNF8 dimerization or RNF8-mediated ubiquitylated further enhances CHD4 recruitment in the DDR. Together, these data show that RNF8 recruits CHD4 to sites of DSBs in a manner that is largely, if not completely, independent of the ubiquitin-ligase activity of RNF8 identifying a new non-catalytic role of RNF8 in the DDR.
Figure 6.
RNF8 regulates recruitment of CHD4 to DSBs independently of its ubiquitin-ligase activity. (A) U2OS cells stably expressing a doxycycline (dox)-inducible shRNA targeting RNF8 (shRNF8) were induced (lower panel) or not (upper panel) with dox for 48–72 h, exposed to laser micro-irradiation and 15 min later co-immunostained with antibodies to γH2AX (red) and CHD4 (green). (B) U2OS/shRNF8 cells engineered to induce WT or *RING alleles of FLAG-tagged, shRNA-resistant RNF8 were incubated with dox (48–72 h), exposed to laser micro-irradiation and 15 min later co-immunostained with antibodies to γH2AX (red) and CHD4 (green). Quantification of the accumulation of (C) endogenous CHD4 or (D) endogenous 53BP1 accumulation at laser-generated DSBs marked by γH2AX in the indicated cell lines. (E) U2OS/shRNF8 cells were treated with PARP inhibitor (10 μM) for 1–2 h and the expression of an shRNA targeting RNF8 was either induced with dox (48–72 h; lower panel) or not (upper panel). Cells were subsequently exposed to laser micro-irradiation and 15 min later co-immunostained with antibodies to γH2AX (red) and CHD4 (green). (F) Quantification of the accumulation of endogenous CHD4 at laser-generated DSBs using the indicated conditions. Graphs represent the relative fluorescence increase in the micro-irradiated area calculated by subtracting the background intensity in the undamaged parts of the nucleus from the intensity in the micro-irradiated area. The relative accumulation of CHD4 (C, F) or 53BP1 (D) in the absence of dox was normalized to 1 for comparison. Values represent the mean of ∼50 cells from at least two independent experiments. Error bars indicate the s.d. Stars (**) indicate highly significant differences (P=3.8 × 10−6 for panel (C), P=9.4 × 10−13 and 8.1 × 10−17 for panel (D) and P=2.8 × 10−4 and 1.9 × 10−4 for panel (F)) based on a t-test.
Although our results demonstrate that RNF8 recruits CHD4 to DSBs, previous studies have reported that CHD4 is recruited to laser-generated DNA lesions in an, at least partially poly(ADP-ribose) polymerase (PARP)-dependent fashion (Chou et al, 2010; Polo et al, 2010). However, in lieu of a quantitative evaluation (Chou et al, 2010; Polo et al, 2010), it is currently unclear to what extent CHD4 recruitment depends on the action of PARP. Analysis of U2OS cells following micro-irradiation in the presence of PARP inhibitors revealed a quantitatively reduced recruitment of CHD4 to DSBs to a similar extent as knockdown of RNF8 (Figure 6E and F). Given that PARP inhibition or RNF8 knockdown equally impairs CHD4 recruitment, this raises the question whether the RNF8- and PARP-dependent recruitment of CHD4 are part of the same pathway. We found that combining PARP inhibition with RNF8 knockdown had an additive inhibitory effect on the recruitment of CHD4 to DSBs (Figure 6E and F), suggesting the presence of distinct PARP- and RNF8-dependent pathways for CHD4 recruitment. This is also consistent with our findings that CHD4 recruitment by tethered RNF8 is not sensitive to PARP inhibition (Figure 3E and G), and that the PAR-binding factor ALC1 is not recruited by RNF8 (Supplementary Figure S4). Thus, CHD4 is recruited to DSBs by mechanistically distinct pathways mediated through either the action of PARP or non-catalytically through RNF8.
