A novel telomere-protection mechanism relies upon nucleostemin-mediated recruitment of PML-IV to telomeres and subsequent recruitment of RAD51 in both ALT and telomerase-active cells.
Abstract
Continuously dividing cells must be protected from telomeric and nontelomeric DNA damage in order to maintain their proliferative potential. Here, we report a novel telomere-protecting mechanism regulated by nucleostemin (NS). NS depletion increased the number of telomere damage foci in both telomerase-active (TA+) and alternative lengthening of telomere (ALT) cells and decreased the percentage of damaged telomeres associated with ALT-associated PML bodies (APB) and the number of APB in ALT cells. Mechanistically, NS could promote the recruitment of PML-IV to SUMOylated TRF1 in TA+ and ALT cells. This event was stimulated by DNA damage. Supporting the importance of NS and PML-IV in telomere protection, we demonstrate that loss of NS or PML-IV increased the frequency of telomere damage and aberration, reduced telomeric length, and perturbed the TRF2ΔBΔM-induced telomeric recruitment of RAD51. Conversely, overexpression of either NS or PML-IV protected ALT and TA+ cells from telomere damage. This work reveals a novel mechanism in telomere protection.
Introduction
Telomeres protect chromosomal ends from replicative attrition and consist of tandem repeats of telomeric DNAs and a multiprotein complex (Greider and Blackburn, 1985, 1996; de Lange, 2005; Songyang and Liu, 2006). The telomere length plays a crucial role in preserving its integrity, and is maintained by the telomerase in more than 80% of the human cancers (Greider and Blackburn, 1989; Shay et al., 2001). In the other 10–15% of human cancers, the telomerase activity is undetectable (Shay and Bacchetti, 1997). Those telomerase-inactive cells are known as alternative lengthening of telomeres (ALT) cells and are thought to use the homologous recombination (HR) mechanism for telomere maintenance (Bryan et al., 1995, 1997; Shay and Bacchetti, 1997; Liu et al., 2007). One unique feature of ALT cells is the formation of the ALT-associated PML body (APB; Yeager et al., 1999; Dunham et al., 2000), which requires the SUMOylation of TRF1 and TRF2 (Potts and Yu, 2007) and several PML-associated proteins, including PML, MRN complex, RAD52, and RPA (Wu et al., 2000; Zhu et al., 2000; Jiang et al., 2007). The biological role of APB remains unclear, but may be linked to the HR event (Grobelny et al., 2000).
A potential molecule that regulates the telomere integrity in cancer and stem cells is nucleostemin (NS). NS is a nucleolar GTP-binding protein preferentially expressed by multiple types of stem cells and human cancers (Tsai and McKay, 2002; Baddoo et al., 2003; Liu et al., 2004; Ohmura et al., 2008; Nomura et al., 2009; Lin et al., 2010). Its function is required for self-renewal maintenance and early embryogenesis (Tsai and McKay, 2002; Liu et al., 2004; Beekman et al., 2006; Zhu et al., 2006). We previously found that NS and its vertebrate paralogue, guanine nucleotide-binding protein-like 3-like (GNL3L), interact with one of the telomeric proteins, telomeric repeat-binding factor 1 (TRF1; Zhu et al., 2006, 2009; Tsai, 2009), which serves several key functions, including chromosomal end protection (Martínez et al., 2009), telomere shortening (van Steensel and de Lange, 1997), mitotic progression (Zhou et al., 2003, 2009), and APB formation (Potts and Yu, 2005; Jiang et al., 2007). Here, we report a novel mechanism by which NS prevents TIF (telomere dysfunction-induced foci) formation and telomere aberration in both ALT and telomerase-active (TA+) cells. NS does so by promoting the association between PML-IV and SUMOylated TRF1, which increases the telomeric recruitment of RAD51 proteins. We propose that continuously dividing cells may use NS as a protective mechanism to maintain their telomere integrity.
Results
Loss of NS triggers telomeric and nontelomeric DNA damage
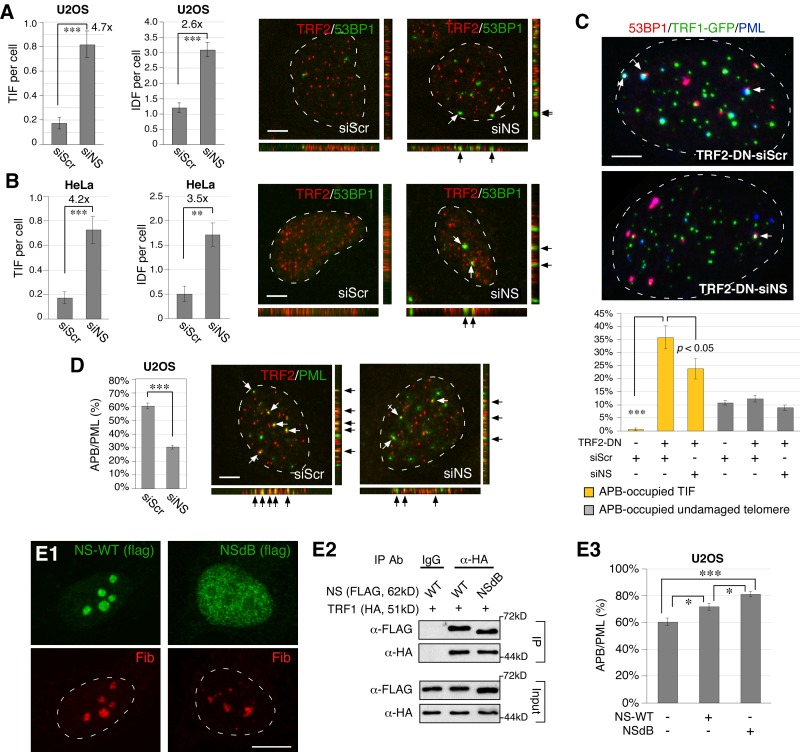
We first investigated the effect of NS depletion on the number of telomeric (TIF) and nontelomeric (IDF, interstitial damage foci) damage foci in ALT (U2OS) and TA+ (HeLa) cells. TIF (53BP1+TRF2+) and IDF (53BP1+TRF2−) were determined by 3D-reconstructed confocal analyses. Depletion of NS was achieved by the siRNA-mediated knockdown approach, which allowed a significant reduction of NS proteins in U2OS and HeLa cells (90% or more; Fig. S1, A and B). We found that knockdown of NS (NS-KD) by siNS induces a significant increase of TIF in both U2OS and HeLa cells (Fig. 1, A and B). NS-KD also increases the number of damage foci on interstitial chromosomes (IDF) in both U2OS and HeLa cells. The increase of IDF by NS-KD (2.6-fold and 3.5-fold) is less than that of TIF (4.7-fold and 4.2-fold) in either cell type.
Figure 1.
