Abstract
During embryonic development, cells must divide to produce appropriate numbers, but later must exit the cell cycle to allow differentiation. How these processes of proliferation and differentiation are co-ordinated during embryonic development has been poorly understood until recently. However, a number of studies have now given an insight into how the cell cycle machinery, including cyclins, CDKs (cyclin-dependent kinases), CDK inhibitors and other cell cycle regulators directly influence mechanisms that control cell fate and differentiation. Conversely, examples are emerging of transcriptional regulators that are better known for their role in driving the differentiated phenotype, which also play complementary roles in controlling cell cycle progression. The present review will summarise our current understanding of the mechanisms co-ordinating the cell cycle and differentiation in the developing nervous system, where these links have been, perhaps, most extensively studied.
Keywords: cell cycle, cyclin-dependent kinase (CDK), development, differentiation, neurogenesis
Abbreviations: Ascl1, achaete-scute homologue 1; bHLH, basic helix–loop–helix; CDK, cyclin-dependent kinase; CDKi, CDK inhibitor; CNS, central nervous system; DP, dimerization partner; Fox, forkhead box; Hes, hairy and enhancer of split; HLH, helix–loop–helix; Id, inhibitor of DNA binding; NSC, neural stem cell; Ngn2, neurogenin 2; Prox1, Prospero-related homeobox 1; Rb, retinoblastoma susceptibility gene; Sox3, Sry-type high mobility group box; TGF-β, transforming growth factor-β
THE CELL CYCLE AND NEURAL FATE DECISIONS
In metazoans, the cell cycle can be divided at the cellular level into two gap phases (G1 and G2) separated by a phase of DNA synthesis (S-phase, between G1 and G2) and cell division/cytokinesis (M-phase, between G2 and G1); see Figure 1 for a summary (also [1]). Transition between the phases is driven by the activity of specific CDKs (cyclin-dependent kinases) bound to their cognate cyclins. It is generally accepted that cell cycle exit precedes differentiation, but the mechanisms co-ordinating these two processes have remained elusive. However, recent advances have uncovered multiple direct links between the cell cycle and differentiation machinery, which we describe in the present review with a focus on the nervous system, where these links have perhaps been most extensively studied.
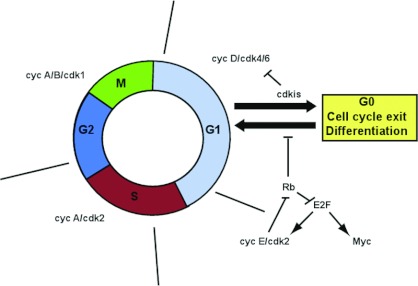
Figure 1. The cell cycle.
The points at which specific cyclin–CDK complexes are active and where inhibitors or positive regulators of the cell cycle act are illustrated. cyc, cyclin.
THE CELL CYCLE DURING NEUROGENESIS: CELL CYCLE LENGTH
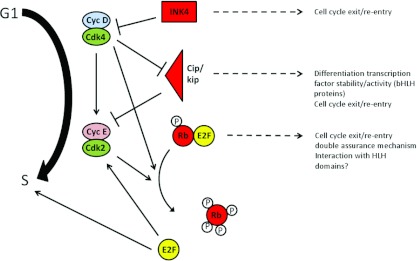
Neural precursors undergo cell cycle arrest prior to, or at least concomitant with, neuronal differentiation. Evidence has emerged demonstrating that the regulation of the length of G1 is pivotal in controlling the balance between progenitor maintenance and generation of differentiated neurons (see Figure 2 for a summary of cell cycle components involved in regulating the length of G1). In general, it is known that down-regulation or inhibition of positive regulators of cell cycle progression leads to increased differentiation and a reduction in the neural stem/progenitor cell pool. This is clearly seen during embryonic development, where CDKis (CDK inhibitors) accumulate in progenitor cells with each successive division, progressively increasing the length of G1 [2–4]. Indeed, addition of olomoucine, a synthetic CDKi, to whole mouse embryos in culture artificially lengthens the cell cycle of neuroepithelial progenitors and causes premature neurogenesis [5]. In vitro, experiments using the rat pheochromocytoma cell line PC12 also show that inhibition of CDKs by chemical inhibition or expression of the CDKi p21cip1 leads to increased neural differentiation [6,7].
Figure 2. G1/S-phase transition.
The role of negative regulators (CDKis and Rb) with respect to both cell cycle progression and development are highlighted. Cyc, cyclin; P, phosphorylation.
A number of studies have reinforced the observation that experimental manipulation of cell cycle length can alter the balance between proliferation and differentiation of neural precursors in vivo and in vitro. Elegant experiments conducted in utero in the cortex of developing mouse embryos demonstrate that shortening the cell cycle has the opposite effect to CDKis; overexpression of cyclin D/CDK4 delays neurogenesis and enhances the basal progenitor population. This appears to be a direct effect of lengthening of the cell cycle as, in contrast, knockdown of cyclin D/CDK4 by shRNA (small hairpin RNA) increases the number of differentiated neurons by 40% [8]. In this study, the authors use a cumulative BrdU (bromodeoxyuridine) labelling technique to estimate the length of each phase of the cell cycle, demonstrating that cyclin D/CDK4 overexpression leads to a shortened G1-phase, which correlates with the decrease in neurogenesis. It is unlikely that this phenomenon is specific to the activity of cyclin D/CDK4, as similar results have been reported in adult progenitors of the dentate gyrus when the length of G1 was modulated by altering expression of CDK6 [9]. Further, overexpression of cyclin A2/CDK2 in Xenopus embryos specifically inhibits epidermal and neural differentiation [10]. Thus it appears to be the overall cell cycle structure and accumulation of the population in G1 that leads to increased differentiation rather than the effects of specific cell cycle regulators. Such observations, however, do not address the questions of the directionality of signalling or the mechanistic links between the cell cycle and differentiation machineries during neurogenesis.
