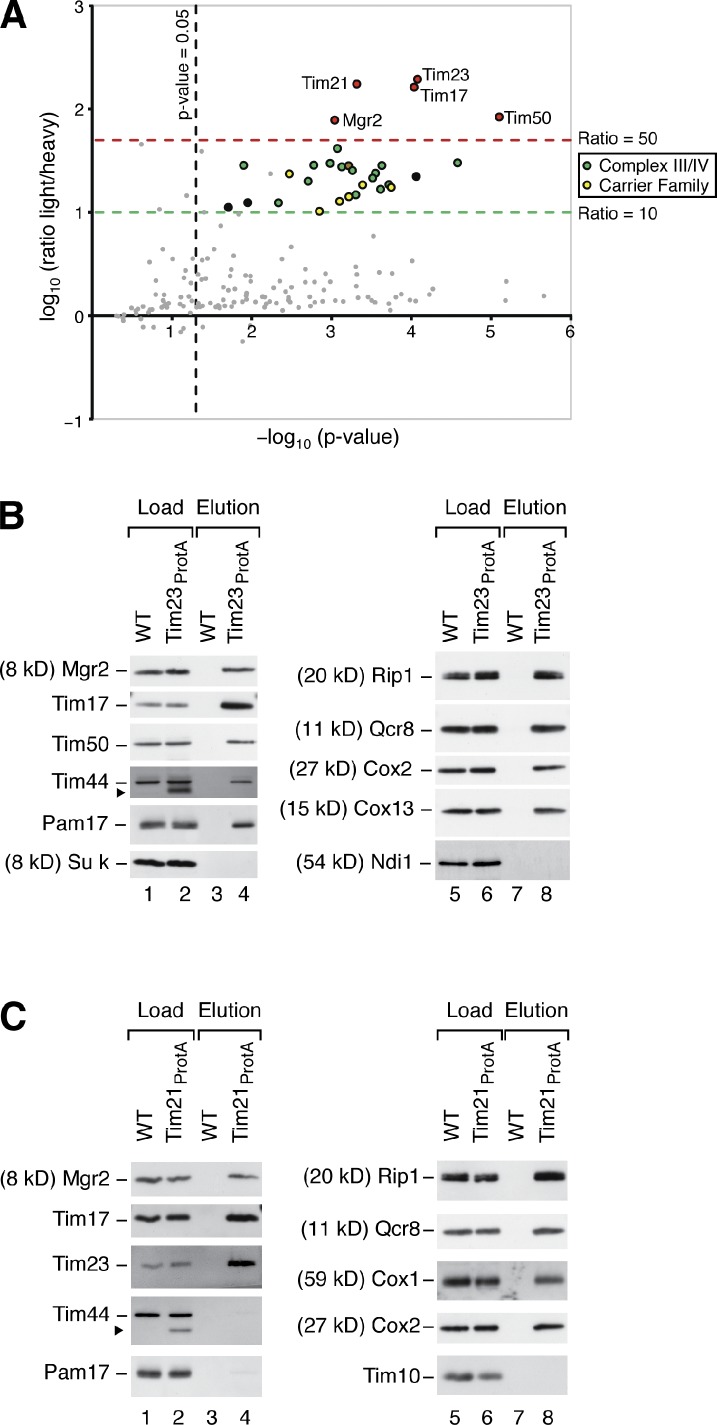
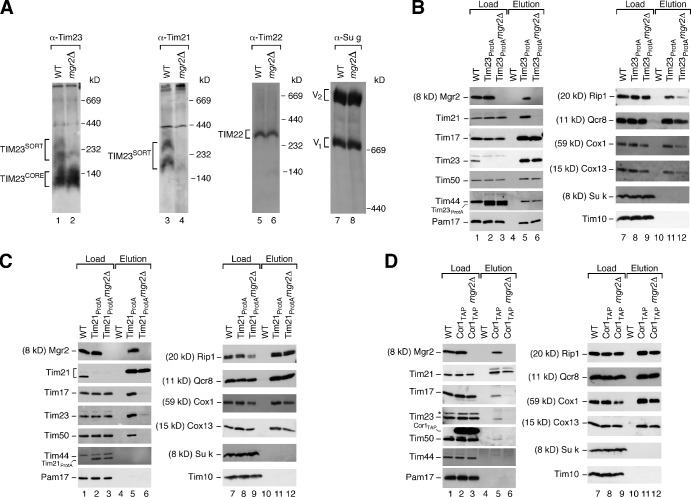
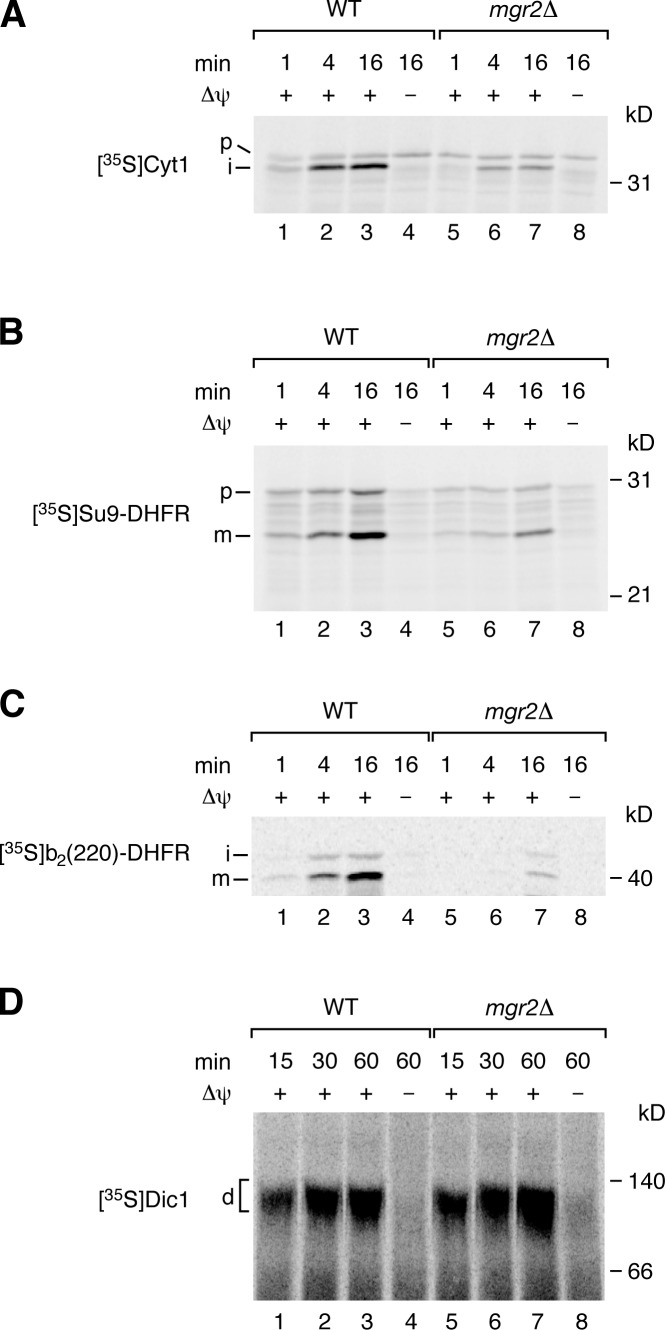
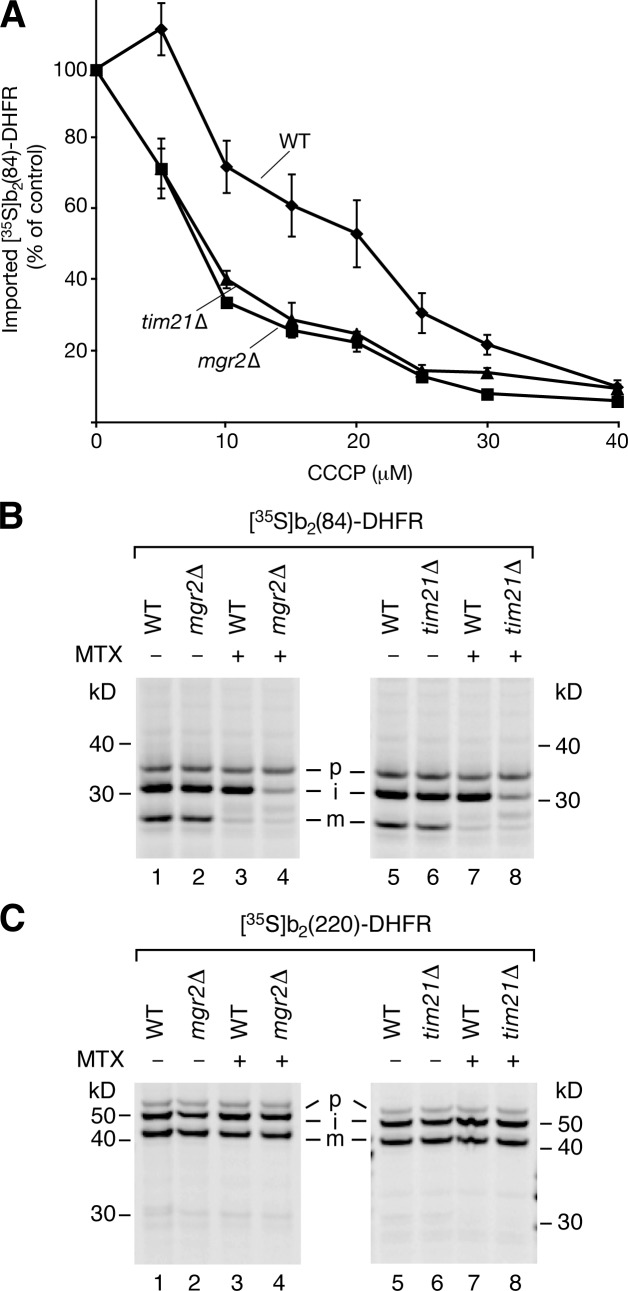
Mgr2 is a new component of the mitochondrial presequence translocase required for efficient coupling of the TIM23 core complex to Tim21, respiratory chain complexes, and the translocase of the outer mitochondrial membrane.