NS depletion triggers telomeric and nontelomeric DNA damage in ALT and TA+ cells, and decreases the formation of APB in ALT cells. (A) Damage on the telomere (TIF) and interstitial chromosome (IDF) was detected by the 53BP1+TRF2+ and 53BP1+TRF2− foci, respectively, on 3D-reconstructed confocal images. The bottom and right images represent the stacked images along the X-Z and Y-Z axis, respectively. NS knockdown (NS-KD) by siNS increases both TIF and IDF in U2OS (ALT) cells. (B) NS-KD also increases TIF and IDF in HeLa (TA+) cells. (C) Transfection of a TRF2-dominant mutant (TRF2-DN, also known as TRF2ΔBΔM) significantly increases the number of TIF and the percentage of TIF associated with APB (PML+53BP1+TRF1-GFP+ over 53BP1+TRF1-GFP+ foci). The TRF2-DN–induced increase of APB-occupied TIF is reduced by NS-KD (yellow bars). In contrast, the percentage of undamaged telomeres occupied by APB is hardly affected by TRF2-DN transfection or NS-KD (gray bars). (D) NS-KD reduces the formation of APB (TRF2+PML+) in U2OS cells. Y-axis indicates the percentage of APB among PML bodies (APB/PML). (E) A nucleoplasmic mutant of NS, NSdB, is distributed predominantly in the nucleoplasm (E1) but still retains the ability to bind TRF1 in coIP experiments (E2). Overexpression of wild-type NS or NSdB can both promote the APB formation in U2OS cells (E3). Fib, fibrillarin. *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Bars, 5 µm.
NS depletion decreases the percentage of TIF associated with APB and the formation of APB
To investigate the mechanism by which NS depletion triggers telomere damage, we chose to study its effect on APB formation, as APB was linked to the HR event (Grobelny et al., 2000) and NS was shown to interact with an essential component of APB, TRF1 (Zhu et al., 2006). First, we used triple-labeled confocal analyses to determine whether APBs are associated with TIF induced by TRF2ΔBΔM transfection in U2OS cells. TRF2ΔBΔM is a TRF2 mutant that triggers telomere damage by destabilizing the telomeric complex (van Steensel et al., 1998). We found that TRF2ΔBΔM dramatically increases the percentage of APB-occupied TIF (PML+53BP1+TRF1-gfp+ foci divided by 53BP1+TRF1-gfp+ foci) from 0.5% to 35.8% (P < 0.0001), but has no effect on the percentage of APB-occupied undamaged telomeres (10.7% vs. 12.3%; P = 0.28; Fig. 1 C). Importantly, knockdown of NS reduces the TRF2ΔBΔM-triggered increase of APB-occupied TIF from 35.8% to 23.7% (P < 0.05). These results support a link between APB and telomere damage in U2OS cells and suggest that NS may have a role in the formation of APB. Indeed, NS-KD significantly reduces the percentage of APB (PML+TRF2+) among PML bodies from 60.3% to 30.4% (P < 0.0001; Fig. 1 D), and NS overexpression shows the opposite effect (Fig. 1, E3). To determine whether the nucleolar localization of NS is required for its APB-regulatory function, we measured the effect of a nucleoplasmic mutant of NS, NSdB, which is deleted of its nucleolar localization signal (NoLS, aa 1–46) at the N terminus (Meng et al., 2006). NSdB is distributed predominantly in the nucleoplasm (Fig. 1, E1), but still retains the ability to bind TRF1 (Fig. 1, E2). Compared with the wild-type NS, NSdB shows a similar but stronger effect in promoting the formation of APB in U2OS cells (Fig. 1, E3), indicating that the telomere function of NS takes place outside the nucleolus. These results suggest that NS may prevent telomere damage by stimulating the formation of APB in ALT cells. Because NS perturbation does not change the number of PML bodies, TRF2 foci, or TRF1-gfp foci (Fig. S1, C and D), we reason that NS may increase APB by promoting the association between telomeres and PML bodies.
NS promotes PML body recruitment to SUMOylated TRF1
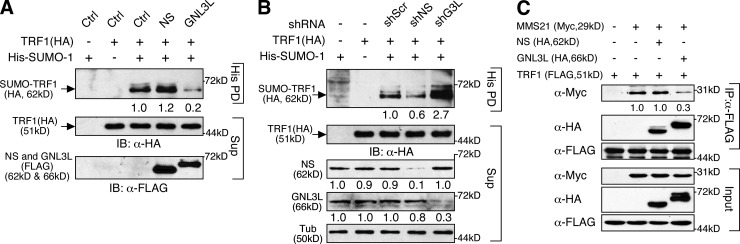
Because APB formation requires SUMOylation on TRF1 (Potts and Yu, 2007), we next determined whether NS regulates TRF1 SUMOylation. Our results showed that NS overexpression increases TRF1 SUMOylation and NS knockdown does the opposite (Fig. 2, A and B). Interestingly, GNL3L perturbation shows the opposite effects on TRF1 SUMOylation compared with NS perturbation. Because TRF1 SUMOylation is catalyzed by an E3 ligase, MMS21 (Potts and Yu, 2007), we examined how NS and GNL3L affect the interaction between TRF1 and MMS21. Coimmunoprecipitation (coIP) showed that the binding between TRF1 and MMS21 is reduced by GNL3L overexpression but not affected by NS overexpression (Fig. 2 C). As NS and GNL3L display opposite effects on APB formation and TRF1 SUMOylation, we tested whether GNL3L depletion causes DNA damage, and found that GNL3L knockdown has no effect on the formation of TIF or IDF in either ALT or TA+ cells (Fig. S2, A and B). These findings suggest that TRF1 SUMOylation by itself may not be a sufficient determinant to explain the TIF-regulatory activity of NS.
Figure 2.
TRF1 SUMOylation is increased by NS and reduced by GNL3L. (A) The amount of SUMOylated TRF1 in the pull-down fraction (His PD) is increased by NS overexpression and decreased by GNL3L overexpression. (B) Conversely, NS-KD (shNS) decreases TRF1 SUMOylation, whereas GNL3L knockdown (shG3L) shows the opposite effect. (C) The coIP efficiency of TRF1 and MMS21 is reduced by GNL3L overexpression but not affected by NS overexpression.