How might the length of the cell cycle regulate the differentiation of neural progenitors mechanistically? One potential way is for cell cycle regulators to directly control the activity of proteins that drive neuronal differentiation. Ngn2 (neurogenin 2) is a tissue-specific bHLH [basic HLH (helix–loop–helix)] protein, which is active as a heterodimer with a ubiquitously expressed bHLH E protein binding partner [11,12], and plays a pivotal role in differentiation of glutamatergic neurons. It has recently been shown that the Ngn2 protein is phosphorylated on multiple sites by CDKs [13]. Intriguingly, the higher the CDK activity, the greater the number of sites that are modified, making Ngn2 quantitatively sensitive to CDK levels. Preventing Ngn2 phoshorylation significantly enhances Ngn2's ability to transcribe downstream target genes that drive neuronal differentiation by promoting DNA binding. In this way, the length of the G1-phase can directly influence neuronal differentiation: when the G1-phase is short, CDKs accumulate rapidly and phosphorylate Ngn2, limiting its ability to drive neuronal differentiation. Conversely, when the G1-phase is long, CDK levels remain low for longer, allowing un(der)phosphorylated Ngn2 to accumulate. This efficiently activates downstream targets that promote the differentiation of mature neurons [13]. Thus the length of G1 can influence a neural progenitor's propensity to differentiate by directly regulating the level of activity of a component of the differentiation machinery.
SPECIFIC ROLES OF CELL CYCLE REGULATORS IN THE CONTROL OF NEUROGENESIS
Far from being uniformly expressed in all neural tissues of the developing embryo, cell cycle regulators frequently show tissue- and developmental stage-dependent patterns of expression that cannot be predicted solely from the cell cycle rate in these areas (e.g. [14]). This indicates potential additional roles for cell cycle regulators in the control of multiple aspects of neurogenesis, and many such roles have been uncovered (summarized in Table 1).
Table 1. Cell cycle components regulating cell fate.
| Component | Roles in cell fate specification | Reference(s) |
|---|---|---|
| CDK2 | May be required for neural stem cell self-renewal properties. | [93] |
| CDKi | General role in promoting cell cycle exit and differentiation. p27kip1 and p57kip2 in mouse and Xic1 in Xenopus promote neurogenesis independent of CDKi activity. | [33,35,36] |
| Cyclin A | Overexpression in Xenopus leads to thickened epidermis and inhibited neurogenesis. | [14] |
| Cyclin E | Overexpression in Xenopus leads to an enlarged cells phenotype. Specification of the NB6-4t lineage in Drosophila. | [10,14,27] |
| Geminin | Required for maintenance of neural precursors in Xenopus and possibly mammals. | [45,48,49] |
| Rb | Part of a general mechanism for the maintenance of cell cycle exit. Interacts with HLH proteins to promote neurogenesis. | [53,56,59] |
Cyclins
In addition to a more general role in influencing G1-phase length, specific D-type cyclins have been shown to have distinct roles in driving progenitor maintenance and cell fate decisions within the nervous system. For instance, cyclin D1 is expressed at high levels during proliferation of cells in the retina and cerebellum, and the cyclin D1−/− mouse has a reduced thickness of retinal cell layers [15,16]. The cyclin D2−/− mouse has a decreased number of granule cells and a complete ablation of stellate interneurons [17], indicating an ability of D-type cyclins to influence neuronal subtype. However, a recent report suggests that cyclin D2 is, in fact, responsible for maintenance of the shared granule cell and stellate interneuron progenitor pool, thus allowing production of later-born cell types as well as regulating their maturation [18]. Meanwhile, cyclin D3 is specifically down-regulated in differentiating Müller glia of the retina [19,20].
Somewhat paradoxically, cyclin D has also been reported to promote neuronal differentiation in a number of cases. During the neural differentiation of PC12 cells, cyclin D expression is up-regulated [7]. A recent study has demonstrated that, in mouse and chick spinal cord, expression of cyclin D1 promotes neural differentiation, whereas cyclin D2 promotes cell cycling [21]. Down-regulation of cyclin D1 in the chick spinal cord reduced the proportion of Lim3+ or NeuroM+/Olig2+ progenitor cells, which are committed to differentiation, as well as the number of newly differentiated HB9+ motor neurons. This knockdown could be rescued by expression of mouse cyclin D1, but not mouse cyclin D2, suggesting that simple regulation of cell cycle progression could not explain this activity. Furthermore, expression of a mutant form of cyclin D1, cyclin D1KE, which cannot interact with CDKs, promoted a significant increase in the number of differentiation-committed progenitors, greater than the expression of wild-type cyclin D1. As non-overlapping expression of cyclin D1 and cyclin D2 is found in the mouse forebrain neuroepithelium [22], this may be a phenomenon that is not confined to the developing spinal cord. This cell-cycle-independent function of cyclin D1 may be mediated by its ability to act as a regulator of transcription [23–26], specifically in the recruitment of CBP histone acetyltransferase to promoters [23], and, in the spinal cord, where its activity was upstream of Hes6 (hairy and enhancer of split 6) activation [21]. Therefore it may be that as CDKi levels increase during development, and the activity of G1 CDKs is inhibited, cyclin D1 begins to act as a positive regulator of transcription rather than a positive regulator of the cell cycle.