Abstract
Many mitochondrial proteins are synthesized with N-terminal presequences in the cytosol. The presequence translocase of the inner mitochondrial membrane (TIM23) translocates preproteins into and across the membrane and associates with the matrix-localized import motor. The TIM23 complex consists of three core components and Tim21, which interacts with the translocase of the outer membrane (TOM) and the respiratory chain. We have identified a new subunit of the TIM23 complex, the inner membrane protein Mgr2. Mitochondria lacking Mgr2 were deficient in the Tim21-containing sorting form of the TIM23 complex. Mgr2 was required for binding of Tim21 to TIM23CORE, revealing a binding chain of TIM23CORE-Mgr2/Tim21–respiratory chain. Mgr2-deficient yeast cells were defective in growth at elevated temperature, and the mitochondria were impaired in TOM-TIM23 coupling and the import of presequence-carrying preproteins. We conclude that Mgr2 is a coupling factor of the presequence translocase crucial for cell growth at elevated temperature and for efficient protein import.
Introduction
Most mitochondrial proteins are nuclear encoded and produced in the cytosol as precursor proteins (Koehler, 2004; Dolezal et al., 2006; Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo et al., 2011). The most frequently found mitochondrial import signals are N-terminal presequences (Neupert and Herrmann, 2007; Chacinska et al., 2009; Vögtle et al., 2009). Presequence-carrying preproteins are recognized by receptors on the mitochondrial surface and transported by the general translocase of the outer membrane (TOM complex) and the presequence translocase of the inner membrane (TIM23 complex; Brix et al., 1999; Abe et al., 2000; Chacinska et al., 2005; Popov-Celeketić et al., 2008; Yamano et al., 2008). The inner membrane potential Δψ activates the TIM23 channel and drives translocation of the positively charged presequences (Truscott et al., 2001; Meinecke et al., 2006).
The TIM23 complex transports preproteins to two different subcompartments, inner membrane and matrix. Preproteins carrying a hydrophobic sorting (stop transfer) signal behind the presequence are laterally released into the inner membrane, whereas preproteins lacking the hydrophobic signal are translocated into the matrix (Glick et al., 1992; Chacinska et al., 2005, 2010; Meier et al., 2005; van der Laan et al., 2007; Popov-Celeketić et al., 2008). The TIM23 complex cooperates with different partner complexes. First, it interacts with the TOM complex to promote preprotein transfer from the outer to the inner membrane (Dekker et al., 1997; Yamamoto et al., 2002; Chacinska et al., 2003, 2005, 2010; Mokranjac et al., 2005; Tamura et al., 2009). Second, the TIM23 complex is coupled to respiratory chain supercomplexes comprising complexes III and IV. This coupling promotes the Δψ-dependent step of preprotein insertion into the inner membrane (van der Laan et al., 2006; Wiedemann et al., 2007; Dienhart and Stuart, 2008; Saddar et al., 2008; Endo et al., 2011). Third, the TIM23 complex cooperates with the presequence translocase-associated motor (PAM) to drive ATP-dependent protein translocation into the matrix (Chacinska et al., 2005; Hutu et al., 2008; Popov-Celeketić et al., 2008, Popov-Čeleketić et al., 2011).
The essential core of the TIM23 complex (TIM23CORE) consists of three proteins: the pore-forming Tim23, Tim17, and Tim50 (Truscott et al., 2001; Geissler et al., 2002; Yamamoto et al., 2002; Meinecke et al., 2006; Gevorkyan-Airapetov et al., 2009; Mokranjac et al., 2009; Tamura et al., 2009; Marom et al., 2011; Qian et al., 2011). A fourth subunit, Tim21, associates with TIM23CORE to form a larger TIM23SORT complex. Tim21 performs regulatory functions; it transiently interacts with the TOM complex as well as respiratory chain supercomplexes, and, thus, TIM23SORT is involved in preprotein transfer from TOM to TIM23 and in Δψ-dependent membrane insertion of preproteins (Chacinska et al., 2005, 2010; Mokranjac et al., 2005; Albrecht et al., 2006; van der Laan et al., 2006, 2007; Wiedemann et al., 2007; Dienhart and Stuart, 2008).
Matrix translocation of preproteins depends on the cooperation of TIM23 with the motor PAM. The key component of PAM is the ATP-dependent mitochondrial heat shock protein 70 (mtHsp70). The import-driving activity of mtHsp70 and its cooperation with TIM23 are regulated by the membrane-bound cochaperones Tim44, Pam18-Pam16, and Pam17 (Neupert and Herrmann, 2007; Chacinska et al., 2009).
For this report, we performed a systematic analysis of the composition of the TIM23 complex and identified a new subunit, the integral inner membrane protein Mgr2 (mitochondrial genome required 2). We show that Mgr2 is required for efficient TOM-TIM23 coupling and the association of Tim21 and respiratory chain complexes with TIM23CORE. Thus, our study identifies Mgr2 as a new coupling factor in the TIM23 complex.
Results and discussion
Mgr2 is a subunit of the TIM23 complex
We purified the yeast TIM23 complex via protein A–tagged Tim21 upon differential stable isotope labeling with amino acids in cell culture (SILAC; Ong et al., 2002). Candidate proteins specifically enriched with Tim21ProtA were obtained based on the measured light-over-heavy peptide ratios (L/H; Gebert et al., 2011; von der Malsburg et al., 2011). As expected, the highest enrichment factors were found for the subunits of the TIM23 complex (Fig. 1 A and Table S1). We observed a similarly strong enrichment for a protein termed Mgr2. MGR2 was identified in a screen for genes required for survival of yeast cells without mitochondrial DNA (Dunn et al., 2006). In higher eukaryotes, the Mgr2 homolog Romo1 (reactive oxygen species modulator 1) is involved in production of reactive oxygen species, apoptosis, and regulation of mitochondrial morphology (Chung et al., 2008, 2009; Zhao et al., 2009; Kim et al., 2010; Lee et al., 2010). The molecular function of Mgr2/Romo1 is unknown.
Figure 1.