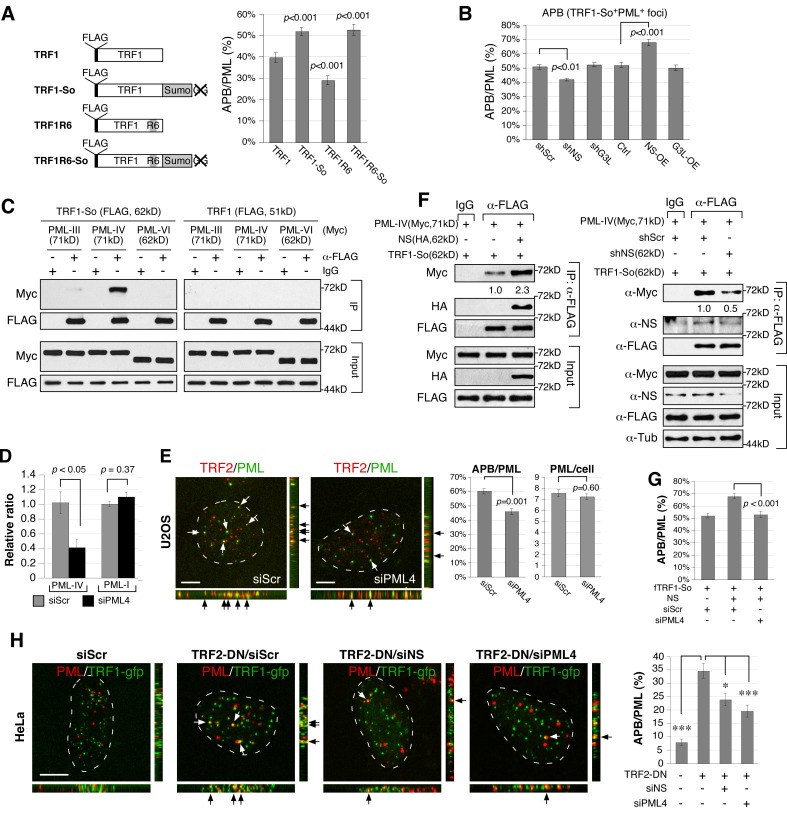
To explore the possibility that NS may regulate APB formation after the TRF1 SUMOylation step, we designed a C-terminally SUMO1-fused TRF1 construct (TRF1-So) to mimic the mono-SUMOylated TRF1 protein (Fig. 3 A, left). This design is based on a similar strategy used to study p53 monoubiquitination (Carter et al., 2007). The six C-terminal residues on the SUMO1 fusion of TRF1-So, including the di-Gly motif, were deleted to avoid direct conjugation of TRF1-So to off-target proteins. A SUMO-compromised mutant (TRF1R6) was also created by mutating the six SUMO-targeting Lys residues to Arg, as described in Potts and Yu (2007). TRF1-So and TRF1R6 display a higher (51.9%) and a lower (29.0%) percentage of APB among PML bodies (APB/PML) than does TRF1 (39.6%; P < 0.001), respectively, and may therefore mimic the SUMOylated and deSUMOylated states of TRF1 (Fig. 3 A, right). The APB/PML percentage of a compound mutant, TRF1R6-So, resembles that of TRF1-So, suggesting that the SUMO conjugate acts dominantly over the R6 mutant. We first confirmed that all mutant proteins colocalize with the endogenous TRF2 signal at the telomere (Fig. S3). NS knockdown reduces the APB/PML percentage of TRF1-So from 50.9 to 42.7% (P < 0.01), and NS overexpression does the opposite (67.9%; P < 0.001; Fig. 3 B). In contrast, GNL3L perturbation has no effect on the APB/PML percentage of TRF1-So. These results indicate that the APB- and TIF-regulatory activities of NS regulate the recruitment of PML bodies to SUMOylated TRF1, whereas the APB activity of GNL3L targets the SUMOylation event of TRF1.
Figure 3.
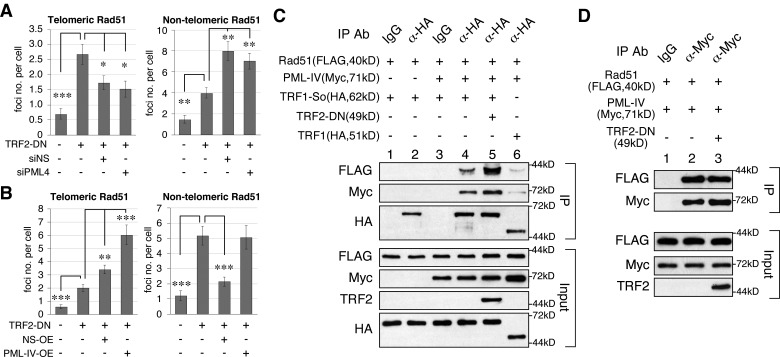
NS promotes the colocalization of PML bodies and SUMOylated TRF1 foci and the binding between PML-IV and SUMOylated TRF1 proteins. (A) A SUMO-conjugated TRF1 (TRF1-So) was created by fusing SUMO1 to the C terminus of TRF1 (left). The SUMO1 fusion was deleted of its six C-terminal residues, including the di-Gly motif indicated by the X-mark. SUMO-compromised TRF1 constructs were created by mutating six Lys residues to Arg on TRF1 (TRF1R6) or TRF1-So (TRF1R6-So). Respectively, TRF1-So and TRF1R6 mutants mimic the SUMOylated and deSUMOylated state of TRF1 in their abilities to form APB in U2OS cells (right). (B) The percentage of APB among PML bodies (APB/PML) of TRF1-So is decreased by NS-KD (shNS) and increased by NS overexpression (NS-OE), but not affected by GNL3L perturbation. (C) CoIP of FLAG-tagged TRF1 (or TRF1-So) and Myc-tagged PML (III, IV, or VI) reveals a specific interaction between PML-IV and TRF1-So. (D) Real-time RT-PCR confirms the knockdown efficiency of PML-IV–specific siRNA (siPML4) on the PML-IV transcript but not on the PML-I transcript in U2OS cells. (E) Knockdown of PML-IV decreases the APB/PML percentage of U2OS cells from 60.3% to 46.1% without changing the total number of PML bodies. (F) Knockdown and overexpression experiments showed that NS plays a role in promoting the binding between SUMOylated TRF1 and PML-IV. (G) PML-IV knockdown completely abolishes the NS-dependent increase of APB formation of SUMOylated TRF1. (H) TRF2-DN–triggered telomere damage induces APB formation in HeLa (TA+) cells. This increase is significantly attenuated by depletion of NS or PML-IV.
NS promotes the interaction between SUMOylated TRF1 and PML-IV
To determine the molecular event mediating the association between PML bodies and SUMOylated TRF1, we performed coIP of TRF1-So and different PML isoforms (Jensen et al., 2001). We found that TRF1-So interacts strongly with PML-IV (X63131) but not with PML-III (S50913) or PML-VI (M80185; Fig. 3 C, left). TRF1 itself shows a detectable but much weaker binding to PML-IV compared with TRF1-So (Fig. 3 C, right; and Fig. S4 A). A PML-IV-specific siRNA was synthesized that can knock down more than 60% of the PML-IV transcript without affecting PML-I, which shares the most sequence identity with PML-IV (Fig. 3 D). Supporting the role of PML-IV in APB regulation, PML-IV knockdown decreases the APB/PML percentage from 60.3 to 46.1% (P < 0.001) without changing the total number of PML bodies per cell (P = 0.60; Fig. 3 E). In addition, NS overexpression significantly increases the binding between TRF1-So and PML-IV (Fig. 3 F, left), and NS knockdown does the opposite (Fig. 3 F, right). For control experiments, we confirmed that NS interacts with TRF1 and TRF1-So equally well (Fig. S4 B) but not with PML-IV (Fig. S4 C), and excluded the possibility that NS and PML-IV bind TRF1-So through its C-terminal SUMO-1 fusion (Fig. S4 D). Furthermore, the NS ability to promote the association between SUMOylated TRF1 (TRF1-So) and PML bodies is completely abolished by knockdown of PML-IV (P < 0.001; Fig. 3 G). As APB is a hallmark of ALT cells, it is unclear whether this NS-regulated recruitment of PML bodies to the telomere occurs in TA+ cells. To address this issue, we examined the formation of APB in HeLa cells (Fig. 3 H). HeLa cells contain few APBs under normal conditions (7.9% of total PML), but show a significant increase of APBs under the TRF2ΔBΔM-induced telomere damage condition (34.6%; P < 0.0001). Notably, knockdown of NS or PML-IV reduces the TRF2ΔBΔM-triggered APB formation to 23.8 and 19.5%, respectively. These results demonstrate that NS promotes the telomeric recruitment of PML bodies by increasing the association between SUMOylated TRF1 and PML-IV in both ALT and TA+ cells.