The ATPase activity of CHD4 promotes ubiquitin conjugation and BRCA1 assembly
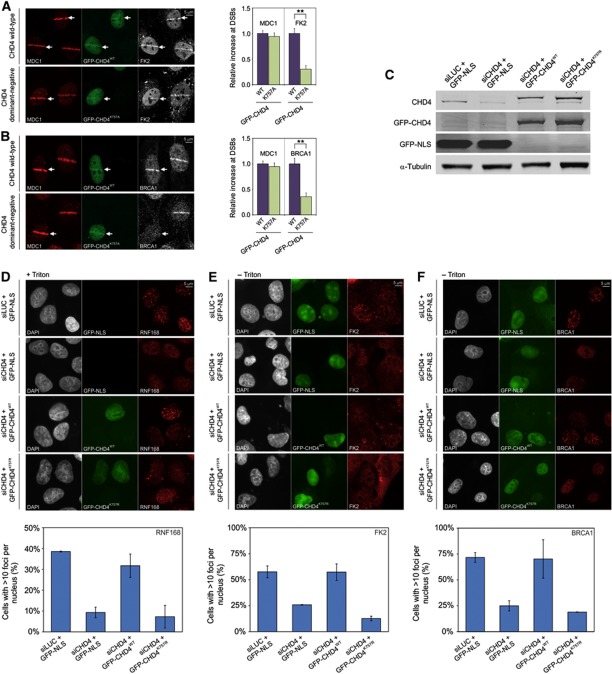
We next sought to gain insight into the function of the RNF8-mediated recruitment of CHD4 to DSBs. Having established that the ATPase activity of CHD4 is required for RNF8-mediated chromatin unfolding, we tested whether catalytically active CHD4 is necessary for RNF8-mediated ubiquitylation. Interestingly, expression of dominant-negative GFP-CHD4K757A markedly impaired ubiquitin conjugation (FK2 staining) at DSBs, while MDC1 recruitment, which occurs upstream of RNF8 signalling, was not affected (Figure 7A). RNF8-mediated ubiquitin conjugates play a central role in the recruitment of BRCA1 to DNA lesions. Accordingly, we found that expression of GFP-CHD4K757A markedly suppressed BRCA1 assembly, whereas MDC1 recruitment in the same cells was not affected (Figure 7B). Notably, over-expression of WT GFP-CHD4 did not affect ubiquitin conjugation (Figure 7A) or recruitment of BRCA1 (Figure 7B). To corroborate these findings and to establish whether the chromatin remodelling activity of CHD4 is required for an efficient DDR, we depleted endogenous CHD4 by RNA interference and complemented the cells with siRNA-resistant GFP-tagged CHD4WT or ATPase-dead CHD4K757R (Larsen et al, 2010). Non-complemented cells were transfected with GFP-NLS as a control. Western blot analysis confirmed that endogenous CHD4 was depleted and replaced with the GFP-tagged versions of CHD4 (Figure 7C). Cells transfected with control siRNAs showed clear accumulation of endogenous RNF168 (Figure 7D), conjugation of ubiquitin (Figure 7E) as well as BRCA1 accumulation (Figure 7F) in ionizing-radiation-induced foci (IRIF) following irradiation with 2Gy. However, all these events were significantly suppressed by depletion of endogenous CHD4 (Figure 7D–F), in line with recent findings (Larsen et al, 2010; Smeenk et al, 2010). Ectopic expression of GFP-CHD4WT in cells depleted for endogenous CHD4 rescued the defects in RNF168 and BRCA1 accumulation as well as ubiquitin conjugation in IRIF to a large extent (Figure 7D–F). Conversely, ectopic expression of CHD4K757R failed to rescue these defects (Figure 7D–F), showing that the ATPase activity of CHD4 is essential to initiate an efficient DDR in chromatin-flanking DSBs. These findings suggest that RNF8-mediated and CHD4-executed changes in chromatin structure promote RNF8-mediated ubiquitylation and the subsequent recruitment of RNF168 and BRCA1 to DSBs.
Figure 7.
The ATPase activity of CHD4 regulates DNA damage-induced ubiquitin conjugation and efficient BRCA1 assembly. (A) U2OS cells expressing either GFP-CHD4WT or dominant-negative GFP-CHD4K757A were micro-irradiated and 15 min later co-immunostained with antibodies to MDC1 (red) and conjugated ubiquitin (FK2; white). Non-transfected cells were also micro-irradiated to serve as an internal control. (B) Similar to (A) except that cells were co-immunostained with antibodies to MDC1 (red) and BRCA1 (white). Stars (**) indicate highly significant differences (P=5.5 × 10−8 for panel (A), P=5.0 × 10−5 for panel (B) based on a t-test. (C) Western blot analysis of U2OS cells transfected with the indicated siRNAs and GFP constructs. Membranes were probed with antibodies to CHD4, GFP and α-tubulin. (D–F) U2OS cells were transfected with the indicated siRNAs and GFP constructs, irradiated with IR (2 Gy) and stained for (D) RNF168, (E) FK2 or (F) BRCA1. Note that cells stained for RNF168 were Triton X-100 extracted before fixation, which resulted in loss of the GFP-NLS signal. Quantification of the percentage of cells with >10 foci per nucleus is shown below the images. Values represent the mean of >100 cells from at least two independent experiments. Error bars indicate the s.d. Figure source data can be found with the Supplementary data.
