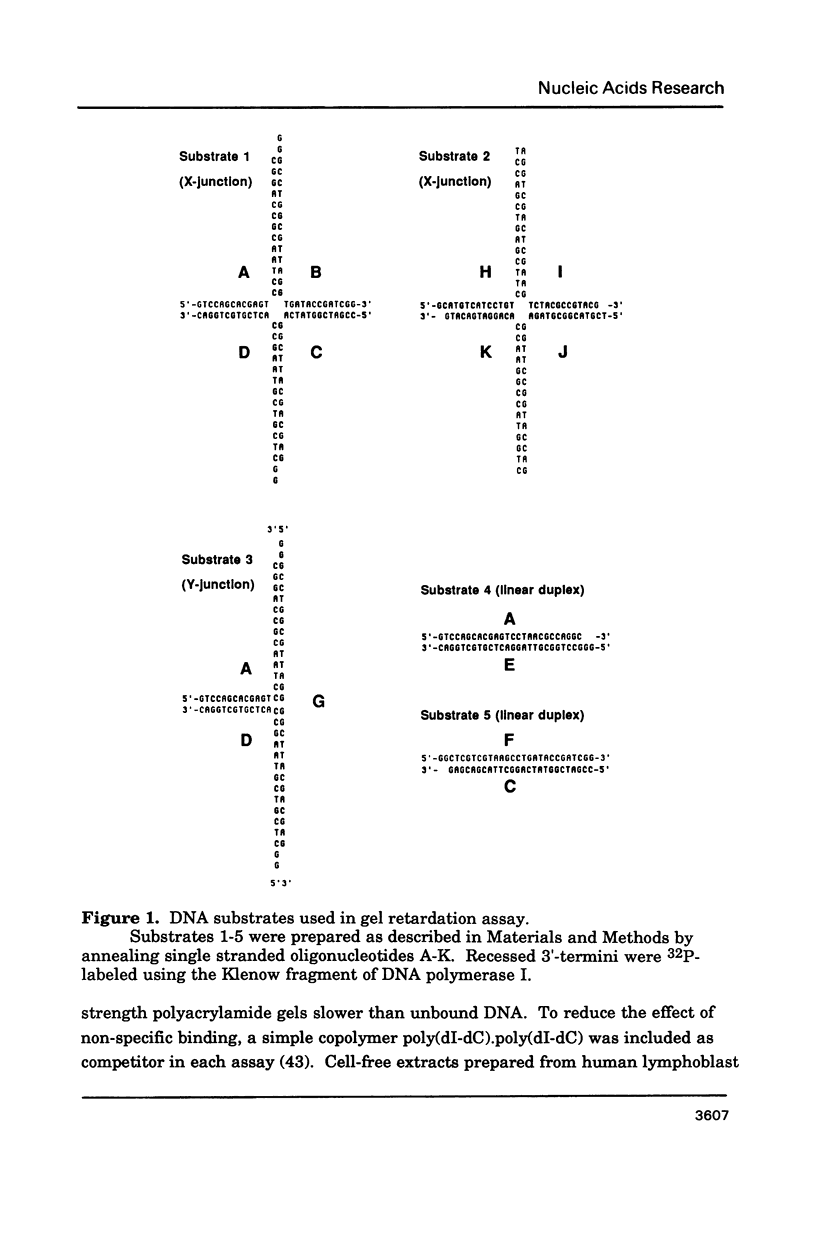
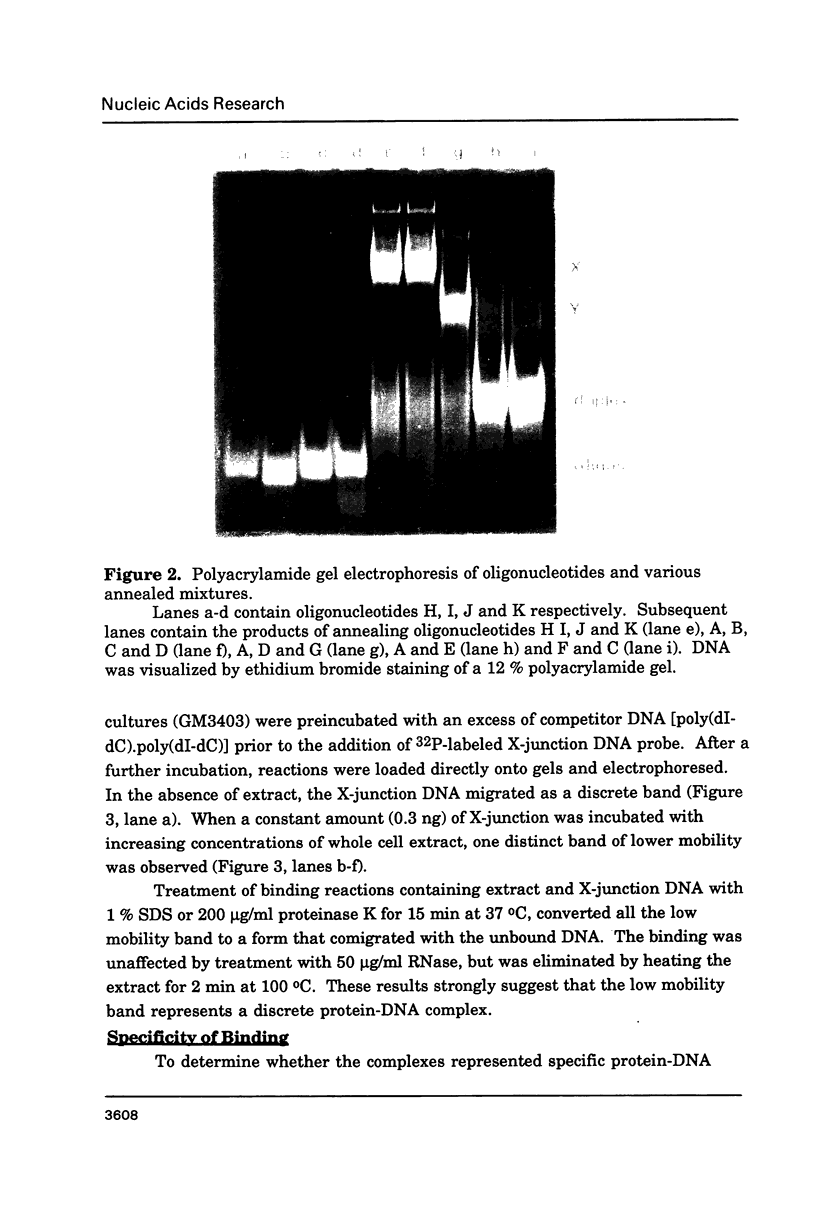
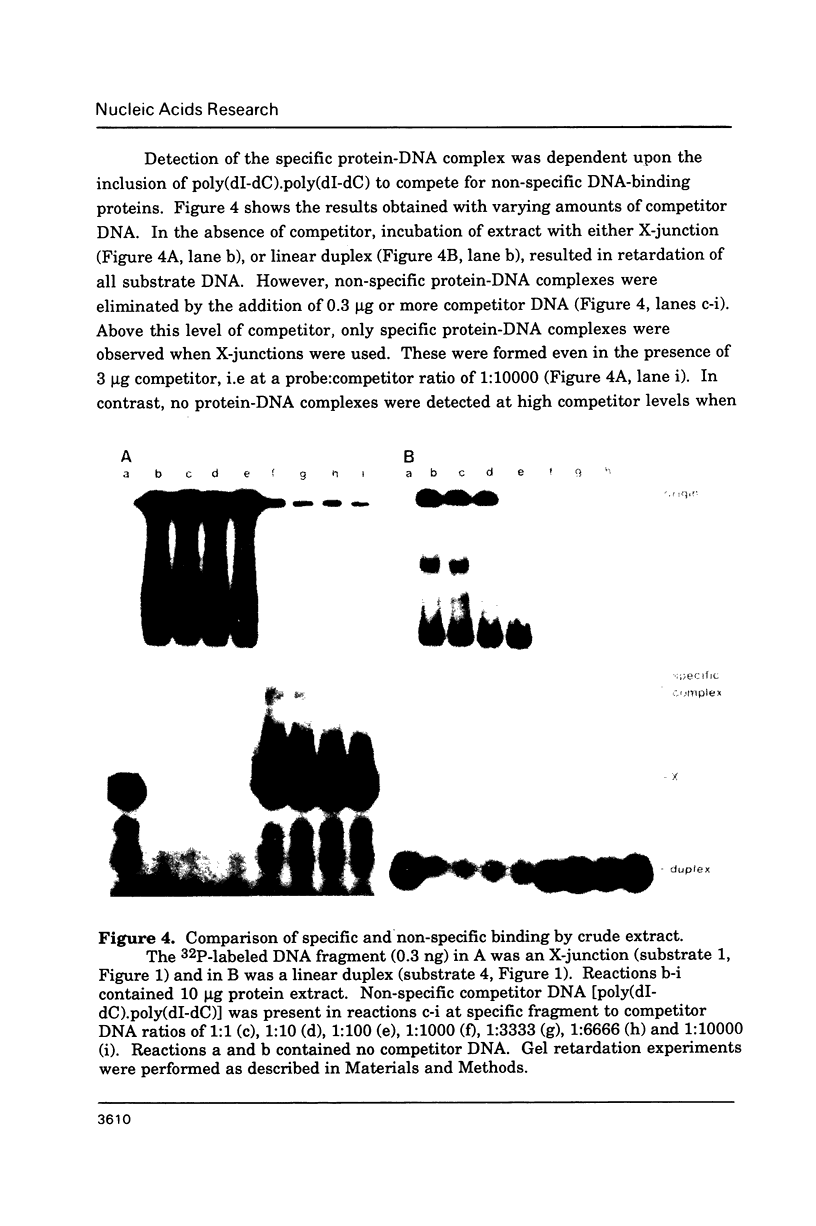
Abstract
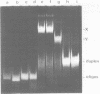
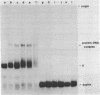
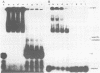
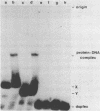
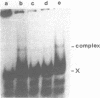
A gel electrophoresis binding assay has been used to probe extracts from cultured human lymphoblasts for proteins that bind cruciform structures in duplex DNA. Proteins have been detected that form complexes with synthetic X- and Y-junctions. Several lines of evidence suggest that binding is specific for DNA structure rather than sequence: (1) X- and Y-structures were bound whereas linear duplexes containing identical DNA sequences were not, (2) Binding occurred with equal efficiency to two X-junctions that were constructed from DNA strands of different sequence, (3) One X-junction successfully competed with another for binding whereas linear duplex DNA did not; and (4) protein-DNA complexes were observed at probe:non-specific competitor DNA ratios of 1:10,000.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. Recombinant DNA molecules of bacteriophage phi chi174. Proc Natl Acad Sci U S A. 1975 Jan;72(1):235–239. doi: 10.1073/pnas.72.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E. Interaction of a protein from rat liver nuclei with cruciform DNA. EMBO J. 1988 Mar;7(3):843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Palindromic base sequences and replication of eukaryote chromosome ends. Nature. 1974 Aug 9;250(5466):467–470. doi: 10.1038/250467a0. [DOI] [PubMed] [Google Scholar]