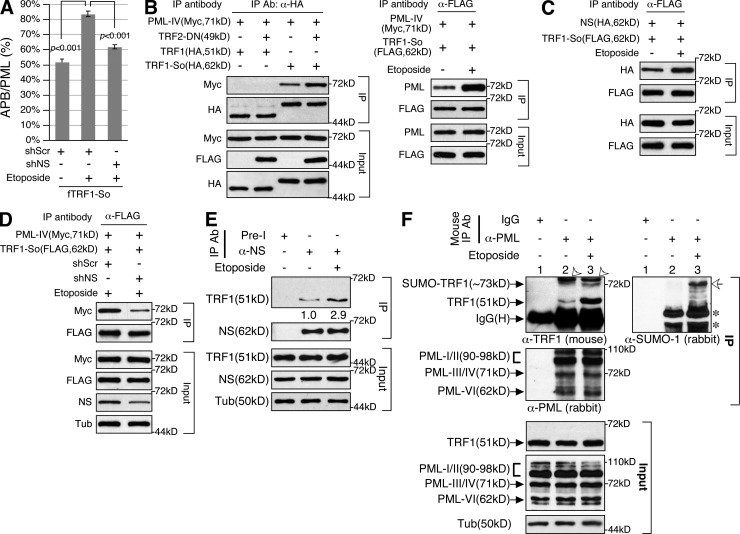
DNA damage increases the interaction between SUMOylated TRF1 and PML-IV in an NS-dependent manner
Because TA+ cells show APB formation only under the TRF2ΔBΔM-transfected condition, we asked whether DNA damage increases the recruitment of PML bodies to SUMOylated TRF1 foci in an NS-dependent manner in U2OS cells. DNA damage was induced by etoposide, which is a topoisomerase II inhibitor capable of inducing telomeric and nontelomeric DNA damage (Yoon et al., 1998; Zhang et al., 2003; Kang et al., 2004). We showed that etoposide can significantly increase the APB/PML percentage of TRF1-So from 51.9 to 83.6% (P < 0.001), and that NS-KD abolishes most of this effect (61.2%; Fig. 4 A). CoIP showed that both TRF2ΔBΔM-induced telomere damage and etoposide-induced DNA damage increase TRF1-So binding to PML-IV (Fig. 4 B), and that etoposide treatment increases the interaction between NS and TRF1-So (Fig. 4 C). Importantly, the increased binding between PML-IV and TRF1-So after etoposide treatment is abrogated by NS-KD (Fig. 4 D). To determine the interaction between native NS and TRF1 proteins under normal and DNA damage conditions, we performed endogenous coIP experiments in HeLa cells. The results confirmed that the endogenous NS and TRF1 proteins bind each other under the undamaged condition and that their interaction is increased by etoposide treatment (Fig. 4 E). Similarly, binding between endogenous TRF1 and PML proteins is significantly increased under the DNA damage condition (Fig. 4 F). More importantly, a high molecular weight product of TRF1 (∼73 kD) is co-purified with PML proteins under the DNA damage condition. This same product is recognized by the anti–SUMO-1 antibody and hence represents the SUMOylated form of TRF1. The ratio of SUMOylated-to-non-SUMOylated TRF1 proteins is significantly greater in the pull-down fraction than in the input lysate, indicating that PML binding enriches for the SUMOylated form of TRF1. These results support that DNA damage increases the interaction between SUMOylated TRF1 and PML proteins.
Figure 4.
NS mediates DNA damage–increased interaction between PML-IV and SUMOylated TRF1. (A) The APB/PML percentage of TRF1-So foci is increased by etoposide (Etop) treatment (from 51.9% to 83.6%). NS-KD abolishes most of the etoposide-induced APB increase (61.2%). (B) CoIP demonstrated that the interaction between PML-IV and TRF1-So is increased by TRF2-DN–induced telomere damage (left) or etoposide-induced DNA damage (right). (C) Etoposide treatment also increases TRF1-So binding to NS. (D) NS-KD (shNS) significantly reduces the etoposide-increased interaction between PML-IV and TRF1-So. (E) Endogenous coIP results confirmed that the native NS and TRF1 proteins bind each other under the undamaged condition. The interaction between these two proteins is increased by etoposide treatment. (F) Immunoprecipitation of endogenous PML proteins by mouse anti-PML antibody (PG-M3) coprecipitates only a minimal amount of TRF1 under the normal condition (lane 2). Under the DNA damage condition, a significant amount of endogenous TRF1 proteins is copurified with PML proteins (lane 3). A high molecular weight product of TRF1 is copurified with PML proteins only under the DNA damage condition. The same TRF1 product can be recognized by the anti–SUMO-1 antibody on the same blot (open arrows). Open arrowheads and asterisks mark IgG (H+L) and SUMO-modified proteins copurified with PML proteins, respectively.
Loss of PML-IV or NS increases the frequency of telomere damage and aberration
To demonstrate the importance of PML-IV in reducing telomere damage, we showed that loss of PML-IV in U2OS cells increases the number of TIF per cell by threefold (from 0.17 to 0.50; P < 0.01) and IDF by 2.4-fold (from 1.2 to 2.9; P < 0.01; Fig. 5 A). In HeLa cells, PML-IV depletion also increases the number of TIF (from 0.17 to 0.54; P < 0.01) and IDF (from 0.49 to 1.24; P < 0.001; Fig. 5 B). To establish the biological significance of NS and PML-IV in maintaining the telomere integrity, we measured their knockdown effects on telomere abnormalities in U2OS cells by telomere fluorescence in situ hybridization (Tel-FISH). Tel-FISH showed that loss of NS increases the frequency of chromosomes with low telomere signals (from 27.2 to 42.3%; P < 0.0001) and the frequency of chromosomal fusion (from 3.1 to 5.9%; P < 0.001; Fig. 5 C). More than 90% of the increased chromosomal fusion occurs between sister chromatids. Similarly, PML-IV depletion increases the frequency of chromosomes with low telomere signals (44.5%; P < 0.0001) but has no effect on the chromosomal fusion event. Low telomere signals indicate that the telomere length is decreased. Quantitative FISH (Q-FISH) analyses showed that the average telomere intensities of NS-KD and PML-KD U2OS cells are significantly lower than that of control cells (Fig. 5 D). These data demonstrate the effect of PML-IV knockdown in causing telomeric and nontelomeric DNA damage and the roles of NS and PML-IV in maintaining the telomere integrity.
Figure 5.
Loss of PML-IV or NS increases the frequency of telomere damage and abnormalities. Knockdown of PML-IV by siPML4 increases both TIF (53BP1+TRF2+) and IDF (53BP1+TRF2−) in U2OS (A) and in HeLa cells (B). (C) The effect of NS and PML-IV knockdown on the telomere integrity was determined by telomere fluorescence in situ hybridization (Tel-FISH), which showed that loss of NS increases the frequency of chromosomes with low telomere signals (Low-TS, arrows) from 27.2% to 42.3% and chromosomal fusion (asterisks) from 3.1% to 5.9%, but has no clear effect on the number of chromosomes with multi-telomeric signals (Multi-TS, M). PML-IV knockdown also increases the frequency of Low-TS chromosomes (44.5%) but does not change the frequency of fusion or Multi-TS event. (D) The telomere length distributions of control (gray), NS-knockdown (green), and PML-IV-knockdown (red) cells were analyzed by Q-FISH. X-axis represents the telomere signal intensity (in arbitrary units). Y-axis indicates the frequency of event. The average telomere intensity (mean) and number of telomeres analyzed (n) were shown. Asterisks in A–C represent P values (see Fig. 1). Bars: (A and B) 5 µm; (C, large panels) 10 µm; (C, small panels) 5 µm.