We next wondered what could underlie the need for CHD4-mediated chromatin remodelling in initiating efficient RNF8 signalling. It is possible that the short retention time of RNF8 at sites of DSBs (∼4.3s; Mailand et al, 2007) is not sufficient to efficiently initiate the subsequent steps in the DDR and that CHD4-mediated chromatin remodelling plays an important role in facilitating these early events. To test if a more stably bound RNF8 still required CHD4 to initiate the DDR, we tethered either WT LacR-RNF8 or an LacR-RNF8 mutant (RNF8(Δ1−109)) that cannot interact with CHD4 and fails to unfold chromatin (Figure 4F and L) in human U2OS cells harbouring a LacO array (Janicki et al, 2004). Due to the high affinity binding of the LacR to LacO sequences in the genome, this approach is expected to result in a significantly prolonged retention time of RNF8 compared with its binding during the DDR (Mailand et al, 2007). Tethering RNF8WT triggered efficient H2A ubiquitylation (Supplementary Figure S5A) as well as recruitment of endogenous BRCA1 (Supplementary Figure S5B) and RNF168 (Supplementary Figure S5C) at the array. However, tethering RNF8(Δ1−109) triggered these events to the same extent (Supplementary Figure S5A–C), suggesting that a more stably bound RNF8 does not require CHD4-mediated chromatin remodelling to recruit downstream repair factors. Moreover, tethering of CHD4, although sufficient to unfold chromatin (Figure 5C), failed to trigger H2A ubiquitylation or BRCA1 recruitment (Supplementary Figure S5A and B), suggesting that an open chromatin structure is not sufficient to initiate the DDR in the targeting system. Corroborating these findings, we found that tethering LacR-RNF8 in control cells or in cells depleted for CHD4 (see Supplementary Figure S5D) triggered efficient H2A ubiquitylation (Supplementary Figure S5E), as well as BRCA1 (Supplementary Figure S5F) and RNF168 accumulation (Supplementary Figure S5G) to the same extent. Our results suggest that the ATPase activity of CHD4 promotes efficient initiation of the DDR by transiently bound RNF8 at damaged chromatin and that artificially prolonging the chromatin retention time of RNF8 bypasses the requirement for CHD4.
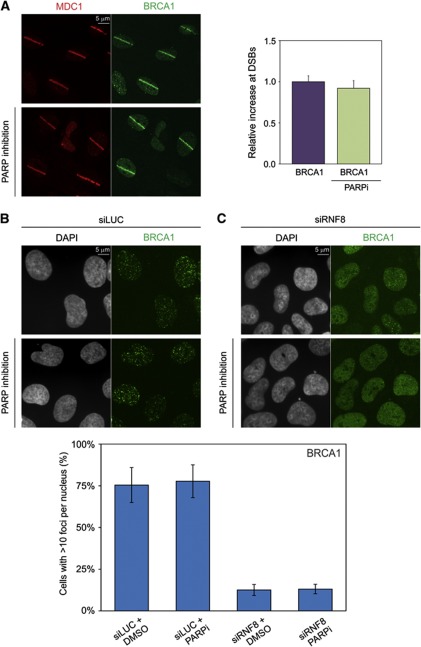
The PARP-dependent recruitment of CHD4 does not contribute to BRCA1 assembly
Besides its recruitment to DSBs in an RNF8-dependent fashion, CHD4 is also partially recruited through a PARP-dependent mechanism (Figure 6E and F; Chou et al, 2010; Polo et al, 2010). This raises the question whether the PARP-dependent recruitment of CHD4 contributes to efficient assembly of BRCA1 at DSBs. Treatment of cells with PARP inhibitors did not affect the recruitment of BRCA1 to laser-induced DSBs compared with control cells (Figure 8A), suggesting that the PARP-dependent recruitment of CHD4 does not promote BRCA1 accumulation. Corroborating these findings, we found that BRCA1 accumulation in IRIF was also not affected by PARP inhibitors (Figure 8B), while RNF8 knockdown significantly suppressed BRCA1 recruitment in IRIF, as expected (Figure 8C; Mailand et al, 2007). Moreover, PARP inhibitors did not further reduce BRCA1 accumulation in cells depleted for RNF8 (Figure 8C). Together, our study suggests that RNF8 recruits the chromatin remodelling activity of CHD4, which subsequently creates a local chromatin environment that promotes the RNF8-dependent initiation of the DDR through regulating efficient RNF168 assembly, DNA damage-induced ubiquitylation and the subsequent recruitment of BRCA1 to sites of DNA damage.
Figure 8.