Cyclin E also has a number of additional roles during neuronal differentiation not linked to its ability to regulate the length of G1. In Drosophila, cyclin E is particularly associated with development of the NB6-4 neuroblast lineage and assignment of asymmetric fate. Cyclin E was identified as being an upstream determinant of prospero and GCM (glial cells missing), which together specify neuronal fate [27]. Cyclin E is down-regulated in the NB6-4 abdominal lineage by AbdA and AbdB Hox proteins, so that it only promotes neurogenesis in the thoracic lineage [27–29]. Furthermore, it appears that cyclin E may play a later developmental role in post-mitotic neuron maintenance, as it has recently been found to constrain CDK5 activity [30]. Cyclin E expression is maintained at a high level in the adult murine brain, in contrast with other organs, and the authors of that study found that virtually all cyclin E is complexed to CDK5 to form a catalytically inactive complex. Inactivation of CDK5 appears to be required for efficient synaptogenesis, as genetic deletion of cyclin E or overexpression of CDK5 in murine hippocampal neurons resulted in a decrease in the number of synapses formed. Interestingly, using mass spectrometric analysis, the authors also identified the CDKi p27kip1 (see below) as a component of the inactive cyclin E–CDK5 complex.
CDKis
In mammals, the functional redundancy between members of the Cip/Kip family has hampered efforts to investigate specific functions of CDKis during neurogenesis over and above their propensity to lengthen the cell cycle. To overcome this problem, Xenopus has proved to be an excellent model system, as the only CDKi expressed during primary neurogenesis is Xic1, potentially a homologue of all three Cip/Kip family members (reviewed in [31]). Xic1 is highly expressed in dorsal tissue at late gastrula and neurula stages and is particularly prominent in the developing myotome (muscle precursors) and neural plate [32]. This is indicative of a specific role for Xic1 during neurogenesis, and indeed it was found that Xic1 was required for differentiation of primary neurons [33]. Overexpression of Xic1 in Xenopus embryos promotes neurogenesis, but only within territories of endogenous proneural gene expression, suggesting an interaction between Xic1 and proneural proteins. Further investigation demonstrated that Xic1 acts in parallel with the proneural protein xNgn2 to regulate neurogenesis and that both Xic1 and Ngn2 expression is down-regulated by Notch signalling [34]. In addition to the studies in Xenopus, studies of CDKis in the developing murine cortex have found that both p27kip1 and p57kip2 promote neurogenesis and enhance neuronal migration when overexpressed [35,36]. In the case of Xic1 and p27kip1, it is clear that enhancement of neurogenesis is independent of, but complimentary to, cell cycle regulatory activity, as CDKis with compromised CDK inhibitory activity still promote neurogenesis. In fact, these CDKis have been shown to stabilize the Ngn2 protein [33,35]. Thus these CDKis bring about cell cycle lengthening and exit while simultaneously stabilizing the proneural protein that will drive the differentiation process, providing co-ordination of these two events within a single molecule [33,35].
Additional functions have also been ascribed to specific CDKis. For instance, although both p27kip1 and p57kip2 promote neurogenesis in the developing mouse cerebral cortex, only p57kip2 is resistant to astrogliogenic signalling by ciliary neurotrophic factor and requires intact cyclin/CDK binding domains to promote neurogenesis, whereas p27kip1 promotes neurogenesis independent of its cell cycle regulatory activity [33,35,36]. Although p57kip2 promotes the cell cycle exit of murine pituitary precursors, p57kip2+/cyclin E+ non-cycling progenitors are found in vivo, suggesting that p57kip2 inhibition of the cell cycle does not induce differentiation in these precursors [37]. Instead, p57kip2 was down-regulated and p27kip1 up-regulated upon precursor differentiation, and loss of p27kip1 allowed cell cycle re-entry of differentiated cells. In Xenopus, Xic1 has been shown to have an additional role independent of its ability to regulate the cell cycle in the developing retina, where it is required for generation of Müller glial cells [38].
Geminin
Geminin was first identified as a protein responsible for the loading of MCM (mini-chromosome maintenance) proteins on to replication origins, and the degradation of geminin by the APC/C at the metaphase/anaphase transition represents an important control to prevent re-synthesis of DNA during M-phase (reviewed in [39–41]). Geminin was also identified in an independent screen for proteins that perturb early neural development in Xenopus [42]. Geminin was found to interact directly with the homeobox transcription factor Six3 in retinal precursors during eye development [43]. Overexpression of geminin phenocopied inactivation of Six3 in the medaka fish and loss of geminin potentiated the Six3 overexpression phenotype, suggesting that geminin and Six3 play antagonistic roles in the regulation of proliferation during retinogenesis.
As well as interacting directly with transcription factors, geminin has been reported to interact with the SWI/SNF chromatin remodelling factor Brg1 in Xenopus embryos and P19 embryonal carcinoma cells [44]. Overexpression of geminin prevented ectopic neurogenesis in the presence of overexpressed proneural proteins and this activity required geminin's ability to bind to Brg1, suggesting that geminin inhibits neural differentiation by antagonizing Brg1 binding to proneural proteins [45]. Geminin has also been reported to bind to Polycomb group proteins, implying that it interacts with several chromatin modifiers to maintain repression of genes driving differentiation [46,47]. Geminin's role in the maintenance of mammalian neural precursors is controversial, with some reports suggesting it is required for regulation of cortical progenitor proliferation [48] and other reports suggesting it is dispensable during neurogenesis [49].
Rb (retinoblastoma susceptibility gene)
Rb functions as an inhibitor of the E2F transcription factor, which is responsible for the up-regulation of a number of genes involved in the G1/S-phase transition, including cyclin E. In its hypophosphorylated form, Rb binds to E2F and its DP (dimerization partner) and converts the complex into a transcriptional repressor by recruiting repressive chromatin-modifying complexes [50]. Phosphorylation by cyclin/CDKs promotes a hyperphosphorylated form of Rb (pRb) that cannot associate with the E2F–DP complex [51]. Regulation of Rb is implicated in a broad range of differentiation events, including a general control mechanism preventing differentiated cells from re-entering the cell cycle. Experiments in differentiating neurons in Drosophila demonstrated that the Rb and p27kip1 homologues contribute to repression of E2F and cyclin E/CDK2 activities in parallel [52]. In order for differentiated cells to re-enter the cell cycle, both E2F and cyclin E had to be supplied [52,53]. Although this double-assurance mechanism seems to apply to diverse cell types, the mechanism inhibiting the feed-forward response between cyclin E and E2F appears to differ between cell types [52]. Despite this, degradation of key E2F targets in the presence of overexpressed E2F or degradation of E2F activator complexes in the presence of overexpressed cyclin E may well be key to the block to cell cycle re-entry [53].