Mgr2 is a subunit of the TIM23 complex. (A) Yeast cells were labeled with [13C6]lysine/[13C6]arginine (wild type) or their 12C-containing counterparts (Tim21ProtA strain). Mitochondria were isolated, mixed, and lysed with digitonin. The TIM23 complex was isolated by IgG affinity purification. The mean log10 L/H ratio was plotted against the corresponding p-value (−log10) of each protein. Red, presequence translocase subunits including Mgr2; green, complexes III and IV; yellow, mitochondrial carrier family; brown, Tim44; black and grey, further proteins. See Materials and methods and Table S1. (B) Wild-type (WT) and Tim23ProtA mitochondria were lysed in digitonin and incubated with IgG Sepharose. Bound complexes were eluted by TEV protease cleavage and analyzed by SDS-PAGE and immunoblotting. Eluate: 100%; load: TIM/PAM subunits 17%, respiratory chain subunits 0.5%, and further proteins 6%. The arrowhead indicates the signal of Tim23ProtA. (C) Wild-type and Tim21ProtA mitochondria were analyzed as described for B. Eluate: 100%; load: TIM/PAM subunits 6%, respiratory chain subunits 0.5%, and further proteins 4%. The arrowhead indicates the signal of Tim21ProtA.
A lower enrichment with Tim21ProtA was observed for several proteins that mainly fall in two groups of inner membrane proteins: respiratory chain complexes III and IV and mitochondrial metabolite carriers (Fig. 1 A and Table S1). These proteins are highly abundant inner membrane proteins, and it has been previously shown for several of them that a small fraction is associated with the TIM23 complex (Geissler et al., 2002; van der Laan et al., 2006, 2007; Wiedemann et al., 2007; Dienhart and Stuart, 2008; Saddar et al., 2008).
Mgr2 was found in the mitochondrial fraction of yeast cells and was not extracted at alkaline pH (Fig. S1 A), indicating that it is an integral mitochondrial membrane protein in agreement with the prediction of two transmembrane segments. Mgr2 behaved as inner membrane protein, as it was not present in outer membrane vesicles, and its precursor was imported into isolated mitochondria in a Δψ-dependent manner and processed to the mature form (Fig. S1, B and C).
Mitochondria containing a protein A–tagged version of Tim23 were lysed with digitonin, and the TIM23 complex was purified by affinity chromatography (Geissler et al., 2002; Chacinska et al., 2005). Mgr2 was efficiently copurified with Tim23 like the known subunits of the TIM23 complex (Fig. 1 B). As expected, the import motor PAM and a fraction of the respiratory complexes III and IV were also coisolated with the TIM23 complex. A copurification with protein A–tagged Tim21 allows to differentiate between TIM23 subunits and PAM subunits. Tim21 is present in the TIM23SORT complex but not associated with the motor PAM (Chacinska et al., 2005, 2010; van der Laan et al., 2007). Mgr2 was efficiently copurified with Tim21ProtA (Fig. 1 C), indicating that Mgr2 is associated with TIM23SORT.
Yeast cells lacking the MGR2 gene were partially impaired in growth on nonfermentable medium at the usual growth temperature of 30°C. At elevated temperature on nonfermentable medium, where a high mitochondrial activity is required, the growth of mgr2Δ cells was strongly inhibited (Fig. S1 D). For the biochemical analysis, we first used mitochondria isolated from cells grown on nonfermentable medium at 30°C (Fig. S1 E). Different forms of the TIM23 complex can be visualized by blue native electrophoresis upon solubilization of mitochondria with digitonin. With wild-type mitochondria, antibodies against Tim23 decorate the TIM23CORE complex and the larger TIM23SORT complexes; Tim21 is only present in the TIM23SORT complexes (Fig. 2 A; Chacinska et al., 2005, 2010; van der Laan et al., 2007). mgr2Δ mitochondria contained the TIM23CORE complex like wild-type mitochondria; however, TIM23SORT was largely absent (Fig. 2 A). Other inner membrane complexes like the carrier translocase (TIM22 complex) or the F1Fo-ATP synthase were not affected (Fig. 2 A). The MGR2 gene is located in the vicinity of the genes MSY1 and AIM43 on Saccharomyces cerevisiae chromosome XVI. The phenotype of MGR2 deletion is not caused by affecting MSY1 or AIM43 for the following reasons: (a) expression of the Aim43 protein was not blocked in the mgr2Δ mutant (Fig. S1 F); (b) msy1 mutant yeast are defective in respiratory growth (Saccharomyces Genome Database) in contrast to mgr2Δ yeast; and (c) reexpression of the MGR2 ORF in mgr2Δ yeast using a plasmid with the phosphoglycerate kinase (PGK) promoter restored growth and formation of the TIM23SORT complexes (Fig. S1 G). Yeast mitochondria contained about 10 pmol Mgr2, 17 pmol Tim21, and 16 pmol Tim23 per milligram of protein (see Materials and methods). For comparison, the motor subunit Pam18 was present in ∼60 pmol/mg and the ADP/ATP carrier (AAC) in ∼280 pmol/mg. In the TIM23SORT complex purified via tagged Tim21, Mgr2 was present in stoichiometric amounts to Tim23 (Fig. S1 H), demonstrating a quantitative association of Mgr2 with this TIM23 form (Pam18 was depleted in TIM23SORT as expected; Chacinska et al., 2005, 2010; van der Laan et al., 2007). Collectively, we conclude that Mgr2 is a subunit of the TIM23 complex.
Figure 2.
Mgr2 is required for binding of Tim21 and respiratory chain complexes to TIM23CORE. (A) Mitochondria were solubilized in digitonin and analyzed by blue native electrophoresis and Western blotting. WT, wild type. (B) Mitochondria were solubilized with digitonin and subjected to IgG affinity purification. Eluates after TEV cleavage were analyzed by SDS-PAGE and immunoblotting. Eluate: 100%; load: TIM/PAM subunits 17%, respiratory chain subunits 0.5%, and further proteins 6%. (C) Mitochondria were analyzed as described for B. Eluate: 100%; load: TIM/PAM subunits 6%, respiratory chain subunits 0.5%, and further proteins 4%. (D) Mitochondria were analyzed as described for B. Eluate: 100%; load: respiratory chain subunits 28% and TIM/PAM subunits and further proteins 6%. The asterisk indicates the nonspecific signal.
Mgr2 is required for coupling of Tim21 to the TIM23CORE complex
Mitochondria were isolated from an mgr2Δ yeast strain expressing protein A–tagged Tim23, lysed with digitonin, and subjected to affinity purification. The subunits of TIM23CORE as well as the subunits of the motor PAM were efficiently purified from wild-type and mgr2Δ mitochondria (Fig. 2 B). However, the copurification of Tim21 with tagged Tim23 was strongly impaired when Mgr2 was lacking. Tim21 is involved in coupling of respiratory supercomplexes to the presequence translocase (van der Laan et al., 2006; Wiedemann et al., 2007). Consequently, the yield of copurification of respiratory complexes III and IV was reduced with mgr2Δ mitochondria (Fig. 2 B).
To study which proteins are associated with Tim21 in mgr2Δ mitochondria, we generated an mgr2Δ yeast strain expressing tagged Tim21. Although the recovery of the bait protein Tim21 was comparable in wild-type and mgr2Δ mitochondria, copurification of the TIM23CORE proteins with Tim21ProtA was almost abolished in the absence of Mgr2 (Fig. 2 C). In contrast, subunits of the respiratory chain complexes III and IV were copurified with Tim21ProtA, with similar efficiency in wild-type and mgr2Δ mitochondria (Fig. 2 C).
To independently demonstrate that Tim21 is associated with the respiratory chain but not the TIM23 complex in mgr2Δ mitochondria, we deleted MGR2 in a yeast strain containing tagged Cor1 (a subunit of complex III). Tim21 was copurified with tagged Cor1 independently of Mgr2; however, the copurification of TIM23CORE components was strongly reduced in the absence of Mgr2 (Fig. 2 D). As a control, the copurification of complex III and IV subunits with Cor1 was not affected by the lack of Mgr2 (Fig. 2 D). The activities of complexes III and IV were not or only mildly affected by the lack of Mgr2 (Fig. S1 I).
We conclude that Mgr2 is selectively required for the binding of Tim21 to TIM23CORE and thus for the formation of TIM23SORT complexes and the efficient recruitment of respiratory chain complexes to the presequence translocase.