NS and PML-IV promotes the telomeric recruitment of RAD51 induced by telomere damage
RAD51 is the core protein in the HR-based repair of damaged telomeres (Kibe et al., 2003; Carneiro et al., 2010). To determine whether NS or PML-IV regulates the recruitment of RAD51 to the telomere under the telomere damage condition, U2OS cells were transfected with TRF2ΔBΔM and measured for the colocalized RAD51 and TRF1-GFP signals. The results showed that the number of telomere-associated RAD51 foci is greatly increased by TRF2ΔBΔM-induced telomere damage (from 0.7 to 2.7 per cell; P < 0.0001), and that knockdown of NS or PML-IV reduces the frequency of this event (1.7 and 1.5 per cell; P < 0.01; Fig. 6 A, left). Conversely, overexpression of NS or PML-IV under the telomere damage condition increases the telomere recruitment of RAD51 from 2.7 to 3.4 (P < 0.001) and 6.0 per cell (P < 0.0001), respectively (Fig. 6 B, left). As NS knockdown increases nontelomeric RAD51 foci (P < 0.001; Fig. 6 A, right) and NS overexpression reduces them (P < 0.0001; Fig. 6 B, right), we reason that the NS function is to promote the telomeric recruitment of RAD51. On the other hand, overexpression of PML-IV increases telomeric RAD51 foci without reducing the number of nontelomeric RAD51 foci (Fig. 6 B), which suggests that overexpression of PML-IV may increase the formation of RAD51 foci. We confirmed this idea by showing that overexpression of PML-IV alone significantly increases the formation of RAD51 foci without triggering DNA damage, and that all RAD51 foci colocalize with PML-IV signals (Fig. S5), which suggests that PML-IV may directly interact with RAD51.
Figure 6.
NS and PML-IV promote telomeric recruitment of RAD51 under the telomere damage condition. (A) Telomeric recruitment of RAD51 was determined by confocal colocalization of TRF1-GFP and endogenous RAD51 foci. The number of telomere-associated RAD51 foci is increased by TRF2-DN transfection (from 0.7 to 2.7 per cell). Knockdown of NS or PML-IV significantly reduces the telomeric recruitment of RAD51 induced by TRF2-DN transfection (1.7 and 1.5 per cell, respectively). Both NS and PML-IV knockdown increase the number of nontelomeric RAD51 foci. Asterisks represent P values (see Fig. 1). (B) Overexpression of NS or PML-IV increases the number of telomere-associated RAD51 foci. Non-telomeric RAD51 foci are decreased by NS overexpression but unchanged by PML-IV overexpression. (C) CoIP confirms that PML-IV mediates the interaction between RAD51 and TRF1-So (lanes 1–4) and that this interaction is enhanced by TRF2-DN–induced telomere damage (lane 5). By comparison, PML-IV shows a much weaker effect in mediating the binding between RAD51 and TRF1 (lane 6). (D) PML-IV binds RAD51 directly and independently of telomere damage.
The interaction between RAD51 and TRF1-So or PML-IV was further investigated by coIP experiments. Regular coIP showed a direct interaction between PML-IV and RAD51 but a very weak binding between TRF1-So and RAD51, with or without PML-IV (unpublished data). The lack of binding between TRF1-So and RAD51 in regular coIP suggests that this event may be transient and highly regulated. Therefore, we used the cross-link coIP approach to delineate the binding relationship between RAD51 and TRF1-So. Cross-link coIP showed that PML-IV is required for the specific interaction between RAD51 (FLAG) and TRF1-So (HA), and that this interaction is increased by TRF2ΔBΔM-induced telomere damage (Fig. 6 C, lanes 1–5). By contrast, PML-IV binds RAD51 directly and independently of telomere damage (Fig. 6 D). These results support that NS promotes the association between RAD51-bound PML-IV and SUMOylated TRF1, and that this mechanism is activated by telomere damage.
NS and PML-IV protect ALT and TA+ cells from telomere damage
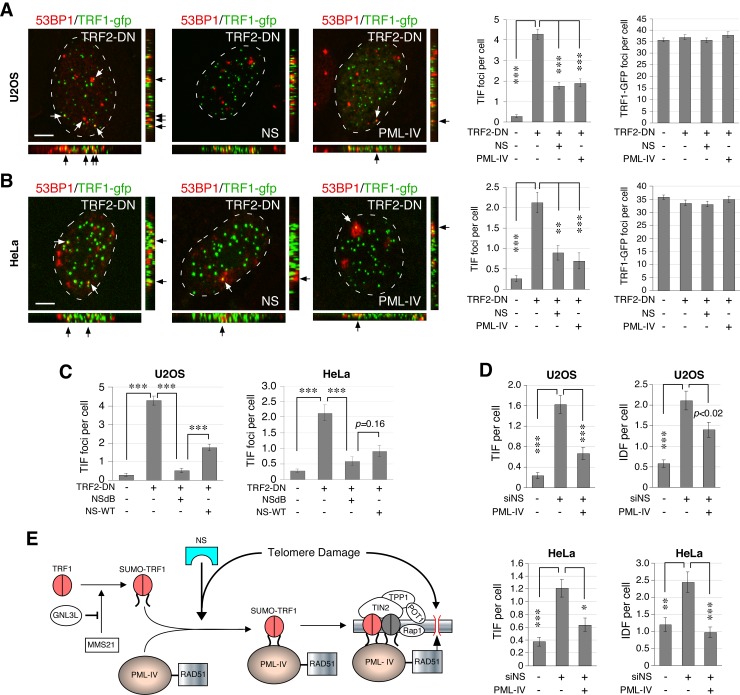
To demonstrate the function of NS and PML-IV in protecting against telomere damage, we measured the effect of NS or PML-IV overexpression on the formation of TRF2ΔBΔM-induced TIF. First, we confirmed that TRF2ΔBΔM transfection significantly increases the amount of TIF in both U2OS (Fig. 7 A) and HeLa cells (Fig. 7 B). In TRF2ΔBΔM-transfected U2OS cells, overexpression of NS reduces the number of TIF from 4.3 to 1.8 per cell (P < 0.0001; Fig. 7 A). Overexpression of PML-IV in TRF2ΔBΔM-transfected U2OS cells shows the same effect (TIF = 1.9 per cell; P < 0.0001). In TRF2ΔBΔM-transfected HeLa cells, overexpression of NS or PML-IV decreases the number of TIF from 2.1 to 0.9 and 0.7 per cell, respectively (P < 0.001; Fig. 7 B). Notably, the nucleoplasmic mutant of NS, NSdB, is also capable of protecting against TRF2ΔBΔM-induced telomere damage in U2OS and HeLa cells (Fig. 7 C). This activity of NSdB is even stronger than that of wild-type NS in U2OS cells. To determine whether PML-IV is also involved in the nontelomeric DNA damage induced by NS-KD, we used PML-IV to rescue the DNA damage phenotype of NS-KD cells (Fig. 7 D). Our results showed that PML-IV is able to partially rescue the telomeric and nontelomeric DNA damage induced by NS-KD in both TA+ and ALT cells. These findings indicate that the NS-regulated TRF1–PML-IV pathway protects both TA+ and ALT cells from telomere damage.
Figure 7.
