Abstract
Eukaryotic cells have quality control systems that eliminate nonfunctional rRNAs with deleterious mutations (nonfunctional rRNA decay, NRD). We have previously reported that 25S NRD requires an E3 ubiquitin ligase complex, which is involved in ribosomal ubiquitination. However, the degradation process of nonfunctional ribosomes has remained unknown. Here, using genetic screening, we identified two ubiquitin-binding complexes, the Cdc48–Npl4–Ufd1 complex (Cdc48 complex) and the proteasome, as the factors involved in 25S NRD. We show that the nonfunctional 60S subunit is dissociated from the 40S subunit in a Cdc48 complex-dependent manner, before it is attacked by the proteasome. When we examined the nonfunctional 60S subunits that accumulated under proteasome-depleted conditions, the majority of mutant 25S rRNAs retained their full length at a single-nucleotide resolution. This indicates that the proteasome is an essential factor triggering rRNA degradation. We further showed that ribosomal ubiquitination can be stimulated solely by the suppression of the proteasome, suggesting that ubiquitin–proteasome-dependent RNA degradation occurs in broader situations, including in general rRNA turnover.
Keywords: quality control, ribosome, RNA
Introduction
Eukaryotic ribosomes are highly stable ribonucleoproteins (RNPs) composed of four rRNAs and roughly 80 ribosomal proteins. Ribosomes involve highly stable interactions between rRNAs and basic proteins (Ban et al, 2000). These static interactions contribute to the physical robustness of the ribosomes, because the stably bound proteins would hinder the access of RNases to the rRNAs (Williamson et al, 1969). As a result, the half-life of ribosomes in the mammalian liver is reported to be as long as 100 h (4 days) (Tsurugi et al, 1974). Ribosomal turnover is also only marginally observed in the growing cells of Saccharomyces cerevisiae. Nevertheless, some cases have been reported in which these highly stable ribosomes are rapidly degraded (Lafontaine, 2010).
One of these cases is ribophagy, a substrate-specific autophagy described in S. cerevisiae (Kraft et al, 2008). Both ribosomal subunits are preferentially degraded in the ribophagy pathway when cells are starved of nitrogen (Kraft et al, 2008). Ribophagy has been shown to be essential for cell survival during nutrition shortage, suggesting that the degradation of excess ribosomes may provide new building blocks to maintain cellular homeostasis. It has been reported that a deubiquitinase complex, consisting of four factors, Ubp3, Bre5, Ufd3, and Cdc48, is required for 60S ribophagy (Kraft et al, 2008; Ossareh-Nazari et al, 2010). This complex is involved in the removal of ubiquitin molecules from ribosomes, suggesting that the ubiquitination status of ribosomes is important for their degradation.
Another reported mechanism of ribosomal degradation is a quality control mechanism, nonfunctional rRNA decay (NRD), which eliminates nonfunctional 18S and 25S rRNAs (LaRiviere et al, 2006). When mutant 18S or 25S rRNAs containing a deleterious point mutation in their decoding centre or peptidyltransferase centre (PTC), respectively, were expressed in S. cerevisiae, the nonfunctional rRNAs are selectively degraded after being incorporated into the 40S and 60S particles, respectively (LaRiviere et al, 2006). Interestingly, it has been shown that 18S NRD requires the same proteins as in no-go mRNA decay (NGD) (Cole et al, 2009), which selectively eliminates aberrant mRNAs containing regions that prevent ribosomal passage (Doma and Parker, 2006), including regions with strong secondary structures, rare codons, depurination sites, etc. (Chen et al, 2010). This supports the idea that 18S NRD and NGD are different sides of the same phenomenon, both initiated by a stalled ribosome on a sense codon. It has also been shown that 25S NRD is a distinct process and requires a distinct set of factors, which are not involved in 18S NRD (Cole et al, 2009; Fujii et al, 2009).
By genetically screening a yeast knock-out (YKO) collection, we have previously found that an E3 ubiquitin ligase complex containing Mms1 and Rtt101 is required for 25S NRD (Fujii et al, 2009). We have shown that ribosomal ubiquitination is induced in an Mms1–Rtt101-dependent manner when nonfunctional 25S rRNA is expressed, indicating a role for ubiquitin in this pathway (Fujii et al, 2009). However, the principle underlying the disassembly of the ribosome, a highly stable RNP, has been unclear, including whether or not the ubiquitin molecules are conjugated selectively to the nonfunctional ribosomes as a degradation tag.
In this study, we identified two ubiquitin-binding complexes, the Cdc48–Ufd1–Npl4 complex and the proteasome, as novel factors required for 25S NRD, by genetically screening the essential genes of S. cerevisiae. We showed that the selectively ubiquitinated nonfunctional 60S subunit dissociates from the intact 40S subunit in a Cdc48 complex-dependent manner, before it is attacked by the proteasome. We also showed that proteasome activity is essential for the initiation of 25S rRNA degradation, suggesting a role for the proteasome in removing a key factor(s) that prevents the access of RNase(s) to the ribosomes. Our results identify a previously unappreciated role of the ubiquitin–proteasome system in the degradation of stable RNAs.
Results
A novel ribosomal purification method was developed to study nonfunctional ribosomes containing a mutant 25S rRNA
We have previously reported that ubiquitin signals are enhanced in the ribosomal fractions of S. cerevisiae cells expressing nonfunctional mutant 25S rRNAs. This ubiquitination is dependent on an E3 ubiquitin ligase complex containing Mms1 and Rtt101, both of which are essential for 25S NRD. Although these results revealed an important role for the ubiquitin ligase in this pathway, the role of ubiquitin in the degradation process remained unclear. In this study, we investigated the principle underlying the degradation of a stable RNP, the ribosome, by identifying the direct role of ubiquitin in 25S NRD. The first question discussed in this paper is whether or not the ubiquitin molecules are conjugated specifically to the nonfunctional ribosomes.
Two other observations regarding ribosomal ubiquitination have been reported. First, the ubiquitination of Rpl28p, which constitutes the largest proportion of total ubiquitinated proteins in S. cerevisiae, is regulated by the cell cycle and is predominantly observed in G1 phase (Spence et al, 2000). Second, Kraft et al (2008) reported that nitrogen starvation stimulates ribosome-specific autophagy (ribophagy), in which the deubiquitination of the ribosomes is involved. The observed ribosomal ubiquitination in 25S NRD (Fujii et al, 2009) might be a consequence of G1 arrest or the modulation of the ribophagy pathway potentially induced by the expression of nonfunctional 25S rRNA. If either of these possibilities is true, then it would predict the general enhancement of ubiquitination in the total ribosomal pool. Another possibility is that ubiquitin molecules are selectively conjugated to nonfunctional ribosomes as tags for their degradation, as occurs in the quality control of damaged proteins. To clarify this issue, we purified nonfunctional and functional ribosomes separately to compare the extent of their ubiquitination.