Mgr2 is required for efficient protein import at elevated temperature
With mitochondria isolated from yeast cells grown at 30°C, Δψ-dependent import of matrix-targeted (Su9–dihydrofolate reductase [DHFR]) and inner membrane–inserted preproteins (cytochrome b2–DHFR and cytochrome c1) was not or only mildly affected by the lack of Mgr2 (Fig. S2, A–C). Thus, under standard import conditions with fully energized mitochondria, protein import can occur into mgr2Δ mitochondria in agreement with the observation that Mgr2 is not strictly essential for growth of yeast, whereas the TIM23CORE components Tim17, Tim23, and Tim50 are essential for cell viability under all growth conditions (Dekker et al., 1993; Emtage and Jensen, 1993; Maarse et al., 1994; Ryan et al., 1994; Geissler et al., 2002; Yamamoto et al., 2002).
As the growth defect of mgr2Δ cells is temperature dependent (Fig. S1 D), we grew cells at 30°C and then shifted them to elevated temperature. Under these conditions, the import of presequence-carrying preproteins into mitochondria was strongly impaired (Fig. 3, A–C). To exclude that mgr2Δ mitochondria isolated under these conditions were unspecifically damaged, we analyzed their basic characteristics. The steady-state levels of TIM and TOM subunits, as well as various control proteins, were not or only mildly affected by the lack of Mgr2 (Fig. S2 D). The membrane potential of the mutant mitochondria was moderately reduced (Fig. S2 E). Immunoprecipitation with antibodies against Tim23 under nondenaturing conditions demonstrated that TIM23CORE subunits as well as PAM subunits were copurified like in wild-type mitochondria (Fig. S2 F). Blue native electrophoresis of mgr2Δ mitochondria revealed that the TIM23SORT complexes were disturbed as expected, whereas the TIM22 complex and the TOM complex were not affected (Fig. S2 G). To exclude that the partial reduction of Δψ caused the import defects, we analyzed the Δψ-dependent import of the dicarboxylate carrier (Dic1) via the TIM22 complex and did not observe an inhibition in mgr2Δ mitochondria (Fig. 3 D), demonstrating that the effect on Δψ cannot explain the strong and selective import defect via the presequence translocase. We conclude that Mgr2 is critical for protein import via the presequence pathway at elevated temperature.
Figure 3.
Mgr2 promotes preprotein import in cells grown at elevated temperature. (A–C) 35S-labeled preproteins were imported into mitochondria isolated from yeast cells grown at 39°C. After proteinase K treatment, samples were analyzed by SDS-PAGE and autoradiography. p, precursor; i, intermediate; m, mature; WT, wild type. (D) Dic1 was imported, as described for A–C, and formation of the mature dimer (d) was analyzed by blue native electrophoresis after solubilization with digitonin.
Lack of Mgr2 impairs Δψ-dependent import and TOM-TIM23 coupling
We asked whether Mgr2 was only required for protein import at elevated temperature or whether it also played a role at the usual temperature of 30°C and thus analyzed distinct import conditions. Mgr2 is part of the TIM23SORT complex that is involved in both coupling of TIM23 to the respiratory chain and preprotein transfer from TOM to TIM23 (Chacinska et al., 2005, 2010; Wiedemann et al., 2007). In case of tim21Δ mitochondria, the defective coupling of TIM23SORT with the respiratory chain leads to a decreased protein import when Δψ is partially reduced (van der Laan et al., 2006; Wiedemann et al., 2007). Preprotein import into mgr2Δ mitochondria was indeed impaired like into tim21Δ mitochondria, when Δψ was gradually reduced by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP; Fig. 4 A), supporting the conclusion that Mgr2 and Tim21 are required for coupling of TIM23SORT to the respiratory chain.
Figure 4.
Mgr2 promotes Δψ-dependent protein import. (A) b2(84)-DHFR was imported into wild-type (WT), tim21Δ, and mgr2Δ mitochondria (cells grown at 30°C) in the presence of CCCP and analyzed by SDS-PAGE and autoradiography. Import without CCCP was set to 100% (control). The mean ± SEM (n = 4) is shown. (B and C) Preprotein import into mitochondria followed by SDS-PAGE. MTX, methotrexate; p, precursor; i, intermediate; m, mature.
To study a possible role of Mgr2 in preprotein transfer from TOM to TIM23 complexes, we selectively accumulated cytochrome b2–DHFR fusion proteins at different import stages in TOM-TIM23 contact sites as two-membrane–spanning translocation intermediates (see Materials and methods). For the short fusion protein b2(84)-DHFR, the N-terminal presequence reaches just far enough into the matrix to become cleaved by the matrix processing peptidase (MPP) in wild-type mitochondria (Fig. 4 B; Rassow et al., 1990). Strikingly, in mgr2Δ as well as in tim21Δ mitochondria, this first processing was impaired, when the preprotein was accumulated in TOM-TIM23 import sites (Fig. 4 B). In contrast, the long fusion protein b2(220)-DHFR that becomes arrested at a later stage of import was efficiently processed first by MPP and subsequently by the inner membrane peptidase in wild-type, mgr2Δ, and tim21Δ mitochondria (Fig. 4 C). We conclude that the lack of Mgr2 or Tim21 impairs efficient TOM-TIM accumulation and processing of the short b2 preprotein.
As several metabolite carriers were associated with the TIM23 complex (Fig. 1 A and Table S1), we asked whether they were connected to the role of Mgr2, using the TOM-TIM accumulation of b2(84)-DHFR as assay. Neither inhibition of AACs by carboxyatractyloside (Wachter et al., 1992) nor lack of further metabolite carriers affected the processing of translocation-arrested b2(84)-DHFR (Fig. S3, A and B). Lack of Mgr2 only mildly affected the copurification of imported carrier proteins with the TIM23 complex and the transport activity of AAC (Fig. S3, C and D). These results suggest that the involvement of Mgr2 in preprotein import cannot be attributed to an indirect effect via metabolite carriers. Moreover, we did not obtain evidence for a connection of Mgr2 to the AAA (ATPases associated with diverse cellular activities) proteases of the inner membrane; the proteases were not specifically enriched with the TIM23SORT complex (Table S1), and characteristic substrates, which are processed by the Yta10/Yta12 protease (Bonn et al., 2011), were not affected by the lack of Mgr2 (Fig. S3, E and F).
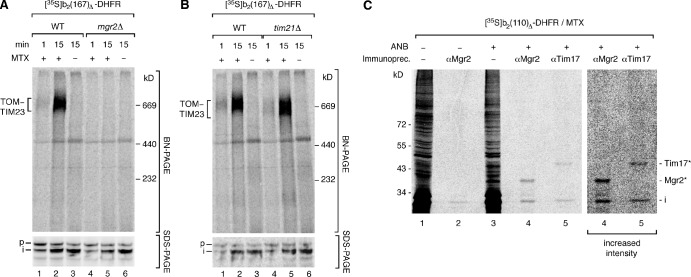
To obtain further evidence for a role of Mgr2 in coordinating efficient TOM-TIM23 cooperation, we used blue native electrophoresis to directly visualize the TOM-TIM23 preprotein connection. A radiolabeled b2-DHFR fusion protein was accumulated in TOM-TIM23 import sites. The mitochondria were lysed by digitonin, and the TOM–TIM23 preprotein supercomplex was detected by autoradiography (Fig. 5 A, top, second lane; Dekker et al., 1997; Chacinska et al., 2003, 2010). In the absence of Mgr2, the supercomplex was not detectable (Fig. 5 A, top, fifth lane). As these mitochondria can import presequence-carrying preproteins across outer and inner membranes (Figs. 5 A [bottom] and S2 [A–C]), a basic TOM–TIM23 cooperation apparently still takes place. However, the stable connection of both translocases that is required for the detection of the supercomplex on native gels is not possible anymore in mgr2Δ mitochondria. Remarkably, in tim21Δ mitochondria, the supercomplex was still detectable, though it migrated faster as a result of the lack of Tim21 (Fig. 5 B, top), indicating that formation of a stable TOM–TIM23 preprotein supercomplex strongly depends on Mgr2 but not on Tim21.
Figure 5.
Mgr2-dependent formation of a TOM–TIM23 preprotein supercomplex. (A and B) b2(167)Δ-DHFR was imported into wild-type (WT), mgr2Δ, and tim21Δ mitochondria in the presence of methotrexate (MTX), as indicated. Samples were lysed with digitonin and analyzed by blue native (BN) electrophoresis or separated by SDS-PAGE. p, precursor; i, intermediate. (C) b2(110)Δ-DHFR with incorporated ANB was imported into wild-type mitochondria in the presence of methotrexate. ANB was activated by UV light. Cross-linking products (asterisks) were identified by immunoprecipitation (Immunoprecip.) and SDS-PAGE.