NS and PML-IV protect both ALT and TA+ cells from TRF2-DN–induced telomere damage. Overexpression of NS or PML-IV protects U2OS cells (A) and HeLa cells (B) from TRF2-DN–induced TIF without affecting the total number of TRF1-GFP foci. (C) Compared with wild-type NS (NS-WT), the nucleoplasmic mutant of NS (NSdB) shows a stronger or similar activity in reducing TRF2-DN–induced TIF in U2OS and HeLa cells, respectively. (D) PML-IV can rescue the NS-KD–induced telomere (TIF) and nontelomeric (IDF) damage in U2OS and HeLa cells. (E) A proposed model for the NS function in protecting ALT and TA+ cells from telomere damage via the recruitment of RAD51-bound PML-IV to SUMOylated TRF1. Bars, 5 µm.
Discussion
NS protects ALT and TA+ cells from telomere damage
In this study, we report the NS function in maintaining the integrity of telomeric and interstitial chromosomes and determine the mechanism underlying the telomere-protecting activity of NS. Even though loss of NS triggers DNA damage on both telomeric and interstitial chromosomes, the NS-KD effect is more notable on the telomere than on the nontelomeric chromosome. The NS role in preventing TIF formation is supported by the findings that NS knockdown triggers spontaneous telomere damage, NS overexpression protects against TRF2ΔBΔM-induced telomere damage, NS depletion reduces the telomere length, and loss of NS increases the frequency of chromosomes with low telomere signals and sister chromatid fusion. Because NS is most abundantly expressed in cancer and stem cells, such a mechanism may allow these cells to quickly respond to and repair telomere damage in order to maintain their self-renewing proliferation. Telomere elongation in embryonic and adult stem cells is normally attributed to the telomerase activity, which alone may not be sufficient over time. Our discovery suggests the possibility that self-renewing cells may be equipped with the NS machinery to safeguard their telomere integrity during the aging process.
NS promotes the association of SUMOylated TRF1 and PML-IV
Our mechanistic investigation on the TIF-regulatory activity of NS was based on its ability to bind TRF1 and increase APB formation. APB is a known feature of ALT cells. It was postulated that APB may serve the function of sequestering low molecular weight telomeric DNAs, as APB contains linear extrachromosomal telomere repeat DNA (Fasching et al., 2007). Others suggested that APB may be linked to the HR function based on its content of DNA damage response and repair proteins and its DNA synthesis activity (Yeager et al., 1999; Wu et al., 2000, 2003; Nabetani et al., 2004). We found that TRF2ΔBΔM-induced telomere damage increases the percentage of APB-occupied TIF but not the percentage of APB-occupied undamaged telomeres, and that NS knockdown reduces the percentage of APB-occupied TIF. These findings suggest a link between APB and telomere damage and a role of NS in APB formation. Indeed, NS can increase APB by promoting the association between telomeres and PML bodies. To define the molecular interaction between telomeres and PML bodies, we discovered a specific binding between PML-IV and SUMOylated TRF1, which can be increased by NS and DNA damage signals. Because PML-IV interacts much more strongly with SUMO-modified TRF1 than with wild-type TRF1 and does not bind NS directly, we propose that NS binding and SUMOylation of TRF1 may trigger a conformational change that favors its association with PML-IV, which then brings PML-IV–tethered proteins (e.g., RAD51) to the telomere under damage conditions (Fig. 7 E). The activity of NS to promote APB formation in U2OS cells or to protect against TRF2ΔBΔM-induced telomere damage in U2OS or HeLa cells does not require its nucleolar localization. In fact, the NSdB mutant performs even more efficiently than the wild-type NS in promoting APB or reducing TIF in U2OS cells, indicating that the telomere function of NS takes place outside the nucleolus. In conjunction with our previous results showing that the NS protein is not recruited to the telomere (Meng et al., 2011), we propose that the NS-mediated regulation on the interaction between TRF1 and PML occurs in the nucleoplasm and extra-chromosomally before they are incorporated into stable large protein complexes on the telomere.
The role of PML-IV in mediating the telomere-protecting function of NS is supported by the knockdown experiments, which show that loss of PML-IV produces the same TIF phenotypes as NS-KD does, and that overexpression of PML-IV or NS can protect against TRF2ΔBΔM-induced telomere damage. In addition, PML-IV is required for the NS-induced association of PML bodies and SUMOylated TRF1 foci, as well as the telomeric recruitment of RAD51 under the telomere damage condition. More importantly, overexpression of PML-IV is able to rescue the telomere damage phenotype of NS-KD in both TA+ and ALT cells. Whereas loss of NS or PML-IV both show increased numbers of chromosomes with low telomere signals, only NS-KD increases the frequency of chromosomal fusion, which raises the possibility that PML-IV may not be the sole mediator for this NS activity. Based on the lack of TRF1 binding by PML-VI and a previous finding that narrows down the TRF1-interactive domain of PML-IV to its C-terminal 154 residues (Yu et al., 2010), one may deduce that TRF1 binding involves the C-terminal 81 residues of PML-IV. Because 84% of this region is found in PML-I, it is possible that PML-I may also interact with SUMOylated TRF1 and play a redundant or complementary role as compared with PML-IV.
The ALT-related activity of NS in TA+ cells and on interstitial chromosomes
As NS knockdown triggers spontaneous DNA damage not only on the telomere of ALT cells but also on the telomere of TA+ cells and on the interstitial chromosome of both TA+ and ALT cells, it raises the question of whether the proposed mechanism of NS (i.e., the telomeric recruitment of PML-IV) works in TA+ cells or on nontelomeric chromosomes. Two lines of evidence support that this mechanism is also in use in TA+ cells. First, in both ALT and TA+ cells, loss of PML-IV induces telomere damage and overexpression of PML-IV rescues the telomere damage effect of NS-KD. Second, APB can be induced by TRF2ΔBΔM-triggered telomere damage in HeLa (TA+) cells, and knockdown of NS or PML-IV significantly decreases the frequency of this event. On the issue of telomeric versus nontelomeric damage, the knockdown and rescue results, too, support the involvement of PML-IV in alleviating nontelomeric DNA damage.
NS and GNL3L differentially regulate APB and TIF
Our data demonstrate that NS and GNL3L exert opposite effects on TRF1 SUMOylation. Given that NS and GNL3L compete against each other for TRF1 binding (Meng et al., 2011), this TRF1 SUMOylation effect of NS may be explained by its ability to displace TRF1-bound GNL3L. On the other hand, only NS but not GNL3L can regulate the association between PML and SUMOylated TRF1. It is noted that even though GNL3L shows the opposite effect on APB regulation (Zhu et al., 2009) and TRF1 SUMOylation compared with NS, it does not exhibit the activity to regulate TIF formation as does NS. Therefore, TRF1 SUMOylation alone may not be sufficient to explain the TIF-regulatory function of NS. Furthermore, the differential activities of NS and GNL3L in APB and TIF regulation also suggest that APB may be heterogeneous. Not all of them are connected to telomere repair. Evolutionarily, NS and GNL3L share the same invertebrate orthologue, GNL3 (Tsai and Meng, 2009). The functional divergence of NS and GNL3L may signify an expansion in the telomere-regulatory modality during vertebrate evolution. Whereas NS extends the proliferative lifespan by providing damaged telomeres a better access to repair proteins, GNL3L may serve the role of stabilizing the telomere structure in differentiated cells.