The PARP-dependent recruitment of CHD4 does not contribute to BRCA1 assembly. (A) U2OS cells were treated with PARP inhibitor (10 μM) for 1–2 h (lower panel) or not (upper panel), exposed to laser micro-irradiation and 15 min later co-immunostained with antibodies to MDC1 (red) and BRCA1 (green). Quantification of the accumulation of DNA damage-induced ubiquitin conjugation or endogenous BRCA1 at laser-generated DSBs using the indicated conditions are shown next to the micrographs. Graphs represent the relative fluorescence increase in the micro-irradiated area calculated by subtracting the background intensity in the undamaged parts of the nucleus from the intensity in the micro-irradiated area. The relative accumulation of BRCA1 in the absence of PARP inhibitors was normalized to 1 for comparison. Values represent the mean of ∼50 cells. Error bars indicate the s.d. (B, C) U2OS cells transfected with (B) siLUC or (C) siRNF8 were treated with PARP inhibitor (10 μM) for 1–2 h or not, irradiated with IR (2 Gy) and stained for BRCA1. Quantification of the percentage of cells with >10 foci per nucleus is shown below the images. Values represent the mean of >300 cells from at least two independent experiments. Error bars indicate the s.d.
Discussion
Chromatin is the primary substrate of all genome-associated processes, including DNA repair, but the wrapping of DNA in chromatin limits the accessibility of repair proteins to lesions. To overcome this physical barrier, different types of chromatin modifiers have been suggested to mediate chromatin changes during DNA repair in order to facilitate the binding of DNA repair proteins (Dinant et al, 2008; Luijsterburg et al, 2009; van Attikum and Gasser, 2009; Venkitaraman, 2010; Luijsterburg and van Attikum, 2011). However, as many different chromatin remodelling factors are recruited to DNA lesions, it is difficult to clearly define the contribution of individual proteins to alterations in chromatin architecture. Moreover, direct assays to monitor DNA damage-induced changes in higher-order chromatin structure are currently limited.
In an attempt to overcome these limitations and gain insights in the individual contribution of DDR proteins to chromatin remodelling, we utilized a system to directly monitor and visualize higher-order chromatin changes triggered by histone ubiquitin ligases (Tumbar et al, 1999; Ye et al, 2001). Inspired by the finding that RNF2-mediated H2A ubiquitylation is linked to tightly compacted chromatin (Simon and Kingston, 2009) and that RNF8-mediated H2A ubiquitylation has recently been linked to transcriptional repression (Shanbhag et al, 2010), we set out to assess the impact of H2A ubiquitin ligases on large-scale chromatin structure. We provide direct evidence that the ubiquitin ligase RNF8, but not RNF2 or RNF168, elicits higher-order chromatin decondensation independently of its ubiquitin-ligase activity by recruiting the NuRD ATPase CHD4. Consistent with these findings, we show that RNF8 directly interacts with CHD4 and regulates its recruitment to DNA lesions in a non-catalytic fashion through a new phospho-independent function of the FHA domain. Although CHD4 is usually considered a transcriptional repressor (Denslow and Wade, 2007), and may therefore be expected to compact chromatin, we show that CHD4 is required for RNF8-mediated chromatin decondensation. Moreover, we show that the ATPase activity of CHD4 promotes DNA damage-induced ubiquitin conjugation and efficient recruitment of RNF168 and BRCA1 to DSBs. Interestingly, direct tethering of RNF168 to the LacO array resulted in efficient poly-ubiquitylation and BRCA1 recruitment (data not shown), even though immobilized RNF168 failed to recruit RNF8 or CHD4 to chromatin. Moreover, while CHD4 was essential to mount an efficient DDR by transiently bound RNF8 at DSBs, CHD4 was not required for the efficient recruitment of repair factors when RNF8 was tethered to the LacO array with high affinity. These findings suggest that the concerted action of RNF8 and CHD4 may promote the initial chromatin ubiquitylation during the DDR to reach a certain threshold sufficient to facilitate stable retention of RNF168 at DSBs, which in turn, amplifies the ubiquitylation signal.
We propose that the initial non-catalytic role of RNF8 in mediating chromatin decondensation in a CHD4-dependent manner results in a local chromatin environment that is amendable to chromatin ubiquitylation, a process that we refer to as ‘chromatin remodelling-assisted ubiquitylation’ (Figure 9). In line with such a model, we show that the chromatin remodelling activity of CHD4 promotes efficient association of RNF168 and the formation of DNA damage-induced ubiquitin conjugates at DSBs, concomitant with efficient assembly of BRCA1. The proposed mechanism bears similarities to the finding that nucleosome remodelling by CHD4 stimulates histone deacetylation by HDAC1/2 in the NuRD complex (Xue et al, 1998). In the latter case, it has been proposed that chromatin decondensation by CHD4 facilitates histone deacetylation by HDAC1/2 resulting in transcriptional repression (Xue et al, 1998). Likewise, our data indicate that CHD4-mediated chromatin remodelling promotes RNF8-catalysed ubiquitin conjugation at DSBs. It is feasible that chromatin unfolding exposes otherwise inaccessible ubiquitylation substrates for RNF8, such as the C-terminal tails of histone H2A. Additionally, we show that RNF8-mediated recruitment of CHD4 promotes RNF168 and BRCA1 assembly, likely by promoting efficient ubiquitin conjugation at DSBs. Whether PARP-dependent recruitment of CHD4 (Chou et al, 2010; Polo et al, 2010) also contributes to promoting ubiquitin conjugating and BRCA1 accumulation at DSBs is currently unclear. However, our data suggest its contribution might be limited, as we found that BRCA1 recruitment to DSBs is not affected by PARP inhibitors. It is tempting to speculate that the PARP-dependent recruitment of CHD4 may play a role in the HDAC1/2-dependent deacteylation of histone H3K56 involved in non-homologous end-joining (Miller et al, 2010; Polo et al, 2010). Whether this process requires the chromatin remodelling activity of CHD4 is currently unclear. It is interesting to note that the activity of RNF8 (and RNF168) in heterochromatin-flanking DSBs regulates the dispersal, rather than the association, of the CHD4-related chromatin remodelling enzyme CHD3 through a KAP-1-dependent mechanism. It is believed that the chromatin-association of the repressive CHD3 remodeller inhibits DNA repair and that the dispersal of CHD3 from heterochromatin may facilitate repair in this condensed chromatin compartment (Noon et al, 2010; Goodarzi et al, 2011).