Thus, we asked whether Mgr2 plays a direct role in protein import and probed for an interaction of Mgr2 with preproteins in transit. Upon incorporation of the photoreactive cross-linking reagent ANB (5-azido-2-nitrobenzoyl), a b2-DHFR preprotein was accumulated in TOM-TIM23 import sites. By immunoprecipitation under stringent conditions, cross-linking of the preprotein to Mgr2 as well as Tim17 was demonstrated (Fig. 5 C). We conclude that Mgr2 is in close vicinity of preproteins in transit and participates in stabilization of the TOM–TIM23 preprotein supercomplex.
Conclusions
Mgr2 is a new subunit of the TIM23 complex. It is in vicinity of preproteins in transit and couples Tim21 to the core translocase, leading to formation of the TIM23SORT complex. Mgr2 promotes interaction of the presequence translocase with the respiratory supercomplex III–IV and the efficient cooperation of TOM and TIM23 complexes in preprotein translocation. Mgr2 is of particular importance for protein import under heat stress conditions when a high mitochondrial activity is needed. Future studies will have to address whether this small hydrophobic protein may play further coupling functions in the mitochondrial inner membrane.
Materials and methods
Yeast strains and growth conditions
A chromosomal deletion of MGR2 in S. cerevisiae was achieved by transformation of a Yep352 plasmid expressing Mgr2 under the control of an MET25 promoter and CYC1 terminator into the YPH499 background. The chromosomal gene was replaced by a kanMX6 cassette, and the plasmid was removed by growth on medium supplied with 5-fluoroorotic acid. Using this approach, the deletion of MGR2 was introduced into the strains Tim21ProtA, Tim23ProtA, and Cor1TAP (YPH499 background; Geissler et al., 2002; Chacinska et al., 2005; van der Laan et al., 2006); the Tim23ProtA strain was also modified such that the tagged protein could be expressed from the chromosome. For isolation of mitochondria, yeast cells were grown in YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] bacto peptone, and 3% [wt/vol] glycerol, pH 5.0) at 30°C continuously or shifted for 24 h to 39°C before harvesting. For reexpression of MGR2 in mgr2Δ cells, the ORF of MGR2 was supplied by the plasmid pPGKMGR2 using the PGK1 promoter. Control transformations were performed with the empty vector pRS426. For SILAC analysis, the strains YPH500 arg8Δ (wild-type) and Tim21ProtA were grown in synthetic defined medium (0.67% bacto-yeast nitrogen base, amino acid mix, and 3% glycerol) supplemented with either [13C6]arginine and [13C6]lysine (Euriso-Top) for wild-type or [12C6]arginine and [12C6]lysine for Tim21ProtA cells (Gebert et al., 2011). Yeast peptone dextrose contained 1% [wt/vol] yeast extract, 2% [wt/vol] bacto peptone, and 2% [wt/vol] glucose, pH 5.0.
Precursor protein import into mitochondria
To study preprotein import into isolated mitochondria, we used several radiolabeled preproteins that are imported via the presequence translocase: cytochrome c1 and model proteins containing N-terminal parts of preproteins (cytochrome b2 and Fo-ATPase subunit 9) fused to the passenger protein DHFR (Chacinska et al., 2005, 2010; van der Laan et al., 2005, 2007; Stojanovski et al., 2007; Hutu et al., 2008). Import of the mitochondrial metabolite carrier Dic1 was monitored by formation of the mature dimer, as detected by blue native electrophoresis (Rehling et al., 2003; Brandner et al., 2005).
Mitochondria were isolated by differential centrifugation. For in vitro synthesis of 35S-labeled precursor proteins, a reticulocyte lysate transcription/translation system (GE Healthcare) was used. For synthesis of [35S]Mgr2, linear DNA templates including an SP6 promoter were generated by PCR and transcribed in vitro (mMESSAGE mMACHINE SP6 kit; Invitrogen); mRNA was purified (MEGAclear kit; Invitrogen) and incubated for translation with yeast cytosolic extract in the presence of [35S]methionine (Garcia et al., 1991). Radiolabeled preproteins were incubated with isolated mitochondria in import buffer (3% [wt/vol] fatty acid–free BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 5 mM methionine, and 10 mM MOPS-KOH, pH 7.2) in the presence of 2 mM ATP and 2 mM NADH at 25°C (Ryan et al., 2001; Stojanovski et al., 2007). For import of Dic1, an ATP regenerating system (5 mM creatine phosphate and 0.1 mg/ml creatine kinase) was included. 5 µM methotrexate was added where indicated. Import reactions were stopped by dissipation of Δψ by adding an AVO mix (8 µM antimycin A, 1 µM valinomycin, and 20 µM oligomycin final concentrations) and transfer on ice. For protease treatment, typically 50 µg/ml proteinase K was added, and samples were incubated for 15 min on ice followed by addition of 2 mM PMSF and further incubation for 10 min on ice. Mitochondria were washed in SEM buffer (10 mM MOPS-KOH, 1 mM EDTA, and 250 mM sucrose, pH 7.2), lysed in SDS lysis buffer (2% [wt/vol] SDS, 10% [vol/vol] glycerol, 60 mM Tris-HCl, pH 6.8, 0.01% bromophenol blue, and freshly added 2 mM PMSF), and subsequently analyzed by SDS-PAGE. For cross-linking, precursor proteins were labeled with the photoreactive cross-linker ANB (tRNA Probes) and, after import, into mitochondria exposed to UV light for 10 min (Devaraneni et al., 2011). For CCCP titration, radiolabeled b2(84)-DHFR was imported into isolated mitochondria (0.7% [wt/vol] BSA in the import buffer) in the presence of 0–40 µM CCCP (to gradually dissipate Δψ) and 20 µM oligomycin (for inhibition of Δψ generation through a reverse action of the F1Fo-ATP synthase) for 10 min at 25°C (Geissler et al., 2000; van der Laan et al., 2006).
For blue native electrophoresis, mitochondria were solubilized in ice-cold blue native lysis buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, and 10% [vol/vol] glycerol) after washing with SEM buffer and incubated for 15 min on ice (Schägger and von Jagow, 1991; Dekker et al., 1997). After a clarifying spin, supernatants were mixed with 0.1 vol of cold blue native loading buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris-HCl, pH 7.0, and 500 mM 6-aminocaproic acid) and subjected to blue native electrophoresis.
Radiolabeled proteins were detected by digital autoradiography (Storm 820 imaging system; GE Healthcare), and files were processed by using ImageQuant software (version 5.2; GE Healthcare). Excision of nonrelevant lanes was performed digitally and is shown by separating white lines.
Accumulation of import intermediates
For the accumulation of translocation intermediates in TOM-TIM23 import sites, different variants of cytochrome b2-DHFR fusion proteins were used. In the presence of the DHFR ligand methotrexate, the folding state of the DHFR moiety is stabilized, and, thus, DHFR does not pass through the TOM channel. The N-terminal b2 portion initiates translocation via TOM and TIM23 and is arrested in a two-membrane–spanning fashion in a TOM–TIM23 supercomplex (Dekker et al., 1997; Chacinska et al., 2003, 2005, 2010; Popov-Celeketić et al., 2008). Cytochrome b2 contains a bipartite presequence that is processed twice, first by MPP and then by the inner membrane peptidase (Glick et al., 1992; Chacinska et al., 2005, 2010; Popov-Celeketić et al., 2008). We used b2-DHFR proteins of different length. b2(84)-DHFR is so short that upon arrest with methotrexate, only the first processing step by MPP can take place (after amino acid residue 31), as the second cleavage site after residue 80 is still in the TOM complex and thus not accessible to the inner membrane peptidase (Rassow et al., 1990). With b2(84)-DHFR, only 60 residues (53 b2 residues plus 7 linker residues) remain in front of the folded and methotrexate-stabilized DHFR moiety for spanning the TOM and TIM23 channels and cleavage by MPP (Rassow et al., 1990), indicating that a close cooperation of TOM and TIM23 complexes is required for efficient accumulation and cleavage of the short preprotein. In the absence of methotrexate, b2(84)-DHFR was cleaved twice by MPP and the inner membrane peptidase. For the long b2(220)-DHFR protein, the b2 portion is imported far enough that even after arrest of the DHFR moiety by methotrexate, both processing steps can take place (Chacinska et al., 2010). In the b2(167)Δ-DHFR preprotein, the hydrophobic stop transfer signal has been inactivated by the deletion of 19 amino acid residues; this preprotein is targeted to the mitochondrial matrix in the absence of methotrexate (Chacinska et al., 2010).