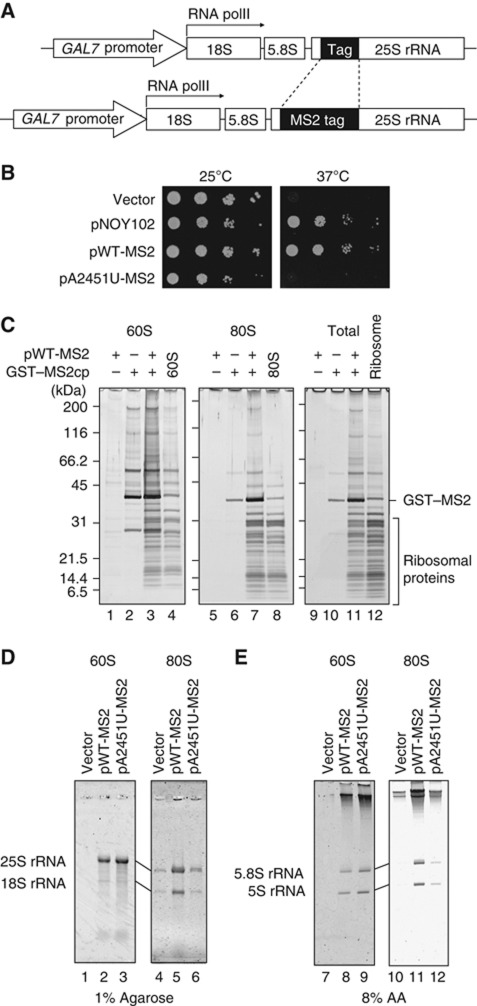
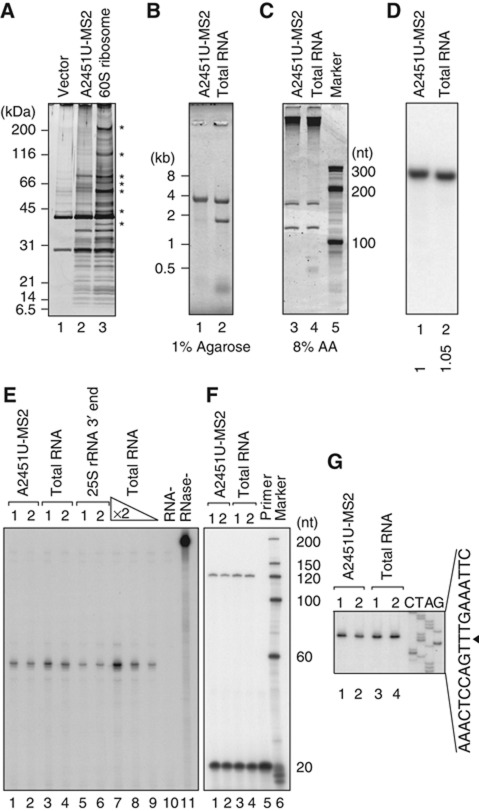
To purify ribosomes containing certain mutant rRNAs, we developed an MS2 tag-based pull-down system. A six-fold (6 × ) repeated MS2 coat protein-binding site was inserted in a non-essential loop of 25S rRNA (Figure 1A). We observed that the expression of the MS2-tagged 25S rRNA from a polymerase II promoter, GAL7 promoter, could rescue the growth of a polymerase I temperature-sensitive (ts) mutant (Nogi et al, 1991) on an SD–galactose plate at the restrictive temperature (Figure 1B), indicating that the 213-nt insertion in this loop did not affect the function of 25S rRNA in vivo. We next examined whether this tag could be used to pull down ribosomes experimentally. A glutathione S-transferase (GST) protein-fused MS2 coat protein (GST–MS2) was co-expressed in a wild-type strain with MS2-tagged 25S rRNA. From this strain, the ribosomal fractions (60S and 80S) were prepared by sucrose density gradient sedimentation and used separately for the affinity purification of the GST fusion protein. As shown in Figure 1C, the ribosomal proteins were efficiently recovered from both the 60S and 80S fractions with this method (compare lanes 3 and 4, 7 and 8, 11 and 12). Lanes 4, 8, and 12 show the ribosomal proteins isolated from the indicated fractions by immunoprecipitation using Rpl28–Flag (Fujii et al, 2009). The recovery of the ribosomal proteins was dependent on the insertion of the MS2 tag into the 25S rRNA, and on the co-expression of the GST–MS2 protein. We confirmed that all the expected rRNAs were observed in the isolated particles (Figure 1D and E). The introduction of a deleterious mutation in PTC, A2451U, into MS2-tagged 25S rRNA reduced the yield of ribosomes pulled down (Figure 1D and E), which is consistent with a reduction in the nonfunctional 25S rRNA by NRD in the wild-type strain. Taking these data together, we concluded that ribosomes with a certain mutation can be purified with the pull-down system developed here.
Figure 1.
Ribosomes containing MS2-tagged rRNAs can be affinity purified with GST–MS2. (A) Schematic representation of the plasmids used for rRNA expression. An 18-nt tag or MS2 tag was inserted into the 5′ region of the 25S rRNA. Both rRNAs were transcribed from the GAL7 promoter by RNA polymerase II. (B) Complementation test of an RNA polymerase I temperature-sensitive (ts) strain. The NOY401 strain containing various plasmids were grown and spotted onto SD–galactose plates after a series of 10-fold dilutions. (C–E) Protein and RNA compositions of affinity-purified ribosomes. (C) Each ribosomal fraction was isolated by sucrose density gradient sedimentation from the strain expressing the indicated plasmids and subjected to GST pull-down. The proteins were visualized by silver staining. Lanes 4, 8, and 12 show the intact ribosomal particles immunopurified with Rpl28–Flag from the fractions. GST–MS2cp: GST–MS2 coat protein. (D, E) RNAs were separated on the indicated gels and visualized with SYBR Gold staining. Figure source data can be found with the Supplementary data.
Nonfunctional ribosomes are selectively ubiquitinated
In addition to the MS2-tagged 25S rRNA described above, we also used another version of 25S rRNA that had an 18-nt insertion in the same position (Figure 1A), to express two different rRNAs in a single cell. These tagged 25S rRNAs, with or without the A2451U mutation, were co-expressed in SD–galactose medium. At mid-log phase, the medium of each culture was replaced with SD–glucose medium to shut off rRNA expression from the GAL7 promoter. The cells were harvested at various time points and the stability of the tagged 25S rRNAs was monitored by northern blotting.
As shown in Figure 2A, we observed that nonfunctional 25S rRNAs were degraded in the wild-type strain with similar kinetics, regardless of the inserted tag sequence (‘A2451U’, upper panel). In contrast, both tagged 25S rRNAs were quite stable when they did not carry the A2451U mutation (‘WT1’, upper panel). Even when they carry the A2451U mutation, both tagged 25S rRNAs were stable in the 25S NRD-defective strain (‘mms1Δ’, lower panel). These results indicate that the inserted tag sequences neither interfered with normal 25S NRD nor induced other types of degradation. Therefore, we next analysed the level of ubiquitination in ribosomes containing the MS2 tag, using GST pull-down eluates from these strains after the co-expression of the GST–MS2 protein (Figure 2B and C).
Figure 2.
Nonfunctional ribosomes are selectively ubiquitinated and degraded in an Mms1-dependent manner. (A) Northern blotting of 18-nt- or MS2-tagged 25S rRNAs. The total RNAs were isolated from cells carrying the indicated plasmids after transcriptional shut-off. These RNAs were cleaved with RNase H (Supplementary Figure S7A). Northern hybridization was performed using a probe that detected both tagged RNAs. (B, C) Immunoblotting of ribosomes purified from cells expressing the indicated tagged rRNAs and Myc–ubiquitin. (B) Ribosomes were Rpl28–Flag immunopurified. In lane 6, an empty vector was used instead of pMyc–Ubi. In lane 7, a wild-type strain with untagged Rpl28 was used. (C) GST–MS2 affinity purification of the ribosomal fraction sedimented by sucrose density gradient centrifugation. In lane 6, an 18-nt-tagged wild-type rRNA was expressed as the negative control. Figure source data can be found with the Supplementary data.