Aside from its more general roles in differentiation, Rb appears to play specific roles in the regulation of neurogenesis. Indeed, Rb was isolated as a gene that was mutated in cases of familial multifocal retinoblastoma, which in itself suggests a tissue-specific function of Rb, as patients appear prone to only certain types of tumour in addition to those of the retina, e.g. osteosarcomas [54]. Rb is strongly expressed in the developing CNS (central nervous system). Rb has also been reported to interact with HLH proteins, key drivers of neuronal differentiation [55] at several levels, possibly via binding to the HLH motif [56], although a more recent NMR study suggests that binding is indirect [57]. It appears that Rb and Id2 (inhibitor of DNA binding 2) can associate, with a requirement for the HLH domain of Id2, and can antagonize each other's activity [56]. Delayed differentiation and apoptosis induced by overexpression of Id2 in cortical progenitors was rescued by co-expression of a constitutively active form of Rb [58]. As well as inhibiting Id proteins, Rb directly enhances the transcriptional activity of NeuroD (neurogenic differentiation) [59]. Thus Rb may interact directly with transcription factors to enhance or repress the transcription of genes, driving differentiation. At present, however, it is unclear as to whether the phosphorylation status of Rb regulates its interaction with these transcription factors and therefore whether the length of the cell cycle, and more specifically cyclin E/CDK2 activity, may regulate differentiation via Rb activity.
Thus it is clear that cell cycle regulators can influence differentiation in the nervous system by diverse mechanisms that require both cell-cycle-dependent and -independent functions. It is also becoming increasingly clear that regulators of differentiation also have direct effects on the cell cycle machinery that are important to co-ordinate these two processes.
REGULATION OF THE CELL CYCLE BY NEURAL TRANSCRIPTION FACTORS
Previous studies have demonstrated that transcription factors traditionally associated with neuronal differentiation can also regulate the cell cycle during neurogenesis. Perhaps the most insightful early studies were investigations of global gene expression profile changes during differentiation [60,61]. These highlighted cell cycle components as a major proportion of differentially expressed genes that were directly down-regulated during differentiation of murine NSCs (neural stem cells) [60]. As the authors of this study were careful to check that the identified genes were indeed enriched in the CNS germinal zone of mice at three different embryonic stages and not simply a feature of proliferating tissues, this suggests a role for the cell cycle machinery during neural differentiation that is not simply linked to self-renewal. The study also identified Sox3 (Sry-type high mobility group box) and FoxM1 (forkhead box M1) as transcription factors enriched in the germinal zone and associated with maintenance of the progenitor state. Both Sox and Fox family members are known to have extensive roles in the regulation of neurogenesis, and both have links to the regulation of the cell cycle.
The Sox family
The evolutionarily conserved Sox family is split into two subgroups: SoxB1 (Sox1–Sox3) and SoxB2 (Sox14 and Sox21). In general, SoxB1 members are thought to maintain the proliferating progenitor state, whereas SoxB2 members counteract the activity of SoxB1 members and promote neuronal differentiation in a variety of systems [62–67]. A study of the downstream targets of Sox3 demonstrated that, in Xenopus, xSox3 can up-regulate the expression of xSox2 and geminin, thus elucidating a direct link to a component of the cell cycle machinery [68]. Although overexpression of either xSox3 or xSox2 in that study caused expansion of the neural plate, and xSox3 overexpression lead to increased cell proliferation, direct links specifically to the regulation of cell cycle length by Sox proteins in Xenopus have not yet been identified.
In mouse neurosphere cultures, Sox1 is required for the maintenance of progenitor cells, and Sox1−/− cells have an elongated cell cycle [69]. The effect on progenitor maintenance appears to be via Sox1 suppression of Prox1 (Prospero-related homeobox 1), which is a factor known to promote neural differentiation and cell cycle exit in mammalian systems [70]: Sox1−/− cells express Prox1 at almost double the level of wild-type cells, leading to more than double the normal number of progenitors exiting the cell cycle [69]. Prox1 also displays cellcycle-phase-specific expression, which is of interest as the cell cycle phase from which progenitors exit is known to determine their final site in the developing cortex [71]. However, although transcriptional activities of Sox family members clearly play a role in cell cycle regulation during neuronal differentiation, there is very little mechanistic evidence showing that this regulation is direct and not mediated by intermediate transcription factors.
The Fox family
The Fox transcription factor classification encompasses a number of subgroups of which three will be highlighted here: FoxM, FoxG and FoxO. FoxM1 is expressed in a number of proliferating tissues and has been identified as a prognostic indicator in cases of medulloblastoma [72], suggesting a role in maintaining the proliferative state. FoxM1 up-regulates Cdc25b (cell division cycle 25b) and cyclin B1 and B3 expression and so promotes G2- to M-phase progression [73–75]. However, in Xenopus, FoxM1 appears to be required both for the proliferation and differentiation of neural progenitors [76]. Knockdown of FoxM1 in Xenopus embryos leads to a reduction in the expression of neural β-tubulin, but an expansion in the expression of xSox2. Ueno et al. [76] therefore concluded that proliferation driven by FoxM1 was actually a requirement for neural differentiation, possibly because FoxM1 expression denotes the final division before differentiation.