IgG affinity purification and immunoprecipitation
Mitochondria were solubilized in ice-cold blue native lysis buffer and incubated at 4°C for 30 min in a head-over-head shaker followed by a clarifying spin. For immunoprecipitation of denatured proteins, mitochondria were lysed in SDS lysis buffer (lacking bromophenol blue); after a clarifying spin, the supernatant was diluted in solubilization buffer (0.3% [vol/vol] Triton X-100, 20 mM Tris/HCl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, and 2 mM PMSF). Supernatants were mixed with purified human IgG (Sigma-Aldrich) coupled to cyanogen bromide–activated Sepharose (IgG affinity purification; GE Healthcare) or specific antibodies coupled covalently to protein A–Sepharose (GE Healthcare) by dimethyl pimelimidate cross-linking (immunoprecipitation) and incubated in a head-over-head shaker for 1–2 h at 4°C. Sepharose beads were reisolated and washed extensively with washing buffer (0.3% [wt/vol] digitonin, 20 mM Tris/HCl, pH 7.4, 0.5 mM EDTA, 60 mM NaCl, and 10% [vol/vol] glycerol). For immunoprecipitation of denatured proteins, solubilization buffer was used as washing buffer. Bound complexes were eluted through cleavage with tobacco etch virus (TEV) protease (Invitrogen) at 4°C overnight under vigorous shaking (IgG affinity purification) or by addition of 0.1 M glycine, pH 2.5 (immunoprecipitation). Elutions were precipitated with trichloroacetic acid and/or mixed with SDS lysis buffer, incubated for 15 min at 37°C under vigorous shaking, and subjected to SDS-PAGE. For immunodecoration, proteins were transferred by semi-dry Western blotting onto a polyvinylidene difluoride membrane (Millipore) and visualized by ECL (GE Healthcare).
SILAC, mass spectrometry (MS), and data analysis
Wild-type S. cerevisiae cells were grown in the presence of 13C-labeled (heavy) arginine and lysine. In parallel, yeast cells expressing protein A–tagged Tim21 were grown with 12C-containing (light) amino acids. Mitochondria were isolated and mixed. After solubilization of mitochondria with digitonin, TIM23 complexes were purified by affinity chromatography and examined by quantitative MS. Based on the ratios of light peptides originating from the Tim21ProtA mitochondria over the heavy peptides from wild-type mitochondria, candidate proteins were determined and further tested for specific association with the TIM23 complex.
After tryptic digestion of affinity-purified and SILAC-labeled protein complexes, peptide mixtures of four independent experiments were analyzed by nano-HPLC/electrospray ionization–MS/MS using an HPLC system (UltiMate 3000; Dionex) connected to a mass spectrometer (LTQ-Orbitrap XL; Thermo Fisher Scientific; Kaller et al., 2011). Peptides were washed and preconcentrated on a C18 µ-precolumn (0.3-mm inner diameter × 5 mm; PepMap; Dionex) using 0.1% (vol/vol) trifluoroacetic acid as a solvent and a flow rate of 30 µl/min. Peptides were subsequently separated on a C18 reversed-phase nano–liquid chromatography column (75-µm inner diameter × 150 mm; PepMap) and eluted applying a linear 150-min gradient ranging from 4–34% (vol/vol) acetonitrile (ACN) in 0.1% (vol/vol) formic acid (FA) and a flow rate of 300 nl/min. The column was subsequently washed with 80% ACN/0.1% FA (3 min) and reequilibrated with 4% ACN/0.1% FA for the next liquid chromatography run. Mass spectrometric data were acquired in a data-dependent mode using the following settings: acquisition of full-scan MS spectra (m/z 300–2,000) at a resolution of 60,000 (at m/z 400) in the Orbitrap, collision-induced dissociation fragmentation of the six most intense multiply charged ions in the linear ion trap at a normalized collision energy of 35%, and a dynamic exclusion time of 45 s.
For data processing, the software MaxQuant (version 1.0.13.13) was used (Cox and Mann, 2008). Proteins were identified by searching peak lists against a decoy version of the Saccharomyces Genome Database using Mascot (version 2.2; Matrix Science). A false discovery rate of <1% was applied for both peptides and proteins. Further criteria for protein identification were as follows: MS and MS/MS mass tolerances of 7 parts per million and 0.5 D, respectively, at least one unique peptide (six or more amino acids), a maximum of two missed cleavages, and methionine oxidation as a variable modification. Protein quantification was performed with [13C6]lysine and [13C6]arginine as heavy labels, unique peptides only, a minimum ratio count of 1, and without low-scoring variants of identified peptides. L/H ratios for proteins were log transformed (log10), and the mean log10 across all experiments, the SEM, and the p-value (determined using a one-sided t test) of all proteins quantified in at least two experiments were calculated. Potential interaction partners were required to be quantified in at least three experiments with a sequence coverage of ≥3%, a posterior error probability of <0.01, and a mean L/H ratio of >50 (category 1), 10–50 (category 2), or 5–10 (category 3).
Cell fractionation and analysis of mitochondrial proteins
The fractionation of yeast cells was performed as described for the isolation of mitochondria (Meisinger et al., 2006; Stojanovski et al., 2007), with the following modifications. For homogenization, a syringe connected to a 0.8 × 22–mm blunt-end cannula (Braun) was used. Cell debris and nuclei were pelleted by low-speed centrifugation at 1,000 g. The resulting supernatant was subjected to centrifugation at 12,000 g, yielding a mitochondrial pellet (P12). The supernatant was further centrifuged (100,000 g), resulting in a supernatant (S100) containing the cytosolic fraction and a pellet (P100) including microsomes. Equal amounts of each fraction were denatured in SDS lysis buffer and subjected to SDS-PAGE and immunoblotting.
For a quantitative assessment of mitochondrial proteins, selected proteins were synthesized by in vitro transcription/translation in chemical amounts (RTS 100 Wheat Germ CECF kit; 5 PRIME; Becker et al., 2011), according to the manufacturer’s instructions, with a C-terminal HIS6 tag. Proteins were purified using Ni–nitrilotriacetic acid agarose (PerfectPro Ni-NTA; 5 PRIME) and eluted with buffer containing 300 mM imidazole. Purified proteins were analyzed on 6–16.5% Tris-Tricine gels in comparison with the Low Molecular Weight Calibration Kit for SDS electrophoresis (multiple lanes with increasing protein concentration; 17-0446-01; GE Healthcare) by colloidal Coomassie staining. The protein concentrations were compared by using the Multi Gauge software (version 3.0; Fujifilm). Mitochondria were solubilized in elution buffer (50 mM NaH2PO4, 300 mM NaCl, 300 mM imidazole, and 0.5% [wt/vol] SDS, pH 7.0), mixed with 4× sample buffer, and boiled for 5 min at 95°C. Defined amounts of solubilized mitochondria and purified proteins (multiple lanes with increasing concentration) were loaded on 6–16.5% Tris-Tricine gels and analyzed by Western blotting. Quantitative assessment of the Western blots was performed using the Multi Gauge software (version 3.0), yielding 9.7 ± 0.4 pmol Mgr2/mg, 17.0 ± 3.6 pmol Tim21/mg, 15.6 ± 2.9 pmol Tim23/mg, 63 ± 16 pmol Pam18/mg, and 279 ± 65 pmol AAC/mg total mitochondrial protein (±SEM). The value for Tim23 agrees with previous determinations of its abundance (∼17–20 pmol/mg; Dekker et al., 1997; Sirrenberg et al., 1997). For each protein, three independent experiments were performed.