The ubiquitination was examined with Myc-tagged ubiquitin (Ellison and Hochstrasser, 1991). First, we purified the ribosomes from the strains with an Rpl28–Flag immunoprecipitation system to confirm whether ribosomal ubiquitination was also induced by the MS2-tagged mutant 25S rRNA. We observed that all the wild-type strains expressing nonfunctional 25S rRNA showed similar levels of ribosomal ubiquitination (Figure 2B, lanes 2–4). However, when only wild-type ribosomes were pulled down using the MS2 system, these signals were greatly reduced and only very faint signals were observed (Figure 2C, lane 4; we will discuss these signals later). In sharp contrast, the nonfunctional ribosomes with the A2451U mutation were highly ubiquitinated (Figure 2C, lanes 2 and 3). These observations led us to conclude that the ubiquitination observed in 25S NRD is not attributable to the universal enhancement of ubiquitination in the whole population of ribosomes. Instead, ubiquitination is highly specific to nonfunctional ribosomes, suggesting that the conjugated ubiquitin might be used as a degradation signal for nonfunctional ribosomes.
We next examined the actual role of this selective ubiquitination. To address this question, we used genetic screening to identify new factors involved in 25S NRD.
Screening for essential genes revealed the involvement of the Cdc48 complex in 25S NRD
We previously identified Mms1 and Rtt101 as factors involved in 25S NRD by screening a YKO collection with a colony northern technique (Fujii et al, 2009). In the present study, to identify more factors, we screened another set of yeast mutants, the yeast Tet-off Hughs collection (yTHC) (Hughes et al, 2000). This collection consists of ∼800 distinct mutant strains. In each strain, the promoter of a certain essential gene is replaced by the Tet-off promoter, making the strain a conditional lethal. When doxycycline (Dox) is added to the medium, it represses the expression of the essential gene, leading to the cessation of growth after several generations. We looked for a mutant strain in which the A2451U mutant rRNA is stabilized after growth is reduced by Dox treatment.
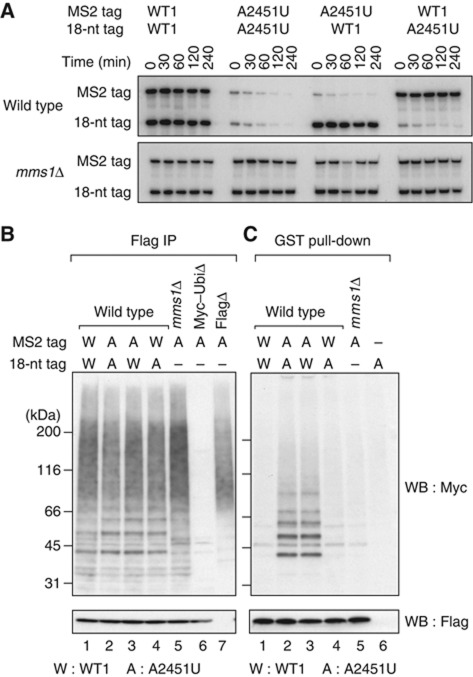
After screening several hundred strains, we noted that Cdc48 and a related factor, Ufd1, are involved in 25S NRD. The growth rates of the strains carrying these mutations decreased at 24 or 12 h after Dox was added to the medium, respectively (Figure 3A). It was indicated that the non-functional 25S rRNAs were stabilized in these strains (Figure 3B and C; Supplementary Figure S1A), whereas the levels of pre-rRNAs containing the PTC mutation A2451U, as well as C2452G and U2585A, persisted unchanged (Supplementary Figure S1B). The A2451U mutant 25S rRNA was degraded with normal kinetics when no Dox was added (Figure 3B and C, Dox– panel) or each repressed gene product was expressed from a plasmid (Figure 3B and C, pCDC48 or pUFD1 panel, respectively; Supplementary Figure S1C and D). These results indicate that Cdc48 and Ufd1 are newly identified factors required for 25S NRD. We also confirmed that Npl4, a binding partner of Ufd1 and Cdc48 (Meyer et al, 2000), is involved in 25S NRD, by showing that nonfunctional mutant 25S rRNA is stabilized in an npl4-1 ts strain (DeHoratius and Silver, 1996) at the semipermissive temperature (Figure 3D; Supplementary Figure S1E).
Figure 3.
Nonfunctional 25S rRNAs were stabilized in Cdc48 complex-deficient strains. (A) Growth curves for CDC48 and UFD1 tet-off strains in the presence (Dox+) and absence (Dox–) of Dox. The cells were grown in SD–galactose medium and A 600 was monitored at the indicated times. The arrows indicate the time points at which the cells were harvested for the following analyses. (B, C) Time course experiments to test the stability of 25S rRNA in CDC48 tet-off (B) and UFD1 tet-off (C) strains. After Dox treatment, the stability of the 18-nt-tagged 25S rRNA was monitored by northern blotting. The pCDC48 plasmid or pUFD1 plasmid was co-introduced into the tet-off strains, as indicated. (D) Stability of nonfunctional 25S rRNA in an npl4-1 mutant strain. The npl4-1 ts strain was grown at the indicated temperature. The pNPL4 plasmid was co-introduced into the strain as indicated. Figure source data can be found with the Supplementary data.
Recently, it has been shown that another Cdc48-containing complex, consisting of Cdc48, Ufd3, Ubp3, and Bre5, is necessary for ribophagy (Ossareh-Nazari et al, 2010). This result prompted us to examine the possibility that the same complex is also involved in 25S NRD. As shown in Supplementary Figure S2A, we initially observed that 25S NRD was inefficient to some extent in the ufd3Δ strain. However, in contrast to ribophagy (Ossareh-Nazari et al, 2010), the overexpression of ubiquitin rescued 25S NRD in this mutant strain (Supplementary Figure S2B). These results suggest that the observed inefficiency of 25S NRD was attributable to the effects of ubiquitin starvation, providing further proof of the requirement for ubiquitination in 25S NRD. Therefore, Ufd3 was excluded from the factors directly involved in 25S NRD. We also observed 25S NRD with normal kinetics in the atg7Δ strain and the atg8Δ strain (Supplementary Figure S2C), in which the autophagy pathway is totally absent (Tsukada and Ohsumi, 1993). Taking these data together, we concluded that 25S NRD and ribophagy are distinct processes catalysed by distinct ubiquitin-binding complexes, which share Cdc48 as a common factor. We also confirmed that no other known binding partners of Cdc48 are required for 25S NRD (Supplementary Figure S2D).
It is well documented that the Cdc48–Ufd1–Npl4 complex is involved in the proteolysis of ubiquitinated proteins by the proteasome in divergent pathways (Ye, 2006). The proteasome might be responsible for the degradation of nonfunctional 25S rRNAs. Therefore, we next examined this possibility.