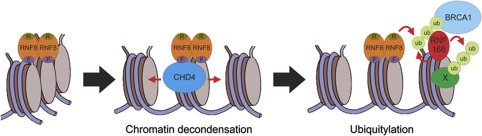
Figure 9.
Model for chromatin remodelling-assisted ubiquitylation in the DDR. An RNF8 dimer (orange oval) is recruited to a DSB by binding to phosphorylated MDC1. The recruited RNF8 dimer binds CHD4 (blue oval) through its FHA domain in a phospho-independent manner, resulting in local chromatin decondensation, which subsequently enhances ubiquitin conjugation at DSBs and association of RNF168 and BRCA1.
In the present study, we have identified a new non-catalytic role of RNF8 in mediating large-scale chromatin decondensation. Our findings implicate chromatin remodelling-assisted ubiquitylation by RNF8 in the efficient assembly of RNF168 and BRCA1 at DSBs. In the light of these findings, it is interesting to note that RNF2 was found to mediate chromatin compaction independently of its ubiquitin-ligase activity (Eskeland et al, 2010). Recently, we (Acs et al, 2011) and others (Meerang et al, 2011) have shown that the ubiquitin-selective AAA-ATPase valosin-containing protein (VCP) is recruited in a ubiquitylation-dependent fashion by RNF8 and plays a critical role in mediating the recruitment of downstream repair factors. Interestingly, VCP stimulates 53BP1 recruitment by the selective extraction of a chromatin-associated target (Acs et al, 2011). Thus, it appears that both ubiquitylation-dependent and -independent activities of RNF8 are responsible for the recruitment of ATPases that affect chromatin structure and composition in a DNA damage-inducible manner. In summary, we propose that RNF8 displays dual non-catalytic and catalytic activities, responsible for chromatin decondensation and histone ubiquitylation, respectively, that are involved in creating a local chromatin environment that is permissive to the assembly of checkpoint and repair machineries. It is feasible, however, that CHD4, while creating a permissive local chromatin environment, may at the same time contribute to the previously reported RNF8-dependent repression of transcription at DNA lesions (Shanbhag et al, 2010).
Materials and methods
Cell lines
NIH2/4 mouse cells containing a 256 × LacO array of ∼10kbp (Soutoglou et al, 2007), human U2OS osteosarcoma cells and human U2OS 2-6-3 cells containing 200 copies of a LacO (256 ×)/TetO (96 ×)-containing cassette of ∼4Mbp (Janicki et al, 2004), were cultured in DMEM, supplemented with antibiotics and 10% fetal calf serum. AO3 hamster cells, containing a 90-Mbps amplification of LacO sequences, the DHFR gene and flanking genomic DNA (Tumbar et al, 1999), were cultured in a 1:1 mixture of DMEM/Ham’s F12 medium, supplemented with antibiotics and 20% fetal calf serum. Human U2OS cells stably expressing doxycline-inducible shRNF8 and FLAG-RNF8 (WT or *RING) (Mailand et al, 2007) were grown in DMEM supplemented with 10% FCS and antibiotics (1μg/ml Puromycin, 2μg/ml, Blasticidin S, and, in case of FLAG-RNF8-expressing cells, 200μg/ml Zeocin). Expression of shRNF8 and FLAG-RNF8 or FLAG-RNF8*RING, or the association of EGFP-rTetR-RNF8 with genomically inserted tetO sequences was induced by the addition of 2 μg/ml doxycycline to culture media for 72–96 h. All media contained glutaMAX (Gibco, Breda, The Netherlands) and all cells were cultured at 37°C in an atmosphere of 5% CO2.