Materials and methods
cDNA constructs and antibodies
Point mutation and fusion of TRF1 were introduced by the stitching PCR strategy (Tsai and McKay, 2005). TRF2ΔBΔM includes amino acids 45–454. Epitope-tagged expression constructs of PML isoforms were made by cloning cDNA fragments PCR amplified from the original PML constructs into the Myc- or FLAG-tagged vector. Primary antibodies include anti-HA (HA.11; Covance), Myc (9E10; Covance), FLAG (M2; Sigma-Aldrich), TRF1 (TRF-78; Santa Cruz Biotechnology, Inc.), TRF2 (4A794; EMD), PML (PG-M3, H-238; Santa Cruz Biotechnology, Inc.), 53BP1 (4937; Cell Signaling Technology), RAD51 (51RAD01; Thermo Fisher Scientific), SUMO-1 (FL-101; Santa Cruz Biotechnology, Inc.), and NS (Ab2438 and Ab138). Secondary antibodies are conjugated to Rhodamine-X, FITC, or peroxidase.
siRNA knockdown
Knockdown was done by transfecting cells with shRNAmir constructs or siRNA duplexes using the Lipofectamine Plus or Oligofectamine reagent, respectively. The sequences targeted by the control (siScr), NS-specific (siNS), GNL3L-specific (siG3L), and siPML-IV-specific (siPML4) siRNA duplexes, as well as the control (shScr), NS-specific (shNS), and GNL3L-specific (shG3L) shRNAmir are as follows. siScr, 5′-TGACGATCAGAATGCGACT-3′; siNS, 5′-GAACTAAAACAGCAGCAGA-3′; siG3L, 5′-AAACGCAGGACCATTGAGA-3′; siPML4, 5′-TGACAATGAAAGTGGGTTC-3′; shScr, 5′-TCTCGCTTGGGCGAGAGTAAG-3′; shNS, 5′-CCTGATATTAAGCCATCAAAT-3′; shG3L, 5′-CCAATCGAGAGGCTGAATTAA-3′.
Immunofluorescence and confocal quantification of TIF, APB, and RAD51 foci
TIF were determined by the colocalization of 53BP1 and TRF2 (or TRF1-GFP) signals. The number of APBs was scored on the basis of colocalized PML and TRF2 (TRF1-GFP, or FLAG-tagged TRF1) signals. To detect RAD51 foci, cells were incubated with the permeabilization solution (10 mM Hepes, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton) for 2 min on ice before the fixation step. Images were acquired on a confocal microscope (LSM510; Carl Zeiss) using a 63x Plan Apochromat oil objective (1.4 NA) at room temperature. Scanning was set with a 512 x 512 frame size, 3x zoom, and <1.0-µm optical thickness. Stacked images of 80–100 randomly chosen cells were collected at 0.5-µm intervals from five independent experiments and analyzed using ImageJ 1.36b software (National Institutes of Health, Bethesda, MD).
In vivo SUMOylation assay
His-tagged SUMO-1 and TRF1 expression plasmids were coexpressed in HEK293 cells. 2 d later, protein lysates were extracted in 6M guanidinium buffer, pulled down by Ni2+-chelating Sepharose in 3 h, and washed carefully to prevent degradation of SUMOylated products.
Coimmunoprecipitation
For regular coIP experiments, protein lysates were extracted in NTEN buffer without cross-linking. For coIP of TRF1-So and RAD51, protein complexes were cross-linked with 1% formaldehyde in PBS for 20 min at 4°C and incubated with 125 mM glycine in PBS before the extraction step. Protein extracts were precleared by microcentrifugation and incubated with primary antibody and protein G–Sepharose (GE Healthcare) for 4 h at 4°C. Immunoprecipitates were washed extensively before SDS-PAGE. For endogenous coIP experiments, the NS and PML protein complexes were immunoprecipitated in HeLa cells by anti-NS (Ab138) or anti-PML (PG-M3) antibody, respectively. Precipitated NS and PML proteins were immunodetected by the Ab2438 or H-238 antibody, respectively.
Telomere fluorescence in situ hybridization (T-FISH) and quantitative-FISH (Q-FISH)
Cells were treated with colcemid (0.1 µg/ml) for 16 h, incubated in hypotonic solution (0.8% sodium citrate), and fixed in methanol/acetic acid (3:1) before spreading onto slides. The T-FISH procedure was conducted by incubating metaphase-spread chromosomes with a Cy3-conjugated telomere-specific peptide nucleic acid (PNA) probe, (C3TA2)2, in hybridization buffer (70% formamide, 10 mM Tris, pH 7.2) for 3 h at 25°C and counterstaining the chromosomes with TO-PRO-3 dyes. High-resolution stacked images were taken on the LSM510 confocal imaging platform (Carl Zeiss) using the 63x Plan Apochromat oil objective, and quantified for the frequency of telomere aberration on each metaphase-spread chromosome. The relative telomere length was measured by Q-FISH, where individual telomere signal intensities of T-FISH were scored using the ImageJ telomeric plug-in program.
Online supplemental material
Fig. S1 shows experiments for the siNS-mediated knockdown efficiency and the effect of NS perturbation on TRF2 foci and PML bodies. Fig. S2 shows that loss of GNL3L does not predispose ALT or TA+ cells to telomere or DNA damage. Fig. S3 shows that GFP- and FLAG-tagged TRF1 proteins colocalize with the endogenous TRF2. Fig. S4 shows control experiments for the binding specificity between TRF1, PML-IV, NS, and/or GNL3L. Fig. S5 shows that overexpression of PML-IV promotes Rad51 foci formation without increasing DNA damage. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201109038/DC1.
Supplementary Material
Acknowledgments
Our gratitude goes to the following investigators for their generous gifts of cDNA constructs: Dr. Zhou Songyang (Baylor College of Medicine, Houston, TX) for TRF2ΔBΔM; Dr. Hongtao Yu (UT Southwestern, Dallas, TX) for MMS21; Dr. Kun-Sang Chang (MD Anderson Cancer Center, Houston, TX) for PML-III and PML-IV; Dr. Edward Yeh (MD Anderson Cancer Center) for Rad51. We also thank Lingjun Meng for his help on western blot and Hsi-Wen Tsai for his unwavering support.