Proteasomal activity is required for the degradation of the 25S NRD substrate
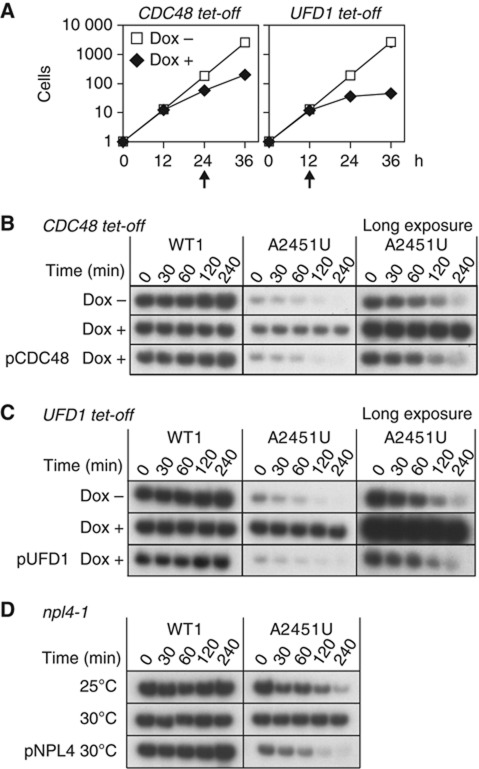
To examine the involvement of the proteasome in 25S NRD, we measured the stability of nonfunctional 25S rRNA after the cellular proteasomal activity was compromised with three different approaches (Figure 4A–C). First, we used a cim3-1 mutant strain (Ghislain et al, 1993), which has a point mutation in Rpt6, a component of the 19S regulatory subunit of proteasome (Glickman et al, 1998). As shown in Figure 4A, the nonfunctional A2451U mutant rRNA was degraded only slowly in this mutant strain, even at the permissive temperature (upper panel). Similarly, 25S NRD was inhibited when another component of the 19S subunit, Rpt2 (Glickman et al, 1998), was depleted with the Tet-off system (Figure 4B; Supplementary Figures S2B and S3A). These instances of inhibition were rescued by a plasmid encoding the corresponding wild-type protein, Cim3/Rpt6 or Rpt2, respectively (Figure 4A and B; Supplementary Figure 3B and C). These results clearly indicate that the 19S proteasomal subunit is essential for 25S NRD.
Figure 4.
Proteasome activity is required for 25S NRD. (A–D) Stability of nonfunctional rRNA in a proteasome-inhibited cell. (A–C) The 18-nt-tagged 25S rRNA was detected by northern blotting. (A) The cim3-1 ts strain was grown at the permissive temperature. (B) Proteasomes were depleted in the RPT2 tet-off strain with Dox treatment for 12 h. (C, D) MG132 was administered at the indicated concentrations to the erg6Δ strain containing the combination of plasmids indicated, to overexpress ubiquitin. (D) The other 16-nt-tagged 18S rRNAs, with or without A1492C mutation, were detected by northern blotting. (E, F) Subcellular localization of 18-nt-tagged 25S rRNAs in various mutant strains. After transcriptional shut-off, the cells were harvested at the indicated time points. (E) The tagged 25S rRNAs were visualized with an in-situ hybridization technique. Scale bar, 4 μm. (F) Cleared lysates were resolved on a 10–40% sucrose gradient and the amounts of tagged rRNA were visualized by northern blotting. All dots of every lane come from the same membrane. Figure source data can be found with the Supplementary data.
Rpt2 and Rpt6 are two of the six AAA-ATPases in the base subcomplex of the 19S subunit (Lander et al, 2012), which are responsible for unwinding ubiquitinated proteins before their degradation by the 20S subunit (Benaroudj et al, 2001; Finley, 2009). To clarify whether the unwinding activity of the 19S subunit or the proteolysis by the 20S subunit is required for 25S NRD, we inhibited the proteasomal activity by treatment with MG132 using the MG132-pearmeable erg6Δ strain. MG132 is a potent inhibitor of the chymotrypsin-like activity of the 20S subunit (Lee and Goldberg, 1996). Figure 4C and Supplementary Figure S3D show that the inhibition of the 20S proteasome by MG132 also interfered with the degradation of nonfunctional 25S rRNAs. The overexpression of ubiquitin did not rescue 25S NRD in this case, indicating that the inhibitory effect of MG132 on 25S NRD is not attributable to the ubiquitin starvation caused indirectly by the drug treatment (Figure 4C: Supplementary Figure S3D). Therefore, we concluded that proteolysis by the 20S proteasome is directly involved in 25S NRD. In contrast to 25S NRD, 18S NRD was not affected by the MG132 treatment (Figure 4D).
Recently, it was proposed that ubiquitin and proteasomes are also involved in ribosome biogenesis in the nucleus (Stavreva et al, 2006). To confirm that the proteasome-dependent process demonstrated here is executed in the cytoplasm, we visualized the nonfunctional 25S rRNA in several mutant strains with an in-situ hybridization technique using a Cy3-labelled oligonucleotide probe (Figure 4E). Four hours after transcriptional shut down, no signals for the nonfunctional A2451U rRNA were detected in the wild-type strain. In contrast, in the mms1Δ, UFD1 tet-off, and RPT2 tet-off strains, the remaining signals were distributed uniformly in the cytoplasm (Figure 4E; Supplementary Figure S3E). These observations indicate that the Cdc48 complex and the proteasome are involved in a cytoplasmic process of 25S NRD.
Considering the involvement of the Cdc48 complex and the proteasome in 25S NRD, we presumed that the observed ubiquitin molecule conjugated to the nonfunctional ribosome was a K48-linked polyubiquitin chain, a common signal for proteasomal degradation. To confirm this, we expressed a nonfunctional mutant 25S rRNA together with Myc–ubiquitin containing either the K48R or K63R mutation and analysed the ribosomal fractions by immunoblotting with anti-Myc antibody. The ladder-like ubiquitin signals were observed when the K63R mutant was used (Supplementary Figure S3F). In contrast, all of these signals were clearly absent when the expressed Myc–ubiquitin contained the K48R mutation, which prevents the formation of the K48-linked polyubiquitin chain. These results indicate that the ubiquitin conjugated to the nonfunctional ribosomes was the K48-linked polyubiquitin chain and supporting the conclusion that it is used as the degradation tag for the proteasome.
Ribosomal remodelling is observed in 25S NRD
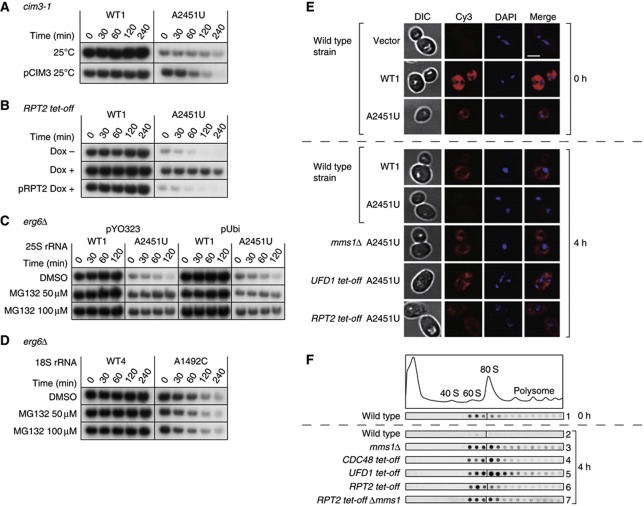
To gain further insight into the details of the 25S NRD pathway, we next analysed the size of the particles containing 25S NRD substrates in each mutant, using a sucrose density sedimentation assay. As shown in Figure 4F, Cdc48 depletion caused nonfunctional 25S rRNA to accumulate mainly in the 80S fraction, with less signal observed in the 60S fraction (panel 4). A similar pattern of accumulation was observed when Ufd1p was depleted (panel 5). In sharp contrast, when proteasomes were compromised by Rpt2 depletion, the 25S NRD substrate mainly accumulated in the 60S fraction (panel 6). Although faint signals were also detected in the 80S fraction, it is clear that the 60S/80S signal ratio in this strain was the inverse of that in strains depleted of the Cdc48 complex (Supplementary Figure S4A). The difference in the nonfunctional 25S rRNA distribution in each mutant is not attributable to the general change in the 60S/80S ratio in the cellular ribosomes, because these mutations essentially do not affect that ratio (Supplementary Figure S4B). Instead, the different pattern of nonfunctional 25S rRNAs in Figure 4F must represent the specific effect of each mutant strain on the nonfunctional 25S rRNAs.