FoxO family members are also broadly expressed, and FoxO1−/− mouse embryos die at E10.5 (embryonic day 10.5) owing to vascular abnormalities [77]. Different FoxO family members are widely believed to up-regulate the same transcriptional targets, one of which is the CDKi p21cip1. In neuroepithelial and glioblastoma cells, FoxO was found to bind specifically to the Smad proteins Smad3 and Smad4 to form a transcriptional activator complex that is targeted to a region of the p21cip1 promoter that contains a consensus forkhead binding element as well as a Smad-binding region [78]. Smad proteins function as part of the highly conserved TGF-β (transforming growth factor-β) signalling pathway: during canonical signalling, the binding of an extracellular ligand of the TGF-β family to TGF receptor 1 leads to the phosphorylation and activation of Smad2 or Smad3 (reviewed in [79]). Activated Smad2/3 is then able to bind to Smad4 and subsequently translocate to the nucleus as a transcriptional activator complex. TGF-β signalling inhibits proliferation in a range of cell types, including NSCs in culture [80]. In epithelial cells, such cytostatic activity results from TGF-β-mediated up-regulation of the CDKis p21cip1 and p15Ink4b and the down-regulation of Id1, Id2 and c-Myc (reviewed in [81]). It would therefore appear that FoxO is a direct mediator of TGF-β cytostatic activity.
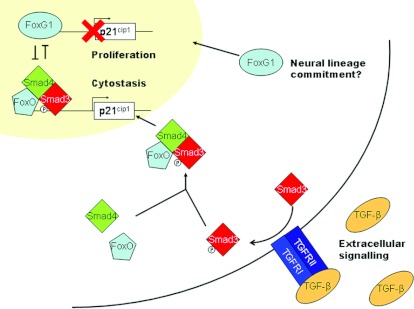
Crucially, the involvement of FoxO in the transcriptional activator complex may provide a mechanism for the specific regulation of cell cycle length in neurons. Another Fox family member, FoxG1, is required specifically for the specification of the ventral telencephalon [82], and FoxG1−/− mice display hypoplasia of the telencephalon and excessive production of Cajal–Retzius neurons, the earliest born neurons in the telencephalon [83]. FoxG1 competes with FoxO for binding to promoter sites and thus acts as a repressor of FoxO activity and a pro-proliferative factor [78]. Interestingly, FoxG1 appears not only to promote the proliferation of neural progenitor cells but also to specify region-specific structures within the developing telencephalon [82] and perhaps even neural lineage identity, as it was recently identified as a factor contributing to the direct reprogramming of fibroblasts to NSC-like cells [84]. As Fox family members constitute a direct link between extracellular signalling, cell cycle control and neuronal differentiation, it is interesting to speculate that the expression of different Fox family members could regulate specific changes in the cell cycle and propensity to differentiate in response to extracellular signalling (see Figure 3).
Figure 3. TGF-β signalling as a cytostatic signal.
A focus on the role of Fox transcription factors in the decision to proliferate. Note how the expression of Fox factors links inhibition of proliferation with extracellular signalling and neural specification. An animation of this Figure is available at http://www.BiochemJ.org/bj/444/0375/bj4440375add.htm.
Proneural genes
The proneural genes constitute a class of bHLH transcription factors which, when overexpressed, potentiate cell cycle exit and neuronal differentiation. Indeed, bHLH factors, such as Ngn2 and Mash1/Ascl1 (achaete-scute homologue 1) are often considered as master regulators of neurogenesis [13,55,85], where they drive neurogenesis cell autonomously, and they are also responsible for the maintenance of the neural progenitor pool via up-regulation of the Notch ligand Delta [86] in a non-cell autonomous manner. Although the level of the CDKi p27Kip1 protein was shown to rise in response to proneural protein-driven neuronal differentiation of P19 embryonal carcinoma cells [85], it is surprising that CDKis such as p27Kip1 have not been shown to be direct transcriptional targets of proneural proteins [87,88]. In proliferating neural precursor cells, Ngn2 is expressed in oscillating waves, driven by a double-negative feedback loop involving Hes1 and the Notch signalling pathway [89]. These oscillations are thought to be essential for maintenance of the progenitor state, whereas cell cycle exit and differentiation coincide with a stable elevation of Ngn2 levels. Although it is currently not known how or whether the transcriptional oscillations in neural precursors are co-ordinated with cell cycling, it is known that Notch signalling also up-regulates cyclin D1 expression and down-regulates the expression of CDKis [90,91], which may have implications for the activity of the Ngn2 protein [13].
Intriguingly, recent data has shown that the proneural protein Mash1/Ascl1 plays a more direct and essential role in both progenitor maintenance and neuronal differentiation. Ascl1 directly transcriptionally regulates both positive regulators of the cell cycle, promoting the transcription of E2F1 and CDK2, as well as drivers and effectors of differentiation, such as MyT1 (myelin transcription factor 1) and neural β-tubulin, although the opposing gene sets are regulated in a temporally distinct manner [88]. It will be important to determine how both cell cycle activating and cell cycle inhibitory functions can be controlled by the same transcription factor, although at present this is unclear. The authors suggest that these opposing functions may be regulated by different events at the promoter (different binding partners or chromatin accessibility) or by direct modification of the Ascl1 protein itself over time [92], and it will clearly be important to investigate this further.
CONCLUSIONS
The processes of differentiation and cell division are often viewed as separate, although it is clear that cross-talk between the two must exist as they are mutually exclusive in the vast majority of cells. In the present review, we have discussed the growing evidence that components of the cell cycle machinery play central roles during neuronal differentiation, while summarising roles that transcription factors, traditionally viewed as part of the differentiation machinery, play in regulating the cell cycle. Although we currently know the most about regulatory mechanisms co-ordinating proliferation and differentiation in the developing nervous system, it is likely that many of the mechanisms outlined are conserved in other tissues. Identification of the mechanistic links between the cell cycle and the differentiation machineries and their subsequent manipulation could lead to clear advances in the fields of cancer therapy and regenerative medicine. This will be an important goal for future research.