- Crews S., Ojala D., Posakony J., Nishiguchi J., Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979 Jan 18;277(5693):192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Wu A. M., Kahn R., Cunningham R. P., Radding C. M. Concerted strand exchange and formation of Holliday structures by E. coli RecA protein. Cell. 1981 Aug;25(2):507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- DeLange A. M., McFadden G. Efficient resolution of replicated poxvirus telomeres to native hairpin structures requires two inverted symmetrical copies of a core target DNA sequence. J Virol. 1987 Jun;61(6):1957–1963. doi: 10.1128/jvi.61.6.1957-1963.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P., Morgan A. R., McFadden G. Cruciform extrusion in plasmids bearing the replicative intermediate configuration of a poxvirus telomere. J Mol Biol. 1987 Aug 5;196(3):541–558. doi: 10.1016/0022-2836(87)90031-3. [DOI] [PubMed] [Google Scholar]
- Evans D. H., Kolodner R. Construction of a synthetic Holliday junction analog and characterization of its interaction with a Saccharomyces cerevisiae endonuclease that cleaves Holliday junctions. J Biol Chem. 1987 Jul 5;262(19):9160–9165. [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Ohmori H., Tomizawa J. DNA gyrase and DNA supercoiling. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):35–40. doi: 10.1101/sqb.1979.043.01.007. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Hoess R., Wierzbicki A., Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Hsu M. T. Electron microscopic evidence for the cruciform structure in intracellular SV40 DNA. Virology. 1985 Jun;143(2):617–621. doi: 10.1016/0042-6822(85)90400-3. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986 Jan;5(1):181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Brown D. T. Function of gene 49 of bacteriophage T4. II. Analysis of intracellular development and the structure of very fast-sedimenting DNA. J Virol. 1976 Jun;18(3):1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mosig G., Shaw M., Garcia G. M. On the role of DNA replication, endonuclease VII, and rII proteins in processing of recombinational intermediates in phage T4. Cold Spring Harb Symp Quant Biol. 1984;49:371–382. doi: 10.1101/sqb.1984.049.01.044. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Resolution of model Holliday junctions by yeast endonuclease is dependent upon homologous DNA sequences. Cell. 1988 Feb 26;52(4):621–629. doi: 10.1016/0092-8674(88)90474-6. [DOI] [PubMed] [Google Scholar]
- Pritchard A. E., Cummings D. J. Replication of linear mitochondrial DNA from Paramecium: sequence and structure of the initiation-end crosslink. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7341–7345. doi: 10.1073/pnas.78.12.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Oka A., Sugisaki H., Takanami M., Nishimura A., Yasuda Y., Hirota Y. Nucleotide sequence of Escherichia coli K-12 replication origin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):575–579. doi: 10.1073/pnas.76.