The observed 60S signals in the Rpt2-depleted strain do not simply represent newly synthesized unused 60S particles. Instead, these 60S signals should correspond to the degradation intermediates, which have undergone a size reduction from 80S particles. We inferred this for two reasons. First, at this time point, only signals stabilized by gene depletion or disruption were detected (Figure 4F, compare panels 3–7 with panel 2), indicating that these signals represent degradation intermediates that accumulated immediately before the steps carried out by those gene products. Second, Rpt2 depletion did not prevent nonfunctional 60S particles from forming 80S particles. When the RPT2 tet-off mms1Δ double mutant was used, the nonfunctional 25S rRNA signals were also observed in the 80S fraction. Essentially, the same pattern was produced in the mms1Δ single-mutant strain (Figure 4F panel 7, compared with panel 3), suggesting that there is no significant delay in the formation of the 80S particle under Rpt2-depleted conditions. Therefore, the reduced signal in the 80S fraction and its enrichment in the 60S fraction of the Rpt2-depleted strain is attributed to the size transition from 80S to 60S of the particles containing nonfunctional 25S rRNA. This 80S to 60S size reduction is dependent on the Cdc48 complex (Figure 4F, panels 4 and 5), which functions in various pathways upstream from the proteasome (Ye, 2006).
Do the observed signals around the 60S fractions represent 60S ribosomal subunits lacking 40S subunits? Or are they incomplete 80S complexes that lack a number of factors from both subunits? To clarify this point, we next purified and analysed the 60S particles that accumulated in the RPT2 tet-off strain, using the MS2 system. As shown in Figure 5A, the accumulated 60S particles clearly retained most of the ribosomal proteins normally found in 60S subunits, when visualized by silver staining after sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). We also confirmed the presence of Rpl3, 5, and 24 and the absence of Rps4 in the particles with immunoblotting (Supplementary Figure S5A). Furthermore, when the RNAs were isolated and analysed, all the expected RNAs (25S, Figures 5B; 5.8S and 5S, Figure 5C) were detected with no apparent size reduction. Conversely, 18S rRNA was completely missing in this intermediate (Figure 5B). From these results, we conclude that nonfunctional 80S particles undergo remodelling from 80S to 60S particles with the Cdc48-dependent dissociation of the 40S subunit, before the process in which proteasomal degradation is involved.
Figure 5.
Nonfunctional 60S particles that accumulated under proteasome-depleted conditions contained most ribosomal proteins and complete sets of all three rRNAs. (A–G) The protein and RNA compositions of nonfunctional ribosomes affinity purified with GST–MS2 from the RPT2 tet-off strain 4 h after transcriptional shut-off. GST pull-down was performed using ∼60S fractions. (A) The eluates were separated and silver stained. In lane 3, WT1–MS2-containing 60S ribosomal particles were purified from the wild-type strain (*non-ribosomal proteins). (B, C) Purified RNAs were separated and stained with SYBR Gold. In lanes 2 and 4, total RNA was purified from the wild-type strain. (D) Northern hybridization was performed using a proof which was designed to bind the 3′-most region of 25S rRNA (position +3251). The number below each lane shows the relative signal intensity. (E) An RNase protection assay was used to examine the 3′ ends of the 25S rRNAs. In lanes 5 and 6, the 3′ region of the 25S rRNA was transcribed in vitro and used. For lanes 7–9, total RNAs diluted two-fold were used. For lane 10, no target RNA was added to the reaction. Lane 11 is the no-RNase control. (F, G) Primer extension showed the 5′ ends of the 25S rRNAs. The primer used was designed to bind the region into which the tags were inserted (Supplementary Figure S7D). (G) The corresponding sequence for the sense strand of the 25S rDNA is shown. The 5′ end of the mature 25S rRNA is indicated by an arrowhead. Figure source data can be found with the Supplementary data.
Proteasomal degradation of key protein(s) is required for the initiation of nonfunctional 25S rRNA degradation
As shown in Figure 5B, almost full-length 25S rRNA was detected in the nonfunctional 60S subunits that accumulated in the Rpt2-depleted strain. A straightforward interpretation is that the protein degradation and RNA degradation are ordered processes; the elimination of certain protein(s) by the proteasome is required for the initiation of RNA degradation by removing key proteins that prevent the access of RNase(s) to the ribosomes. Another interpretation is that the protein degradation facilitates the processivity of unidentified exonuclease(s) by continuously removing proteins that hinder the path of the enzyme(s). If the former is true, then we should see 25S rRNA with complete ends preserved in the nonfunctional 60S particles in the Rpt2-depleted strain. Conversely, if the latter is true, then degradation intermediates that lack some nucleotides or fragments at their ends should be observed. To determine which is the case, we next analysed the accumulated nonfunctional 25S rRNAs in the Rpt2-depleted strain in detail.
When the 3′ ends of the accumulated nonfunctional 25S rRNAs were examined by northern hybridization (Figure 5D) and an RNase protection assay (Figure 5E; Supplementary Figures S5B and S7C), we found that intact 3′ ends were precisely preserved in the accumulated nonfunctional 25S rRNAs. Similarly, we confirmed by primer extension that the 5′ ends were also intact (Figure 5F and G; Supplementary Figure S7D). Based on these results, we concluded that the proteasome is required for the initiation of 25S rRNA degradation in NRD, but not for the processivity of the RNase(s).
Ribosomes are continuously degraded by the ubiquitin–proteasome system
Is the ubiquitin–proteasome-assisted degradation of ribosomal particles described in this paper limited to 25S NRD? Or is it observed in other, more general cases of ribosomal degradation? To address this question, we next evaluated whether ribosomal ubiquitination is enhanced when ribosomal degradation is artificially induced by drug treatments.