Footnotes
Abbreviations used in this paper:
- ALT
- alternative lengthening of telomeres
- APB
- ALT-associated PML body
- coIP
- coimmunoprecipitation
- GNL3L
- guanine nucleotide-binding protein-like 3-like
- HR
- homologous recombination
- IDF
- interstitial damage foci
- NS
- nucleostemin
- PML
- promyelocytic leukemia
- TA+
- telomerase active
- TIF
- telomere dysfunction-induced foci
- TRF
- telomeric repeat-binding factor
References
- Baddoo M., Hill K., Wilkinson R., Gaupp D., Hughes C., Kopen G.C., Phinney D.G. 2003. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J. Cell. Biochem. 89:1235–1249 10.1002/jcb.10594 [DOI] [PubMed] [Google Scholar]
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J.C. 2006. Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 26:9291–9301 10.1128/MCB.01183-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14:4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271–1274 10.1038/nm1197-1271 [DOI] [PubMed] [Google Scholar]
- Carneiro T., Khair L., Reis C.C., Borges V., Moser B.A., Nakamura T.M., Ferreira M.G. 2010. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature. 467:228–232 10.1038/nature09353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K.H. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9:428–435 10.1038/ncb1562 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19:2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- Dunham M.A., Neumann A.A., Fasching C.L., Reddel R.R. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447–450 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- Fasching C.L., Neumann A.A., Muntoni A., Yeager T.R., Reddel R.R. 2007. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 67:7072–7077 10.1158/0008-5472.CAN-07-1556 [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 43:405–413 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 337:331–337 10.1038/337331a0 [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H. 1996. Telomeres, telomerase and cancer. Sci. Am. 274:92–97 10.1038/scientificamerican0296-92 [DOI] [PubMed] [Google Scholar]
- Grobelny J.V., Godwin A.K., Broccoli D. 2000. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J. Cell Sci. 113:4577–4585 [DOI] [PubMed] [Google Scholar]
- Jensen K., Shiels C., Freemont P.S. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 20:7223–7233 10.1038/sj.onc.1204765 [DOI] [PubMed] [Google Scholar]
- Jiang W.Q., Zhong Z.H., Henson J.D., Reddel R.R. 2007. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene. 26:4635–4647 10.1038/sj.onc.1210260 [DOI] [PubMed] [Google Scholar]
- Kang M.R., Muller M.T., Chung I.K. 2004. Telomeric DNA damage by topoisomerase I. A possible mechanism for cell killing by camptothecin. J. Biol. Chem. 279:12535–12541 10.1074/jbc.M309779200 [DOI] [PubMed] [Google Scholar]
- Kibe T., Tomita K., Matsuura A., Izawa D., Kodaira T., Ushimaru T., Uritani M., Ueno M. 2003. Fission yeast Rhp51 is required for the maintenance of telomere structure in the absence of the Ku heterodimer. Nucleic Acids Res. 31:5054–5063 10.1093/nar/gkg718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Meng L., Li Y., Tsai R.Y. 2010. Tumor-initiating function of nucleostemin-enriched mammary tumor cells. Cancer Res. 70:9444–9452 10.1158/0008-5472.CAN-10-2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.J., Cai Z.W., Liu Y.J., Dong M.Y., Sun L.Q., Hu G.F., Wei Y.Y., Lao W.D. 2004. Role of nucleostemin in growth regulation of gastric cancer, liver cancer and other malignancies. World J. Gastroenterol. 10:1246–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Bailey S.M., Okuka M., Muñoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A., et al. 2007. Telomere lengthening early in development. Nat. Cell Biol. 9:1436–1441 10.1038/ncb1664 [DOI] [PubMed] [Google Scholar]
- Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J.M., Fernández-Capetillo O., Tarsounas M., Blasco M.A. 2009. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23:2060–2075 10.1101/gad.543509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Yasumoto H., Tsai R.Y. 2006. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 119:5124–5136 10.1242/jcs.03292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Hsu J.K., Zhu Q., Lin T., Tsai R.Y. 2011. Nucleostemin inhibits TRF1 dimerization and shortens its dynamic association with the telomere. J. Cell Sci. 124:3706–3714 10.1242/jcs.089672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Yokoyama O., Ishikawa F. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J. Biol. Chem. 279:25849–25857 10.1074/jbc.M312652200 [DOI] [PubMed] [Google Scholar]
- Nomura J., Maruyama M., Katano M., Kato H., Zhang J., Masui S., Mizuno Y., Okazaki Y., Nishimoto M., Okuda A. 2009. Differential requirement for nucleostemin in embryonic stem cell and neural stem cell viability. Stem Cells. 27:1066–1076 10.1002/stem.44 [DOI] [PubMed] [Google Scholar]
- Ohmura M., Naka K., Hoshii T., Muraguchi T., Shugo H., Tamase A., Uema N., Ooshio T., Arai F., Takubo K., et al. 2008. Identification of stem cells during prepubertal spermatogenesis via monitoring of nucleostemin promoter activity. Stem Cells. 26:3237–3246 10.1634/stemcells.2008-0506 [DOI] [PubMed] [Google Scholar]
- Potts P.R., Yu H. 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25:7021–7032 10.1128/MCB.25.16.7021-7032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P.R., Yu H. 2007. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 14:581–590 10.1038/nsmb1259 [DOI] [PubMed] [Google Scholar]
- Shay J.W., Bacchetti S. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer. 33:787–791 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
- Shay J.W., Zou Y., Hiyama E., Wright W.E. 2001. Telomerase and cancer. Hum. Mol. Genet. 10:677–685 10.1093/hmg/10.7.677 [DOI] [PubMed] [Google Scholar]
- Songyang Z., Liu D. 2006. Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit. Rev. Eukaryot. Gene Expr. 16:103–118 [DOI] [PubMed] [Google Scholar]
- Tsai R.Y. 2009. Nucleolar modulation of TRF1: a dynamic way to regulate telomere and cell cycle by nucleostemin and GNL3L. Cell Cycle. 8:2912–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991–3003 10.1101/gad.55671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168:179–184 10.1083/jcb.200409053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., Meng L. 2009. Nucleostemin: a latecomer with new tricks. Int. J. Biochem. Cell Biol. 41:2122–2124 10.1016/j.biocel.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature. 385:740–743 10.1038/385740a0 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell. 92:401–413 10.1016/S0092-8674(00)80932-0 [DOI] [PubMed] [Google Scholar]
- Wu G., Lee W.H., Chen P.L. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275:30618–30622 10.1074/jbc.C000390200 [DOI] [PubMed] [Google Scholar]
- Wu G., Jiang X., Lee W.H., Chen P.L. 2003. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen Breakage Syndrome 1. Cancer Res. 63:2589–2595 [PubMed] [Google Scholar]
- Yeager T.R., Neumann A.A., Englezou A., Huschtscha L.I., Noble J.R., Reddel R.R. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175–4179 [PubMed] [Google Scholar]
- Yoon H.J., Choi I.Y., Kang M.R., Kim S.S., Muller M.T., Spitzner J.R., Chung I.K. 1998. DNA topoisomerase II cleavage of telomeres in vitro and in vivo. Biochim. Biophys. Acta. 1395:110–120 [DOI] [PubMed] [Google Scholar]
- Yu J., Lan J., Wang C., Wu Q., Zhu Y., Lai X., Sun J., Jin C., Huang H. 2010. PML3 interacts with TRF1 and is essential for ALT-associated PML bodies assembly in U2OS cells. Cancer Lett. 291:177–186 10.1016/j.canlet.2009.10.009 [DOI] [PubMed] [Google Scholar]
- Zhang P., Chan S.L., Fu W., Mendoza M., Mattson M.P. 2003. TERT suppresses apoptotis at a premitochondrial step by a mechanism requiring reverse transcriptase activity and 14-3-3 protein-binding ability. FASEB J. 17:767–769 [DOI] [PubMed] [Google Scholar]
- Zhou X.Z., Perrem K., Lu K.P. 2003. Role of Pin2/TRF1 in telomere maintenance and cell cycle control. J. Cell. Biochem. 89:19–37 10.1002/jcb.10496 [DOI] [PubMed] [Google Scholar]
- Zhu X.D., Küster B., Mann M., Petrini J.H., de Lange T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347–352 10.1038/77139 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R.Y. 2006. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 26:9279–9290 10.1128/MCB.00724-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Meng L., Hsu J.K., Lin T., Teishima J., Tsai R.Y. 2009. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 185:827–839 10.1083/jcb.200812121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.