Plasmids
The mCherry-LacR gene fused to a nuclear localization signal (Soutoglou and Misteli, 2008) was amplified and inserted into mCherry-C1. To generate mCherry-LacR-tagged fusion proteins, RNF2 (WT or *RING/C90S), RNF8 (WT, *RING/C403S, *FHA/R42A or *RING/*FHA) (Mailand et al, 2007) and RNF168 (*MIU/A179G, A450G and *MIU/*RING/C16S) (Doil et al, 2009) were generated and inserted in mCherry-LacR. RNF8 fragments 1–161 (containing R42A), 162–323 and 324–485 (containing C403S), or deletion mutants RNF8Δ112–161 (containing C403S) or RNF8Δ1–109 (either containing C403S or not) were generated by PCR and inserted into mCherry-LacR. CHD4 (WT, 719–1174, 719–1174 ATPase inactive/K757A or 1–1094) were inserted into mCherry-LacR. The latter deletion mutant lacks the conserved helicase motifs V and VI. The gene encoding the reverse TetR was amplified, fused to an NLS sequence and inserted into EGFP-C1. The RNF8 coding region was subsequently inserted in this vector yielding EGFP-rTetR-RNF8. siRNA-resistant cDNAs of either CHD4WT or CHD4K757R containing silent mutations at positions G1617A, C1620T and T1623C were generated and inserted into pcDNA4TO-GFP (Larsen et al, 2010). Mutations were introduced by site-directed mutagenesis. All constructs were verified by sequencing.
Transfections
Cells were transfected with plasmid DNA using Lipofectamine 2000 according to the manufacturer’s instructions. Cells were typically imaged 24 h after transfection. siRNA oligonucleotides (Qiagen or MWG Biotech) were synthesized to the following mouse sequence: CHD4-1 (5′-CCC GAT CGT GGT GGA AGA CAA-3′), or human sequence CHD4-3 (5′-GAG CGG CAG UUC UUU GUG AUU-3′) (Larsen et al, 2010) or RNF8 (5′-GAG GGC CAA UGG ACA AUU A-3′). AllStars Neg Control siRNA or Luciferase (5′-CGU ACG CGG AAU ACU UCG A-3′) siRNA were used as negative controls. All siRNA transfections were performed with 40 nM (double siRNA tranfection) or 100 nM (single siRNA transfection) siRNA duplexes using Lipofectamine 2000 (Invitrogen). Cells were typically transfected twice with siRNAs (at 0 and 16 h), or with siRNAs (0 h) and plasmid DNA (16 h), and analysed 70 h after the first transfection.
Western blotting
Cell extracts were generated by cell lysis, boiled in sample buffer, separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose or PVDF membranes (Millipore). Expression of CHD4 was analysed by immunoblotting with rabbit anti-CHD4 (Xue et al, 1998) at 1:1000 or mouse monoclonal antibodies against CHD4 (Abcam, 1:1000) followed by a secondary antibody (goat anti-rabbit 1:5000) and ECL detection, or followed by secondary antibodies donkey anti-rabbit 700CW at 1:10.000 and donkey anti-mouse 800CW at 1:5000 and detection using the Odyssey infrared imaging scanning system (LI-COR Biosciences, Lincoln, Nebraska USA).
Immunoprecipitation
U2OS cells were transfected with plasmids encoding GFP-tagged RNF8WT or RNF8*FHA/*RING. For immunoprecipitation, cells were lysed in EBC buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktails. The cleared lysates were subjected to GFP immunoprecipitation with GFP Trap beads (Chromotek), and the beads were then washed four times with EBC buffer and boiled in sample buffer. Bound proteins were resolved by SDS-PAGE and immunoblotted with mouse monoclonal antibodies against CHD4 (Abcam, 1:1000) and GFP (Roche, 1:1000).
GFP-CHD4-RNF8-binding assay
HEK293T cells transfected with GFP constructs for 24 h were lysed in denaturing buffer (20 mM Tris, pH 7.5; 50 mM NaCl; 0.5% NP-40; 1% sodium deoxycholate; 1% SDS; 1 mM EDTA) containing protease and phosphatase inhibitor cocktails (Sigma) and subjected to immunoprecipitation with GFP-TRAP beads (Chromotek) for 2 h at 4°C. The beads were then washed extensively in a buffer (20 mM Tris, pH 7.5; 50 mM NaCl; 0.5% NP-40; 0.5% sodium deoxycholate; 0.5% SDS; 1 mM EDTA) that disrupts protein-protein interactions, followed by two washes in EBC buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 0.5% NP-40; 1 mM EDTA), and incubated with 100 ng bacterially purified, recombinant His6-RNF8 for 2 h at 4°C. The beads were then washed thoroughly in EBC buffer and processed for immunoblotting. Antibodies used were rabbit polyclonal against RNF8 (Mailand et al, 2007) and mouse monoclonal against GFP (Santa Cruz).