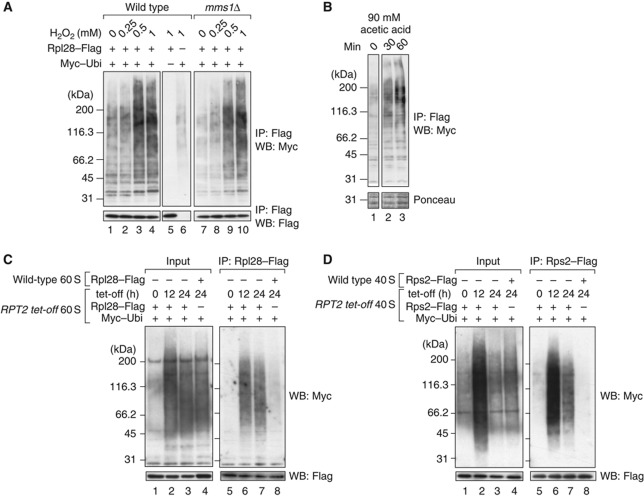
It has been reported that the extensive decay of rRNAs can be artificially induced by the treatment of yeast cells with several compounds, including H2O2, acetic acid, and methylmethanesulphonate (Supplementary Figure S6A; Mroczek and Kufel, 2008; Thompson et al, 2008). We added these drugs to the cultures to induce rRNA decay. During the lag period before rRNA degradation, we immunopurified the ribosomes using Rpl28–Flag and analysed the ubiquitination status of those ribosomes using an Myc–ubiquitin system (Figure 6A and B; Supplementary Figure S6B). In all the cases examined, ribosomal ubiquitination was enhanced by the drug treatment in a dose-dependent manner. The distribution of the ubiquitination signals shifted to higher molecular weights. The total intensity of the signals also increased. These results clearly show that ribosomal ubiquitination occurs when ribosomal degradation is triggered by chemical stressors. The ubiquitination described here was also observed in the mms1Δ strain (Figure 6A, lanes 7–10), suggesting that multiple ubiquitin ligases are involved in the ubiquitination of the damaged ribosomes.
Figure 6.
Ribosomes are continuously ubiquitinated and degraded by proteasomes. (A, B) Ubiquitination signals induced by stress. (A) Wild-type and mms1Δ cells were grown in SD–glucose medium and treated for 2 h with 0, 0.25, 0.5, and 1 mM H2O2. In lane 5, an empty vector was used instead of pMyc–Ubi. In lane 6, another empty vector was used instead of pRpl28–Flag. (B) Wild-type cells were treated with acetic acid at a final concentration of 90 mM. Ribosomes were isolated at the indicated time points. (C) Ubiquitinated 60S ribosomal particles accumulated under Rpt2p-depleted conditions. The RPT2 tet-off strain was treated for the indicated times with 10 μg/ml Dox. The 60S ribosomal fractions were collected by 10–40% sucrose gradient sedimentation containing 40 mM EDTA. The 60S particles from the RPT2 tet-off strain were immunoprecipitated (lanes 5–7). Before the pull-down assay, untagged wild-type 60S fractions were mixed with the RPT2-tet-off strain-derived 60S fractions. To evaluate the non-specific binding of ubiquitinated proteins to ribosomes, as a control, 60S particles from the wild-type strain were immunopurified after they were mixed with the 60S fraction from RPT2 tet-off cells (lane 8). The ubiquitinated proteins in the mixture of 60S fractions are shown in lanes 1–4. (D) Ubiquitinated 40S ribosomal particles under Rpt2-depleted conditions. A similar set of experiments to (C) was performed using the 40S fractions and Rps2–Flag. Figure source data can be found with the Supplementary data.
Interestingly, during these experiments, we noticed that ribosomes from untreated cells always displayed some level of ubiquitination signal (Figure 6A, lanes 1 and 7; see also Figure 2B, lane 1). These faint signals were also observed in MS2-purified wild-type ribosomes (Figure 2C, lanes 1, 4, and 5). We hypothesized that ribosomes might be slowly degraded by the ubiquitin–proteasome system, even in normally growing cells. If this is the case, then the ribosomal ubiquitination will be enhanced without drug treatments when proteasomal activity is compromised. To investigate this possibility, we examined whether Rpt2 depletion influenced the ubiquitination status of the ribosomes.
As shown in Figure 6C, Rpt2p depletion markedly elevated the ubiquitination of the 60S ribosomal subunit (compare lane 5 with lanes 6 and 7). In this assay, the Rpl28–Flag-containing complex was immunopurified from the 60S fraction prepared from the RPT2 tet-off strain expressing Rpl28–Flag and Myc–Ubi (Supplementary Figure S6C), and ubiquitination was detected with anti-Myc antibody after SDS–PAGE. When Rpl28–Flag and Myc–Ubi were expressed separately in different strains and the 60S fractions from those strains were mixed and used for immunoprecipitation, almost no ubiquitination signals were recovered (lane 8), showing that the ubiquitinated proteins in lanes 6 and 7 are ribosomal proteins or proteins physiologically associated with the ribosomes. These results suggest that cellular ribosomes are more or less continuously ubiquitinated and degraded, even in normally growing cells. A similar result was obtained when 40S subunits were analysed (Figure 6D, compare lane 5 with lanes 6 and 7). Therefore, we conclude that the degradation of stable RNPs by the ubiquitin–proteasome system is not specific to 25S NRD but occurs more widely, as ribosomal ubiquitination is observed in the degradation of drug-treated ribosomes and the natural turnover of normally growing cells.
Discussion
In this study, we have shown that the Cdc48–Ufd1–Npl4 complex and the proteasome are the essential factors involved in the late steps of 25S NRD. The 40S subunit dissociated from the nonfunctional 60S subunit in a Cdc48 complex-dependent manner. The affinity purification of nonfunctional 60S particles that accumulated in a proteasome-depleted strain indicated that the proteasome is required for the initiation of rRNA degradation in 25S NRD. Moreover, we have provided evidence that the ubiquitin–proteasome system is involved in stress induced ribosomal degradation and ribosomal turnover.
Novel affinity purification system for eukaryotic ribosomes containing a certain mutation
We have described here a newly developed affinity purification method for ribosomes. Using this system, ribosomes with a certain mutation in their 25S rRNA were biochemically isolated. To date, there have been a number of reports characterizing eukaryotic ribosomes containing mutant 25S rRNAs (Macbeth and Wool, 1999; Panopoulos et al, 2004; Rakauskaite and Dinman, 2008). However, in those studies, entire populations of cellular ribosomes were replaced by the mutant ribosome to be characterized. This limits the variety of mutants that can be characterized, because nonfunctional ribosomes with a critical mutation cannot be prepared with this approach. With the MS2-based purification method described here, any mutant ribosomes containing a deleterious mutation(s) in the 25S rRNA can be isolated, because the growth of the cells is supported by cellular ribosomes in this case. This offers a new avenue for the biochemical characterizations of eukaryotic ribosomes. X-ray crystallography has recently begun to reveal the detailed structures of these ribosomes at atomic resolution (Ben-Shem et al, 2010; Rabl et al, 2011).
Role of the Cdc48 complex in the disassembly of ribosomes
We have shown that the depletion of the Cdc48–Npl4–Ufd1 complex causes nonfunctional 25S rRNAs to accumulate in the 80S particle. When a downstream step was interrupted by proteasomal depletion, those nonfunctional 25S rRNAs were detected in the 60S subunit. These results indicate that the Cdc48 complex is essential for the segregation of the 40S subunit from the nonfunctional 60S subunit. The dissociated 40S subunit will probably be reused for the next round of translation, because the dissociated 40S subunit should be intact, and recycling the 40S particle will reduce cellular energy consumption during the reconstruction of the massive ribosome particle.