ACKNOWLEDGEMENTS
We thank Fahad Ali, Ana Almeida, Christelle Fiore-Heriche, Francois Guillemot, Chris Hurley, Ali Jones, Marc Kirschner, Romana Kucerova, Gary McDowell, Jan Pruszak and Ryan Roark for helpful discussions and technical assistance.
FUNDING
C.H. was supported by a Cancer Research UK Graduate Studentship during the preparation of this paper. A.P. was supported by a MRC Research Grant [grant number G0700758].
References
- 1.Murray A. W., Hunt T. The Cell Cycle: An Introduction. Oxford: Oxford University Press; 1993. [Google Scholar]
- 2.Durand B., Gao F. B., Raff M. Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. EMBO J. 1997;16:306–317. doi: 10.1093/emboj/16.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fero M. L., Rivkin M., Tasch M., Porter P., Carow C. E., Firpo E., Polyak K., Tsai L. H., Broudy V., Perlmutter R. M., et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 4.Kiyokawa H., Kineman R. D., Manova-Todorova K. O., Soares V. C., Hoffman E. S., Ono M., Khanam D., Hayday A. C., Frohman L. A., Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 5.Calegari F., Huttner W. B. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J. Cell Sci. 2003;116:4947–4955. doi: 10.1242/jcs.00825. [DOI] [PubMed] [Google Scholar]
- 6.Dobashi Y., Shoji M., Kitagawa M., Noguchi T., Kameya T. Simultaneous suppression of cdc2 and cdk2 activities induces neuronal differentiation of PC12 cells. J. Biol. Chem. 2000;275:12572–12580. doi: 10.1074/jbc.275.17.12572. [DOI] [PubMed] [Google Scholar]
- 7.Yan G. Z., Ziff E. B. NGF regulates the PC12 cell cycle machinery through specific inhibition of the Cdk kinases and induction of cyclin D1. J. Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lange C., Huttner W. B., Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Beukelaers P., Vandenbosch R., Caron N., Nguyen L., Belachew S., Moonen G., Kiyokawa H., Barbacid M., Santamaria D., Malgrange B. Cdk6-dependent regulation of G1 length controls adult neurogenesis. Stem Cells. 2011;29:713–724. doi: 10.1002/stem.616. [DOI] [PubMed] [Google Scholar]
- 10.Richard-Parpaillon L., Cosgrove R. A., Devine C., Vernon A. E., Philpott A. G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissue-specific differentiation in Xenopus. Development. 2004;131:2577–2586. doi: 10.1242/dev.01121. [DOI] [PubMed] [Google Scholar]
- 11.Ma Q., Kintner C., Anderson D. J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 12.Vosper J. M., McDowell G. S., Hindley C. J., Fiore-Heriche C. S., Kucerova R., Horan I., Philpott A. Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis. J. Biol. Chem. 2009;284:15458–15468. doi: 10.1074/jbc.M809366200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali F., Hindley C., McDowell G., Deibler R., Jones A., Kirschner M., Guillemot F., Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernon A. E., Philpott A. The developmental expression of cell cycle regulators in Xenopus laevis. Gene Expression Patterns. 2003;3:179–192. doi: 10.1016/s1567-133x(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 15.Ma C., Papermaster D., Cepko C. L. A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9938–9943. doi: 10.1073/pnas.95.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sicinski P., Donaher J. L., Parker S. B., Li T., Fazeli A., Gardner H., Haslam S. Z., Bronson R. T., Elledge S. J., Weinberg R. A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 17.Huard J. M., Forster C. C., Carter M. L., Sicinski P., Ross M. E. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development. 1999;126:1927–1935. doi: 10.1242/dev.126.9.1927. [DOI] [PubMed] [Google Scholar]
- 18.Leto K., Bartolini A., Di Gregorio A., Imperiale D., De Luca A., Parmigiani E., Filipkowski R. K., Kaczmarek L., Rossi F. Modulation of cell-cycle dynamics is required to regulate the number of cerebellar GABAergic interneurons and their rhythm of maturation. Development. 2011;138:3463–3472. doi: 10.1242/dev.064378. [DOI] [PubMed] [Google Scholar]
- 19.Dyer M. A., Cepko C. L. Control of Müller glial cell proliferation and activation following retinal injury. Nat. Neurosci. 2000;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- 20.Sun W., Lee D. K., Lee C. C., Kim K. Differential expression of D-type G1 cyclins during mouse development and liver regeneration in vivo. Mol. Reprod. Dev. 1996;43:414–420. doi: 10.1002/(SICI)1098-2795(199604)43:4<414::AID-MRD2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Lukaszewicz A. I., Anderson D. J. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11632–11637. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glickstein S. B., Alexander S., Ross M. E. Differences in cyclin D2 and D1 protein expression distinguish forebrain progenitor subsets. Cereb. Cortex. 2007;17:632–642. doi: 10.1093/cercor/bhk008. [DOI] [PubMed] [Google Scholar]
- 23.Bienvenu F., Jirawatnotai S., Elias J. E., Meyer C. A., Mizeracka K., Marson A., Frampton G. M., Cole M. F., Odom D. T., Odajima J., et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu M., Rao M., Bouras T., Wang C., Wu K., Zhang X., Li Z., Yao T. P., Pestell R. G. Cyclin D1 inhibits peroxisome proliferator-activated receptor γ-mediated adipogenesis through histone deacetylase recruitment. J. Biol. Chem. 2005;280:16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 25.Fu M., Wang C., Rao M., Wu X., Bouras T., Zhang X., Li Z., Jiao X., Yang J., Li A., et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J. Biol. Chem. 2005;280:29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 26.Ratineau C., Petry M. W., Mutoh H., Leiter A. B. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J. Biol. Chem. 2002;277:8847–8853. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 27.Berger C., Pallavi S. K., Prasad M., Shashidhara L. S., Technau G. M. A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat. Cell Biol. 2005;7:56–62. doi: 10.1038/ncb1203. [DOI] [PubMed] [Google Scholar]