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. Replication and resolution of telomeres in yeast. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1187–1194. doi: 10.1101/sqb.1983.047.01.134. [DOI] [PubMed] [Google Scholar]
- Tapper D. P., Clayton D. A. Mechanism of replication of human mitochondrial DNA. Localization of the 5' ends of nascent daughter strands. J Biol Chem. 1981 May 25;256(10):5109–5115. [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Delta sequences and double symmetry in a yeast chromosomal replicator region. J Mol Biol. 1982 Apr 5;156(2):293–307. doi: 10.1016/0022-2836(82)90330-8. [DOI] [PubMed] [Google Scholar]
- West S. C., Countryman J. K., Howard-Flanders P. Enzymatic formation of biparental figure-eight molecules from plasmid DNA and their resolution in E. coli. Cell. 1983 Mar;32(3):817–829. doi: 10.1016/0092-8674(83)90068-5. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Parsons C. A., Picksley S. M. Purification and properties of a nuclease from Saccharomyces cerevisiae that cleaves DNA at cruciform junctions. J Biol Chem. 1987 Sep 15;262(26):12752–12758. [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Kaufmann G., Martin R. G. Mammalian DNA enriched for replication origins is enriched for snap-back sequences. J Mol Biol. 1984 Nov 15;179(4):577–586. doi: 10.1016/0022-2836(84)90156-6. [DOI] [PubMed] [Google Scholar]