An interesting possibility is that the Cdc48 complex directly catalyses this segregation. Cdc48 belongs to the AAA-ATPase family (Ogura and Wilkinson, 2001). It has been reported that the Cdc48–Npl4–Ufd1 complex is involved in the segregation of misfolded proteins from the ER to the cytoplasm during ERAD (Ye et al, 2001). In general, the heterodimer Npl4p–Ufd1p directly binds ubiquitin (Meyer et al, 2002), and the mechanical force for the segregation is provided by a hexamer of Cdc48p (Rape et al, 2001; Ye et al, 2001). Using the ubiquitinated protein on the 60S subunit as a handle, this complex might physically dissociate the 40S subunit from the nonfunctional 80S particle. Another possibility is that the Cdc48 complex indirectly assists the function of other factor(s) involved in subunit dissociation (Kurata et al, 2010; Pisarev et al, 2010; Shoemaker et al, 2010). An in-vitro reconstitution system for Cdc48–Npl4–Ufd1 activity (Shcherbik and Haines, 2007) would effectively address this issue.
Use of a variety of RNP degradation mechanisms
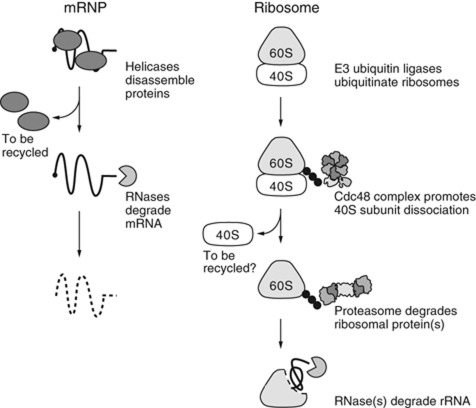
It has recently been shown that the RNA helicase Upf1 is involved in the disassembly of mRNPs in nonsense-mediated mRNA decay (Franks et al, 2010). Using RNA helicase for the disassembly of relatively unstable mRNPs is reasonable, because such disassembly occurs quite often and RNA-binding proteins can be recycled in this situation (Bono and Gehring, 2011). A similar mechanism is also observed in the degradation of the intron lariat complex, where the RNA helicase Prp43 is involved in the disassembly (Arenas and Abelson, 1997; Tsai et al, 2005; Yoshimoto et al, 2009). In contrast, we have shown in this study that the disassembly of a stable RNP, the 60S ribosomal subunit, is processed by a distinct mechanism, a proteasomal degradation dictated by the specific ubiquitination of nonfunctional ribosomes (Figure 7). Why do cells not recycle proteins after the disassembly of stable RNPs?
Figure 7.
A model of RNP degradation. In mRNP degradation, mRNPs are disassembled by RNA helicases (Upf1 in NMD). This RNP disassembly step is essential for mRNA degradation by RNases. During ribosomal degradation, E3 ubiquitin ligase first ubiquitinates the ribosomal proteins. Using these ubiquitins as tags, the Cdc48 complex promotes the dissociation of the 40S subunit. The proteasome degrades key protein(s) from the 60S particles and then triggers the initiation of RNA decay.
There seem to be at least two reasons. First, we infer that it is because recycling ribosomal proteins would be risky for cells. Most of the ribosomal proteins are highly basic and chemically likely to aggregate with cellular RNAs (Jakel et al, 2002). Ribosomal protein(s) should be degraded before RNA degradation to avoid the potential risk of harmful ribosomal proteins dispersed in the cytoplasm. Second, it could be because the ribosomes are composed of a number of components. Most ribosomal proteins and the entire sequences of rRNAs are well conserved, from yeasts to humans (Lecompte et al, 2002), implying that any change in these conserved components might harm the ribosomal function. The identification of the aberrant component in a large nonfunctional ribosome might cost more than the degradation and reconstruction of the total complex, when we consider that cellular stress must randomly induce various ribosomal damages to each component. One such stress is the oxidative stress examined in Figure 6A. In this case, as was shown, various types of ribosomal ubiquitination were induced, suggesting that multiple components were damaged by this treatment.
We have shown in this study that the ubiquitination of ribosomes (both the 60S and 40S subunits) occurs in various situations in which ribosomes are degraded, except in 18S NRD (Figure 4D). How far can we apply this principle of RNP degradation to other stable RNPs? There are a number of stable RNPs in eukaryotic cells. These stable RNPs must also eventually be degraded, for various reasons, including quality control and changes in RNPs’ repertoires during differentiation, apoptosis, etc. It will be very interesting to clarify whether ubiquitin–proteasome-directed RNP disassembly is also involved in the degradation of these stable RNPs.
Materials and methods
Plasmids, yeast strains, and growth conditions
Please see the Supplemental Experimental Procedures. Plasmids and yeast strains are listed in Supplementary Tables S1 and S2, respectively. All oligonucleotides used in this study are listed in Supplementary Table S3.
RNA purification and analysis
To extract the total RNA from yeast cells, we used the MasterPure Yeast RNA Purification Kit (Epicentre Biotechnologies). Sepasol-RNA1 Super (Nacalai Tesque) was used to extract the RNA from cell lysates or purified ribosomes. In a northern analysis of the stability of the tagged rRNAs, 500 ng of total RNA was loaded into the lane at time 0. The amount of RNA loaded into the other lanes was increased according to the growth of the cells, measured as the absorbance at 600 nm (A 600), leaving the signals of the stable RNA species unchanged.
RNase H digestion was performed with 1.5 μg of total RNA and 0.25 μM oligonucleotide DNA (Kota031), according to Uyeno et al (2004). Complete digestion produced an ∼220-nt fragment containing the 5′ end of the 18-nt-tagged 25S rRNAs and an ∼440-nt fragment of the MS2-tagged RNAs.
Ribosome purification and immunoblotting
The purification of total ribosomes using the Flag-tagged ribosomal protein Rpl28 has been described elsewhere (Fujii et al, 2009). To isolate the ribosomes containing plasmid-derived rRNAs, MS2-tagged 25S rRNA and GST–MS2 were co-expressed. Approximately 800 A 600 unit cells were harvested and disrupted in liquid nitrogen in a mortar (Inada et al, 2002). The lysate was dissolved in 1 ml IPP150 (10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40) supplemented with Complete Protease Inhibitor Cocktail (Roche) and 20 mM N-ethylmaleimide. The ribosomes containing MS2-tagged rRNA and GST–MS2 were pulled down with glutathione Sepharose beads from the 60S and 80S fractions of a sucrose density gradient. The ribosomes were recovered after elution with 100 mM reduced glutathione. To immunoblot the ubiquitinated ribosomes, equal amounts of ribosomes in the eluates were subjected to 5–20% gradient SDS–PAGE and transferred onto a nitrocellulose membrane with a semidry blotting apparatus. To analyse the co-sedimented proteins, the proteins were visualized by silver staining after 10–20% gradient SDS–PAGE.
In-situ hybridization
All the cells were grown in SD–raffinose to A 600=0.5. The medium was then replaced with SD–galactose to induce the expression of the tagged rRNAs from the GAL7 promoter. Six hours after induction, the medium was replaced again with SD–glucose to shut-off transcription. The cells were harvested onto a glass slide and analysed 4 h after transcriptional shut-off. The specimens were prepared as described elsewhere (Fujii et al, 2009). Microscopic analyses were performed using Olympus BX61 and UAPON 150XO microscopes (NA=1.45). A series of 15–20 Z-stuck was captured for each picture and processed with AutoQuant deconvolution using MetaMorph. The best-focused picture for each specimen was selected and presented.