Determination of ADP uptake
Reconstitution of detergent-solubilized mitochondrial membrane proteins was performed by a dilution approach similar to the procedure described by Trentmann et al. (2007). Membrane proteins from frozen purified mitochondria were solubilized with 1% n-dodecyl-β-maltoside (Glycon Biomedicals). 200 µg (in 50 µl) of the solubilized mitochondrial proteins was mixed with 400 µl of a homogeneous emulsion of l-α-phosphatidylcholine (125 mg/ml, type IV-S; Sigma-Aldrich) in buffer medium TG1 (100 mM tricine and 30 mM potassium gluconate, pH 7.5). For loading with 10 mM substrate, liposomes were supplemented with 50 µl ATP (100 mM). 50 µl TG1 was added to nonloaded liposomes. Subsequently, the protein-lipid mixture (500 µl) was vigorously mixed and frozen in liquid nitrogen. Proteoliposomes were thawed on ice, sonified for 20 s (Branson Sonifier 250; Emerson Electric Co.) at the lowest output level and 50% duty cycle. For removal of external ATP and to replace external TG1 by TG2 (10 mM tricine and 150 mM potassium gluconate, pH 7.5), the lipid vesicles were applied to an NAP-5 column (GE Healthcare) equilibrated with TG2. Proteoliposomes were eluted from the chromatography column with 1 ml TG2 buffer.
α-[32P]ADP was enzymatically generated from α-[32P]ATP (PerkinElmer) through a hexokinase reaction (Tjaden et al., 1998). 150 µCi α-[32P]ATP, 1 mM glucose, 10 µM nonlabeled ATP, 5 mM MgCl2, and 50 mM Hepes, pH 7.5, were incubated with 3 U hexokinase for 1 h. The reaction was stopped by denaturation of hexokinase (10 min at 100°C), and successful generation of α-[32P]ADP (as a result of a transfer of the γ-phosphate group from α-[32P]ATP to glucose) was analyzed by thin-layer chromatography. For uptake measurements, 100 µl of the respective differently loaded proteoliposomes was incubated at 30°C in the presence of 50 µM α-[32P]ADP. At the indicated time points, samples were applied to anion exchange chromatography columns (DOWEX 1X8 Cl−, 200–400 mesh; Sigma-Aldrich) to remove exterior (nonimported) ADP and to terminate the transport. Liposomes were eluted from the chromatography columns with 1.5 ml Tricine (200 mM, pH 7.5), and radioactivity in the liposome samples was quantified by scintillation counting (Canberra-Packard).
Carbonate extraction
For alkaline extraction, 150 µg YPH499 mitochondria (protein amount) was resuspended in 0.1 M sodium carbonate of the indicated pH and incubated on ice for 30 min (Stojanovski et al., 2007). Mitochondrial membrane sheets were isolated by ultracentrifugation (125,000 g), and the protein content was compared with the supernatant containing extracted proteins by SDS-PAGE and Western blotting. Totals represent 75 µg YPH499 mitochondria (protein amount) directly subjected to SDS-PAGE.
Miscellaneous
Measurement of ubiquinol/cytochrome c reductase activity was performed according to Palsdottir and Hunte (2003), with minor changes. Freeze-thawed mitochondria were assayed in 40 mM potassium phosphate buffer, pH 7.4, 1 mM NaN3, 50 µM horse heart cytochrome c, and 80 µM decylubiquinol at room temperature for ubiquinol-dependent cytochrome c reductase activity. A stable baseline was measured at 550 nm. The mitochondria were diluted into the assay to a final concentration of 30 µg protein/ml to start the reaction. The increase at 550 nm was monitored as an indication of the reduction of cytochrome c, and the rate of cytochrome c reduction was calculated using an extinction coefficient of 19.4 mM−1 cm−1.
For measurement of cytochrome c oxidase activity (Horváth et al., 2000), isolated mitochondria were solubilized in 0.5% (vol/vol) Triton X-100 and assayed in 40 mM potassium phosphate buffer, pH 7.4, and 100 µM fully reduced horse heart cytochrome c at room temperature. A stable baseline was measured at 550 nm. The solubilized mitochondrial proteins were diluted into the assay to a final concentration of 5 µg/ml to start the reaction. The decrease at 550 nm was monitored as an indication of the oxidation of cytochrome c, and the rate was calculated using an extinction coefficient of 19.4 mM−1 cm−1.
Online supplemental material
Fig. S1 shows basic characteristics of the mitochondrial inner membrane protein Mgr2 and of the mgr2Δ mutant. Fig. S2 shows basic characteristics of mitochondria isolated from mgr2Δ yeast cells grown at normal and elevated temperature. Fig. S3 shows control import reactions in mgr2Δ mitochondria. Table S1 shows proteins identified and quantified by SILAC-based affinity purification–MS experiments using Tim21 as bait. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110047/DC1.
Supplementary Material
Acknowledgments
We thank Drs. C. Meisinger, N. Wiedemann, F.-N. Vögtle, A.E. Johnson, D. Stojanovski, S. Rospert, R. Feuerstein, and B. Gissler for discussion and materials and I. Perschil and B. Schönfisch for expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Excellence Initiative of the German Federal and State Governments (EXC 294 Centre for Biological Signalling Studies; GSC-4 Spemann Graduate School), Graduiertenkolleg 845, Bundesministerium für Bildung und Forschung (Dynamo), Landesforschungspreis Baden-Württemberg, Gottfried Wilhelm Leibniz Program, and a Boehringer Ingelheim Fonds predoctoral fellowship (to M. Bohnert).
Footnotes
Abbreviations used in this paper:
- AAC
- ADP/ATP carrier
- ACN
- acetonitrile
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- DHFR
- dihydrofolate reductase
- FA
- formic acid
- MPP
- matrix processing peptidase
- MS
- mass spectrometry
- PAM
- presequence translocase-associated motor
- PGK
- phosphoglycerate kinase
- TEV
- tobacco etch virus
References
- Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 100:551–560 10.1016/S0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- Albrecht R., Rehling P., Chacinska A., Brix J., Cadamuro S.A., Volkmer R., Guiard B., Pfanner N., Zeth K. 2006. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 7:1233–1238 10.1038/sj.embor.7400828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Wenz L.S., Thornton N., Stroud D., Meisinger C., Wiedemann N., Pfanner N. 2011. Biogenesis of mitochondria: Dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J. Mol. Biol. 405:113–124 10.1016/j.jmb.2010.11.002 [DOI] [PubMed] [Google Scholar]
- Bonn F., Tatsuta T., Petrungaro C., Riemer J., Langer T. 2011. Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J. 30:2545–2556 10.1038/emboj.2011.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K., Rehling P., Truscott K.N. 2005. The carboxyl-terminal third of the dicarboxylate carrier is crucial for productive association with the inner membrane twin-pore translocase. J. Biol. Chem. 280:6215–6221 10.1074/jbc.M412269200 [DOI] [PubMed] [Google Scholar]
- Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., Pfanner N. 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274:16522–16530 10.1074/jbc.274.23.16522 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Rehling P., Guiard B., Frazier A.E., Schulze-Specking A., Pfanner N., Voos W., Meisinger C. 2003. Mitochondrial translocation contact sites: Separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22:5370–5381 10.1093/emboj/cdg532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., Lind M., Frazier A.E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H.E., Truscott K.N., Guiard B., et al. 2005. Mitochondrial presequence translocase: Switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120:817–829 10.1016/j.cell.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. 2009. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 138:628–644 10.1016/j.cell.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A., van der Laan M., Mehnert C.S., Guiard B., Mick D.U., Hutu D.P., Truscott K.N., Wiedemann N., Meisinger C., Pfanner N., Rehling P. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30:307–318 10.1128/MCB.00749-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.S., Lee S.B., Park S.H., Kang S.T., Na A.R., Chang T.S., Kim H.J., Yoo Y.D. 2009. Mitochondrial reactive oxygen species originating from Romo1 exert an important role in normal cell cycle progression by regulating p27(Kip1) expression. Free Radic. Res. 43:729–737 10.1080/10715760903038432 [DOI] [PubMed] [Google Scholar]
- Chung Y.M., Lee S.B., Kim H.J., Park S.H., Kim J.J., Chung J.S., Yoo Y.D. 2008. Replicative senescence induced by Romo1-derived reactive oxygen species. J. Biol. Chem. 283:33763–33771 10.1074/jbc.M805334200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Dekker P.J.T., Keil P., Rassow J., Maarse A.C., Pfanner N., Meijer M. 1993. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 330:66–70 10.1016/0014-5793(93)80921-G [DOI] [PubMed] [Google Scholar]