- 28.Berger C., Kannan R., Myneni S., Renner S., Shashidhara L. S., Technau G. M. Cell cycle independent role of Cyclin E during neural cell fate specification in Drosophila is mediated by its regulation of Prospero function. Dev. Biol. 2010;337:415–424. doi: 10.1016/j.ydbio.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Kannan R., Berger C., Myneni S., Technau G. M., Shashidhara L. S. Abdominal-A mediated repression of Cyclin E expression during cell-fate specification in the Drosophila central nervous system. Mech. Dev. 2010;127:137–145. doi: 10.1016/j.mod.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Odajima J., Wills Z. P., Ndassa Y. M., Terunuma M., Kretschmannova K., Deeb T. Z., Geng Y., Gawrzak S., Quadros I. M., Newman J., et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev. Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philpott A. Division versus differentiation in the early Xenopus embryo. SEB Exp. Biol. Ser. 2008;59:145–165. [PubMed] [Google Scholar]
- 32.Vernon A. E., Philpott A. A single cdk inhibitor, p27Xic1, functions beyond cell cycle regulation to promote muscle differentiation in Xenopus. Development. 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- 33.Vernon A. E., Devine C., Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 34.Vernon A. E., Movassagh M., Horan I., Wise H., Ohnuma S., Philpott A. Notch targets the Cdk inhibitor Xic1 to regulate differentiation but not the cell cycle in neurons. EMBO Rep. 2006;7:643–648. doi: 10.1038/sj.embor.7400691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen L., Besson A., Heng J. I., Schuurmans C., Teboul L., Parras C., Philpott A., Roberts J. M., Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tury A., Mairet-Coello G., DiCicco-Bloom E. The cyclin-dependent kinase inhibitor p57Kip2 regulates cell cycle exit, differentiation, and migration of embryonic cerebral cortical precursors. Cereb. Cortex. 2011;21:1840–1856. doi: 10.1093/cercor/bhq254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilodeau S., Roussel-Gervais A., Drouin J. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Mol. Cell Biol. 2009;29:1895–1908. doi: 10.1128/MCB.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohnuma S., Philpott A., Wang K., Holt C. E., Harris W. A. p27Xic1, a Cdk inhibitor, promotes the determination of glial cells in Xenopus retina. Cell. 1999;99:499–510. doi: 10.1016/s0092-8674(00)81538-x. [DOI] [PubMed] [Google Scholar]
- 39.Madine M., Laskey R. Geminin bans replication licence. Nat. Cell Biol. 2001;3:E49–E50. doi: 10.1038/35055158. [DOI] [PubMed] [Google Scholar]
- 40.McGarry T. J., Kirschner M. W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 41.Seo S., Kroll K. L. Geminin's double life: chromatin connections that regulate transcription at the transition from proliferation to differentiation. Cell Cycle. 2006;5:374–379. doi: 10.4161/cc.5.4.2438. [DOI] [PubMed] [Google Scholar]
- 42.Kroll K.L, Salic A. N., Evans L. M., Kirschner M. W. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- 43.Del Bene F., Tessmar-Raible K., Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 44.Seo S., Richardson G. A., Kroll K. L. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 45.Seo S., Herr A., Lim J. W., Richardson G. A., Richardson H., Kroll K. L. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim J. W., Hummert P., Mills J. C., Kroll K. L. Geminin cooperates with Polycomb to restrain multi-lineage commitment in the early embryo. Development. 2011;138:33–44. doi: 10.1242/dev.059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo L., Yang X., Takihara Y., Knoetgen H., Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 48.Spella M., Kyrousi C., Kritikou E., Stathopoulou A., Guillemot F., Kioussis D., Pachnis V., Lygerou Z., Taraviras S. Geminin regulates cortical progenitor proliferation and differentiation. Stem Cells. 2011;29:1269–1282. doi: 10.1002/stem.678. [DOI] [PubMed] [Google Scholar]
- 49.Schultz K. M., Banisadr G., Lastra R. O., McGuire T., Kessler J. A., Miller R. J., McGarry T. J. Geminin-deficient neural stem cells exhibit normal cell division and normal neurogenesis. PLoS ONE. 2011;6:e17736. doi: 10.1371/journal.pone.0017736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 51.Du W., Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buttitta L. A., Katzaroff A. J., Perez C. L., de la Cruz A., Edgar B. A. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell. 2007;12:631–643. doi: 10.1016/j.devcel.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 53.Buttitta L. A., Katzaroff A. J., Edgar B. A. A robust cell cycle control mechanism limits E2F-induced proliferation of terminally differentiated cells in vivo. J. Cell Biol. 2010;189:981–996. doi: 10.1083/jcb.200910006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chinnam M., Goodrich D. W. RB1, development, and cancer. Curr. Top. Dev. Biol. 2011;94:129–169. doi: 10.1016/B978-0-12-380916-2.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertrand N., Castro D. S., Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 56.Iavarone A., King E. R., Dai X. M., Leone G., Stanley E. R., Lasorella A. Retinoblastoma promotes definitive erythropoiesis by repressing Id2 in fetal liver macrophages. Nature. 2004;432:1040–1045. doi: 10.1038/nature03068. [DOI] [PubMed] [Google Scholar]