RNase protection assay and primer extension
An RNase protection assay was performed using an RNase cocktail (Ambion). The template RNAs (10 ng) and probes (1 × 104 c.p.m.) were hybridized overnight at 45 °C in 1 × hybridization buffer (40 mM PIPES (pH 6.8), 1 mM EDTA (pH 8.0), 0.4 mM NaCl, 80% formamide). The RNase digestion mixture (300 mM NaCl, 10 mM Tris–HCl (pH 7.4), 5 mM EDTA, 0.5 U/μl RNase A, 20 U/μl RNase T1) was added to digest the single-stranded RNAs in the reaction. After the reaction was incubated at 30 °C for 60 min, the RNases were inactivated by the addition of sodium dodecyl sulphate and proteinase K. The RNase-resistant hybrids were then analysed by gel electrophoresis and autoradiography (Sambrook, 2001). The probe used in this study was synthesized in vitro with T7 RNA polymerase (Promega). The PCR product generated with primers Kota379 and Kota383 was used as the template. This probe contains a 150-nt region complementary to the 3′ end of 25S rRNA and overhangs both the 5′ end (30-nt) and 3′ end (8-nt). The control RNA used to indicate the 3′ end of the mature 25S rRNA was also transcribed in vitro with the T7 RNA polymerase system, using the Kota381–Kota382 PCR product as the template. For primer extension, 0.5 μg of the purified RNAs and 0.25 pmol of 32P-labelled primer were mixed and treated with reverse transcriptase (ReverTra Ace, Toyobo), following manufacturer’s instructions. The reaction was then analysed by autoradiography after separation on an 8% acrylamide gel containing 7 M urea.
Analysis of the ubiquitination of 40S and 60S ribosomal particles in the RPT2 tet-off strain
To purify 40S and 60S ribosomal particles, Rps2–Flag and Rpl28–Flag were expressed, respectively. The RPT2 tet-off strain expressing Myc–Ubi was grown in SD–glucose. When the cells reached A 600=0.5, they were diluted in the same medium containing 10 μg/ml Dox, to maintain the log phase for the indicated times. Approximately 500 A 600 unit cells were harvested and disrupted in liquid nitrogen in a mortar (Inada et al, 2002). The lysate was fractionated on a sucrose density gradient containing 40 mM EDTA and the 40S and 60S fractions were collected. The ribosomal particles from the RPT2 tet-off strain were immunoprecipitated with anti-Flag agarose and washed with IPP150 (10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40). To elute the ribosomes, IPP150 was used with 3 × Flag peptides (Sigma) at a concentration of 0.25 mg/ml. Before pull-down, the untagged wild-type 40S and 60S fractions were mixed with the RPT2 tet-off strain-derived 40S and 60S fractions, respectively. To evaluate the non-specific binding of ubiquitinated peptides to ribosomes, as a control, the Flag-tagged ribosomal particles from the wild-type strain were immunopurified after 40S or 60S fractions from RPT2 tet-off cells, without Flag-tag expression, were mixed. The purified ribosomes were separated by 5–20% SDS–PAGE and the ubiquitinated proteins were detected with an anti-Myc polyclonal antibody.
Supplementary Material
Acknowledgments
We thank Dr Pamela A Silver for generously providing the yeast strain, Dr Kaori Shinmyouzu and Dr Akira Nakamura for the mass-spectrometric analysis, and Dr Hideji Yoshida for technical advice and discussions. We also thank Tokie Sakai and Kodai Sano for their technical assistance. This work was supported by PRESTO, JST, in a grant to MK, and a Grant-in-Aid for Scientific Research on Innovative Areas ‘RNA regulation’ (no. 20112006) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to MK and MO. KF and TS are Japan Society for the Promotion of Science Research Fellows.
Author contributions: MK and KF designed the experiments. KF performed most experiments. MK and TS performed some experiments. MK and KF wrote the manuscript, which was subsequently improved by the members. MK obtained a part of the funding. MO organized the research group and obtained the main part of the funding.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arenas JE, Abelson JN (1997) Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M (2010) Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Tarcsa E, Cascio P, Goldberg AL (2001) The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie 83: 311–318 [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring NH (2011) Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol 8: 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H (2010) Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ (2009) A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 34: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoratius C, Silver PA (1996) Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell 7: 1835–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M (1991) Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem 266: 21150–21157 [PubMed] [Google Scholar]
- Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J (2010) Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M (2009) A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev 23: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ Jr., Yates JR 3rd, Schieltz D, Sachs AB (2002) One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8: 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 21: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kurata S, Nielsen KH, Mitchell SF, Lorsch JR, Kaji A, Kaji H (2010) Ribosome recycling step in yeast cytoplasmic protein synthesis is catalyzed by eEF3 and ATP. Proc Natl Acad Sci USA 107: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DL (2010) A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci 35: 267–277 [DOI] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ (2006) A late-acting quality control process for mature eukaryotic rRNAs. Mol Cell 24: 619–626 [DOI] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O (2002) Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res 30: 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Goldberg AL (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem 271: 27280–27284 [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Wool IG (1999) Characterization of in vitro and in vivo mutations in non-conserved nucleotides in the ribosomal RNA recognition domain for the ribotoxins ricin and sarcin and the translation elongation factors. J Mol Biol 285: 567–580 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (2000) A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19: 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S, Kufel J (2008) Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res 36: 2874–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M (1991) Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA 88: 3962–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C (2010) Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11: 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos P, Dresios J, Synetos D (2004) Biochemical evidence of translational infidelity and decreased peptidyltransferase activity by a sarcin/ricin domain mutation of yeast 25S rRNA. Nucleic Acids Res 32: 5398–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV (2010) The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736 [DOI] [PubMed] [Google Scholar]
- Rakauskaite R, Dinman JD (2008) rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance. Nucleic Acids Res 36: 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Sambrook J (2001) Molecular Cloning: A Laboratory ManualJoseph Sambrook, David W. Russell Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, [Google Scholar]
- Shcherbik N, Haines DS (2007) Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol Cell 25: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D (2000) Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102: 67–76 [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Kawasaki M, Dundr M, Koberna K, Muller WG, Tsujimura-Takahashi T, Komatsu W, Hayano T, Isobe T, Raska I, Misteli T, Takahashi N, McNally JG (2006) Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol Cell Biol 26: 5131–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169–174 [DOI] [PubMed] [Google Scholar]
- Tsurugi K, Morita T, Ogata K (1974) Mode of degradation of ribosomes in regenerating rat liver in vivo. Eur J Biochem 45: 119–126 [DOI] [PubMed] [Google Scholar]
- Uyeno Y, Sekiguchi Y, Sunaga A, Yoshida H, Kamagata Y (2004) Sequence-specific cleavage of small-subunit (SSU) rRNA with oligonucleotides and RNase H: a rapid and simple approach to SSU rRNA-based quantitative detection of microorganisms. Appl Environ Microbiol 70: 3650–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R, Lanyon G, Paul J (1969) Preferential degradation of ‘messenger RNA’ in reticulocytes by ribonuclease treatment and sonication of polysomes. Nature 223: 628–630 [DOI] [PubMed] [Google Scholar]
- Ye Y (2006) Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol 156: 29–40 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Kataoka N, Okawa K, Ohno M (2009) Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res 37: 891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.