GST-CHD4-RNF8-binding assay
A series of overlapping GST-CHD4 fragments (Urquhart et al, 2011) were immobilized on GSH Sepharose and incubated in binding buffer (50 mM Tris, pH 7.5; 250 mM NaCl; 1 mM EDTA; 0.5% NP-40; 1 mM DTT) supplemented with 0.3 mg/ml BSA for 1 h at 4°C. Bacterially purified, recombinant His6-RNF8 (10 ng per reaction) was added, and incubated with the immobilized GST-CHD4 fragments for an additional 30 min. The beads were then washed five times in binding buffer, and bound complexes were resolved on SDS-PAGE and immunoblotted with anti-RNF8 antibody.
Immunofluorescent labelling
Immunofluorescent labelling of cells was carried out as described previously (Luijsterburg et al, 2010). Cells were either directly fixed or pre-extracted with 0.5% Triton X-100 (Serva, Heidelberg, Germany) in microscopy medium (137 mM NaCl, 5.4 mM KCl, 1,8 mM CaCl2, 0.8 mM MgSO4, 20 mM D-glucose and 20 mM HEPES, pH 7) on ice for 5 min and subsequently fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min at 4°C. Alternatively, cells were pre-extracted with 0.25% Triton X-100 in cytoskeletal (CSK) buffer (10 mM Hepes, 300 mM sucrose, 100 mM NaCl and 3 mM MgCl2) on ice for 2 min when using primary antibodies against RNF168. Cells were subsequently treated with 100 mM glycine in PBS for 10 min to block unreacted aldehyde groups. Cells were rinsed with PBS and equilibrated in WB (PBS containing 0.5% BSA, and 0.05% Tween 20; Sigma-Aldrich, St Louis, MO, USA). Antibody steps and washes were in WB. The primary antibodies were incubated overnight at 4°C. Primary antibodies were rabbit anti-CHD4 (Xue et al, 1998) at 1:500-1:1000 (a gift of Dr W Wang), mouse anti-ubiquitylated H2A at 1:100 (Upstate), mouse anti-FK2 at 1:100 (ENZO), rabbit anti-53BP1 at 1:200 (Novus Biologicals, Cambridge, UK), mouse anti-γH2AX at 1:1000 (Millipore, Billerica, MA, USA), rabbit anti-MDC1 at 1:1000 (Abcam), rabbit anti-RNF168 at 1:350 (a gift of Dr D Durocher; Stewart et al, 2009), and mouse anti-BRCA1 at 1:100 (Santa Cruz). Detection was done using goat anti-mouse or goat anti-rabbit Ig coupled to Alexa 488, 546 or 647 (1:1000; Invitrogen Molecular probes). Samples were mounted in Mowiol, and images were acquired on a Zeiss LSM 510 confocal microscope (see Microscopic analysis for details).
Microscopy analysis
Live-cell imaging and analysis of fixed samples was performed on Zeiss LSM 510 META confocal microscope, equipped with a 37°C climate chamber, a × 63 Plan-A (1.4 NA) oil immersion lens (Zeiss, Oberkochen, Germany), a 60-mW Argon laser (488 and 514 nm), a 5-mW Helium–neon 1 (543 nm) laser, a 15-mW Helium–neon 2 (633) laser, two photomultiplier tubes (PMT) and a META detector. Images were recorded using Zeiss LSM imaging software in multi-track mode (Zeiss, Oberkochen, Germany). Living cells were examined in culture medium at 37°C in an atmosphere of 5% CO2. An argon-ion laser (60 mW) was used for excitation at 488 nm, passed onto the sample by a 490-nm dichroic mirror and emission light was filtered by a 505–550 nm emission filter. A helium–neon laser (5 mW) was used for excitation at 543 nm, passed onto the sample by a 543-nm dichroic mirror and emission light was filtered by a 560–615 nm emission filter. A helium–neon laser (15 mW) was used for excitation at 633 nm, passed onto the sample by a 633-nm dichroic mirror and emission light was filtered by a 650 long-pass emission filter. Images were quantified using Image J software. Image J was also used to convert 8-bit greyscale images into coloured images using a look-up table based on the pixel intensities in the greyscale image. The look-up table is shown next to the images and utilizes colours ranging from black to white to represent pixel intensities ranging from 0 (black) to 256 (white). Histogram plots were generated using Igor Pro software.