- 57.Smialowski P., Singh M., Mikolajka A., Majumdar S., Joy J. K., Nalabothula N., Krajewski M., Degenkolbe R., Bernard H. U., Holak T. A. NMR and mass spectrometry studies of putative interactions of cell cycle proteins pRb and CDK6 with cell differentiation proteins MyoD and ID-2. Biochim. Biophys. Acta. 2005;1750:48–60. doi: 10.1016/j.bbapap.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 58.Toma J. G., El-Bizri H., Barnabe-Heider F., Aloyz R., Miller F. D. Evidence that helix-loop-helix proteins collaborate with retinoblastoma tumor suppressor protein to regulate cortical neurogenesis. J. Neurosci. 2000;20:7648–7656. doi: 10.1523/JNEUROSCI.20-20-07648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Batsché E., Moschopoulos P., Desroches J., Bilodeau S., Drouin J. Retinoblastoma and the related pocket protein p107 act as coactivators of NeuroD1 to enhance gene transcription. J. Biol. Chem. 2005;280:16088–16095. doi: 10.1074/jbc.M413427200. [DOI] [PubMed] [Google Scholar]
- 60.Karsten S. L., Kudo L. C., Jackson R., Sabatti C., Kornblum H. I., Geschwind D. H. Global analysis of gene expression in neural progenitors reveals specific cell-cycle, signaling, and metabolic networks. Dev. Biol. 2003;261:165–182. doi: 10.1016/s0012-1606(03)00274-4. [DOI] [PubMed] [Google Scholar]
- 61.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 62.Bylund M., Andersson E., Novitch B. G., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-Sox3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 63.Collignon J., Sockanathan S., Hacker A., Cohen-Tannoudji M., Norris D., Rastan S., Stevanovic M., Goodfellow P. N., Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 64.Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 65.Pevny L., Placzek M. SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 66.Sandberg M., Kallstrom M., Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- 67.Uchikawa M., Kamachi Y., Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 68.Rogers C. D., Harafuji N., Archer T., Cunningham D. D., Casey E. S. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech. Dev. 2009;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elkouris M., Balaskas N., Poulou M., Politis P. K., Panayiotou E., Malas S., Thomaidou D., Remboutsika E. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells. 2011;29:89–98. doi: 10.1002/stem.554. [DOI] [PubMed] [Google Scholar]
- 70.Misra K., Gui H., Matise M. P. Prox1 regulates a transitory state for interneuron neurogenesis in the spinal cord. Dev. Dyn. 2008;237:393–402. doi: 10.1002/dvdy.21422. [DOI] [PubMed] [Google Scholar]
- 71.McConnell S. K., Kaznowski C. E. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 72.Priller M., Pöschl J., Abrão L., von Bueren A. O., Cho Y. J., Rutkowski S., Kretzschmar H. A., Schüller U. Expression of FoxM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin. Cancer Res. 2011;17:6791–6801. doi: 10.1158/1078-0432.CCR-11-1214. [DOI] [PubMed] [Google Scholar]
- 73.Leung T. W., Lin S. S., Tsang A. C., Tong C. S., Ching J. C., Leung W. Y., Gimlich R., Wong G. G., Yao K. M. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- 74.Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 75.Wang I. C., Chen Y. J., Hughes D., Petrovic V., Major M. L., Park H. J., Tan Y., Ackerson T., Costa R. H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ueno H., Nakajo N., Watanabe M., Isoda M., Sagata N. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development. 2008;135:2023–2030. doi: 10.1242/dev.019893. [DOI] [PubMed] [Google Scholar]
- 77.Hosaka T., Biggs W. H., 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., Arden K. C. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seoane J., Le H. V., Shen L., Anderson S. A., Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 79.Feng X. H., Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell. Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 80.Wachs F. P., Winner B., Couillard-Despres S., Schiller T., Aigner R., Winkler J., Bogdahn U., Aigner L. Transforming growth factor-β1 is a negative modulator of adult neurogenesis. J. Neuropathol. Exp. Neurol. 2006;65:358–370. doi: 10.1097/01.jnen.0000218444.53405.f0. [DOI] [PubMed] [Google Scholar]
- 81.Siegel P. M., Massagué J. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 82.Martynoga B., Morrison H., Price D. J., Mason J. O. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 83.Hanashima C., Li S. C., Shen L., Lai E., Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- 84.Lujan E., Chanda S., Ahlenius H., Südhof T. C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farah M. H., Olson J. M., Sucic H. B., Hume R. I., Tapscott S. J., Turner D. L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 86.Chitnis A., Henrique D., Lewis J., Ish-Horowicz D., Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 87.Sun Y., Meijer D. H., Alberta J. A., Mehta S., Kane M. F., Tien A. C., Fu H., Petryniak M. A., Potter G. B., Liu Z., et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906–917. doi: 10.1016/j.neuron.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castro D. S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L. G., Hunt C., et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimojo H., Ohtsuka T., Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 90.Das D., Lanner F., Main H., Andersson E. R., Bergmann O., Sahlgren C., Heldring N., Hermanson O., Hansson E. M., Lendahl U. Notch induces cyclin-D1-dependent proliferation during a specific temporal window of neural differentiation in ES cells. Dev. Biol. 2010;348:153–166. doi: 10.1016/j.ydbio.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 91.Georgia S., Soliz R., Li M., Zhang P., Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev. Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 92.Castro D. S., Guillemot F. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle. 2011;10:4026–4031. doi: 10.4161/cc.10.23.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jablonska B., Aguirre A., Vandenbosch R., Belachew S., Berthet C., Kaldis P., Gallo V. Cdk2 is critical for proliferation and self-renewal of neural progenitor cells in the adult subventricular zone. J. Cell Biol. 2007;179:1231–1245. doi: 10.1083/jcb.200702031. [DOI] [PMC free article] [PubMed] [Google Scholar]