Micro-irradiation
U2OS grown on 25 mm coverslips were treated with 10 μM BrdU for 24 h. PARP inhibitors at a final concentration of 10 μM were added for 1–2 h before micro-irradiation was performed. For micro-irradiation, the cells were placed in a Chamlide TC-A live-cell imaging chamber that was mounted on the stage of a Leica DMI 6000B microscope stand (Leica, Wetzlar, Germany) integrated with a pulsed nitrogen laser (Micropoint Ablation Laser System; Photonic Instruments, Inc., Belfast, Ireland). The pulsed nitrogen laser (20 Hz, 364 nm) was directly coupled to the epifluorescence path of the microscope and focused through a Leica × 40 HCX PL APO/1.25–0.75 oil immersion objective. The laser output was set to 16% to generate strictly localized sub-nuclear DNA damage. Typically, an average of 50 cells was micro-irradiated (150 × 1 pixel) within 10–15 min. Each experiment was performed at least two times. The growth medium was replaced by CO2-independent microscopy medium (137 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 0.8 mM MgSO4, 20 mM D-glucose and 20 mM HEPES, pH 7) and cells were kept at 37°C.
Generation of DSBs
DSBs were induced by IR, which was delivered by an x-ray generator (200 kV; 4 mA; 1.1 Gy/min dose rate; YXLON International).
Statistical analyses
Statistical significance was calculated by a two-sample equal variance t-test assuming a two-tailed distribution.
Supplementary Material
Acknowledgments
We acknowledge Drs R Tsien, T Misteli, PJ Verschure, A Belmont, J Lukas, W Vermeulen, E Seto, L Mullenders, P Varga-Weisz, S Wilson, Y Zhang, W Wang, MJ Ausserlechner, Lars-Gunnar Larsson and S Janicki for sharing reagents; Dr Titia Sixma for helpful comments and Dr FA Salomons for technical assistance. This work was supported by the Swedish Cancer Society (NPD), the European Community Network of Excellence RUBICON (Project no LSHC-CT-2005-018683) (NPD), the Swedish Research Council (NPD), the Netherlands Organization for Scientific Research (NWO Rubicon grant; MSL, NWO-VIDI grant; HvA), the European Molecular Biology Organization (EMBO long-term fellowship; MSL), the Federation of European Biochemical Societies (FEBS long-term fellowship; MSL) and the Human Frontier Science Program (HFSP-CDA; HvA).
Author contributions: MSL, KA and LA generated constructs and performed the LacR targeting experiments; MSL and KA performed the micro-irradiation experiments; SBJ and NM performed the RNF8–CHD4 interaction experiments; DHL generated the siRNA-resistant CHD4 constructs; MSL and WWW performed the ionizing-radiation-induced foci experiments; KKK contributed the GST–CHD4 fusion proteins; MSL, HvA, NM and NPD designed and analysed the experiments; MSL and NPD wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol 18: 1345–1350 [DOI] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ (2009) Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86sup pp 81-12 [DOI] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP (2006) DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev 20: 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (2001) Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol 8: 833–837 [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Dinant C, Houtsmuller AB, Vermeulen W (2008) Chromatin structure and DNA damage repair. Epigenetics Chromatin 1: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C, Luijsterburg MS, Hofer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, van Driel R (2009) Assembly of multiprotein complexes that control genome function. J Cell Biol 185: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA (2010) Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL (2004) Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839 [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769 [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC (2009) Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA 106: 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Georgel PT (2007) CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85: 463–476 [DOI] [PubMed] [Google Scholar]
- Huang A, Hibbert RG, de Jong RN, Das D, Sixma TK, Boelens R (2011) Symmetry and asymmetry of the RING-RING Dimer of Rad18. J Mol Biol 410: 424–435 [DOI] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Chen J (2010) Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 35: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL (2004) From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, Yasui A (2010) The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40: 976–987 [DOI] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, Lukas C, Bartek J, Andersen JS, Lukas J (2010) The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J (2010) A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 29: 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, van Attikum H (2011) Chromatin and the DNA damage response: the cancer connection. Mol Oncol 5: 349–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Hofer T (2010) Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol 189: 445–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W (2009) Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol 186: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 13: 1376–1382 [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J (2006) Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J 25: 3986–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol 11: 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L (2011) UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol 31: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP (2010) Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29: 3130–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA (2009) The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci USA 106: 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T (2007) Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T (2008) Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320: 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145: 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kaneda Y, Hanaoka F (2001) ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AJ, Gatei M, Richard DJ, Khanna KK (2011) ATM mediated phosphorylation of CHD4 contributes to genome maintenance. Genome Integr 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR (2010) Modifying chromatin architecture during the response to DNA breakage. Crit Rev Biochem Mol Biol 45: 2–13 [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R (2005) In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol 25: 4552–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS (1997) Characterization of the CHD family of proteins. Proc Natl Acad Sci USA 94: 11472–11477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861 [DOI] [PubMed] [Google Scholar]
- Ye Q, Hu YF, Zhong H, Nye AC, Belmont AS, Li R (2001) BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol 155: 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.