- Dekker P.J.T., Martin F., Maarse A.C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., Pfanner N. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408–5419 10.1093/emboj/16.17.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraneni P.K., Conti B., Matsumura Y., Yang Z., Johnson A.E., Skach W.R. 2011. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell. 146:134–147 10.1016/j.cell.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienhart M.K., Stuart R.A. 2008. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell. 19:3934–3943 10.1091/mbc.E08-04-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P., Likic V., Tachezy J., Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science. 313:314–318 10.1126/science.1127895 [DOI] [PubMed] [Google Scholar]
- Dunn C.D., Lee M.S., Spencer F.A., Jensen R.E. 2006. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell. 17:213–226 10.1091/mbc.E05-06-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage J.L., Jensen R.E. 1993. MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J. Cell Biol. 122:1003–1012 10.1083/jcb.122.5.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Yamano K., Kawano S. 2011. Structural insight into the mitochondrial protein import system. Biochim. Biophys. Acta. 1808:955–970 10.1016/j.bbamem.2010.07.018 [DOI] [PubMed] [Google Scholar]
- Garcia P.D., Hansen W., Walter P. 1991. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 194:675–682 10.1016/0076-6879(91)94049-I [DOI] [PubMed] [Google Scholar]
- Gebert N., Gebert M., Oeljeklaus S., von der Malsburg K., Stroud D.A., Kulawiak B., Wirth C., Zahedi R.P., Dolezal P., Wiese S., et al. 2011. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol. Cell. 44:811–818 10.1016/j.molcel.2011.09.025 [DOI] [PubMed] [Google Scholar]
- Geissler A., Krimmer T., Bömer U., Guiard B., Rassow J., Pfanner N. 2000. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b(2) modulates the deltapsi-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell. 11:3977–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A., Chacinska A., Truscott K.N., Wiedemann N., Brandner K., Sickmann A., Meyer H.E., Meisinger C., Pfanner N., Rehling P. 2002. The mitochondrial presequence translocase: An essential role of Tim50 in directing preproteins to the import channel. Cell. 111:507–518 10.1016/S0092-8674(02)01073-5 [DOI] [PubMed] [Google Scholar]
- Gevorkyan-Airapetov L., Zohary K., Popov-Celeketic D., Mapa K., Hell K., Neupert W., Azem A., Mokranjac D. 2009. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J. Biol. Chem. 284:4865–4872 10.1074/jbc.M807041200 [DOI] [PubMed] [Google Scholar]
- Glick B.S., Brandt A., Cunningham K., Müller S., Hallberg R.L., Schatz G. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 69:809–822 10.1016/0092-8674(92)90292-K [DOI] [PubMed] [Google Scholar]
- Horváth A., Berry E.A., Huang L.S., Maslov D.A. 2000. Leishmania tarentolae: A parallel isolation of cytochrome bc(1) and cytochrome c oxidase. Exp. Parasitol. 96:160–167 10.1006/expr.2000.4564 [DOI] [PubMed] [Google Scholar]
- Hutu D.P., Guiard B., Chacinska A., Becker D., Pfanner N., Rehling P., van der Laan M. 2008. Mitochondrial protein import motor: Differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol. Biol. Cell. 19:2642–2649 10.1091/mbc.E07-12-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M., Liffers S.T., Oeljeklaus S., Kuhlmann K., Röh S., Hoffmann R., Warscheid B., Hermeking H. 2011. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol. Cell. Proteomics. 10.1074/mcp.M111.010462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.J., Lee S.B., Park J.K., Yoo Y.D. 2010. TNF-α-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X(L). Cell Death Differ. 17:1420–1434 10.1038/cdd.2010.19 [DOI] [PubMed] [Google Scholar]
- Koehler C.M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335 10.1146/annurev.cellbio.20.010403.105057 [DOI] [PubMed] [Google Scholar]
- Lee S.B., Kim J.J., Kim T.W., Kim B.S., Lee M.S., Yoo Y.D. 2010. Serum deprivation-induced reactive oxygen species production is mediated by Romo1. Apoptosis. 15:204–218 10.1007/s10495-009-0411-1 [DOI] [PubMed] [Google Scholar]
- Maarse A.C., Blom J., Keil P., Pfanner N., Meijer M. 1994. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 349:215–221 10.1016/0014-5793(94)00669-5 [DOI] [PubMed] [Google Scholar]
- Marom M., Dayan D., Demishtein-Zohary K., Mokranjac D., Neupert W., Azem A. 2011. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286:43809–43815 10.1074/jbc.M111.261040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S., Neupert W., Herrmann J.M. 2005. Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J. Cell Biol. 170:881–888 10.1083/jcb.200505126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M., Wagner R., Kovermann P., Guiard B., Mick D.U., Hutu D.P., Voos W., Truscott K.N., Chacinska A., Pfanner N., Rehling P. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 312:1523–1526 10.1126/science.1127628 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfanner N., Truscott K.N. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39 [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Popov-Celeketić D., Hell K., Neupert W. 2005. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280:23437–23440 10.1074/jbc.C500135200 [DOI] [PubMed] [Google Scholar]
- Mokranjac D., Sichting M., Popov-Celeketić D., Mapa K., Gevorkyan-Airapetov L., Zohary K., Hell K., Azem A., Neupert W. 2009. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol. Biol. Cell. 20:1400–1407 10.1091/mbc.E08-09-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 1:376–386 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- Palsdottir H., Hunte C. 2003. Purification of the cytochrome bc1 complex from yeast. Membrane Protein Purification and Crystallization: A Practical Guide. Second edition. Hunte C., von Jagow G., Schägger H., Elsevier Science Publishing Co. Inc, New York: 191–203 [Google Scholar]
- Popov-Celeketić D., Mapa K., Neupert W., Mokranjac D. 2008. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27:1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov-Čeleketić D., Waegemann K., Mapa K., Neupert W., Mokranjac D. 2011. Role of the import motor in insertion of transmembrane segments by the mitochondrial TIM23 complex. EMBO Rep. 12:542–548 10.1038/embor.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Gebert M., Höpker J., Yan M., Li J., Wiedemann N., van der Laan M., Pfanner N., Sha B. 2011. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J. Mol. Biol. 411:513–519 10.1016/j.jmb.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J., Hartl F.U., Guiard B., Pfanner N., Neupert W. 1990. Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett. 275:190–194 10.1016/0014-5793(90)81469-5 [DOI] [PubMed] [Google Scholar]
- Rehling P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H.E., Kühlbrandt W., Wagner R., Truscott K.N., Pfanner N. 2003. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 299:1747–1751 10.1126/science.1080945 [DOI] [PubMed] [Google Scholar]
- Ryan K.R., Menold M.M., Garrett S., Jensen R.E. 1994. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol. Biol. Cell. 5:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.T., Voos W., Pfanner N. 2001. Assaying protein import into mitochondria. Methods Cell Biol. 65:189–215 10.1016/S0091-679X(01)65012-X [DOI] [PubMed] [Google Scholar]
- Saddar S., Dienhart M.K., Stuart R.A. 2008. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J. Biol. Chem. 283:6677–6686 10.1074/jbc.M708440200 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres M., Becker K., Bauer M.F., Walther E., Neupert W., Brunner M. 1997. Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J. Biol. Chem. 272:29963–29966 10.1074/jbc.272.47.29963 [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Pfanner N., Wiedemann N. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80:783–806 10.1016/S0091-679X(06)80036-1 [DOI] [PubMed] [Google Scholar]
- Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., Endo T. 2009. Tim23–Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184:129–141 10.1083/jcb.200808068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J., Schwöppe C., Möhlmann T., Quick P.W., Neuhaus H.E. 1998. Expression of a plastidic ATP/ADP transporter gene in Escherichia coli leads to a functional adenine nucleotide transport system in the bacterial cytoplasmic membrane. J. Biol. Chem. 273:9630–9636 10.1074/jbc.273.16.9630 [DOI] [PubMed] [Google Scholar]
- Trentmann O., Horn M., van Scheltinga A.C.T., Neuhaus H.E., Haferkamp I. 2007. Enlightening energy parasitism by analysis of an ATP/ADP transporter from chlamydiae. PLoS Biol. 5:e231 10.1371/journal.pbio.0050231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Chacinska A., Lind M., Perschil I., Sickmann A., Meyer H.E., Guiard B., Meisinger C., Pfanner N., Rehling P. 2005. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 25:7449–7458 10.1128/MCB.25.17.7449-7458.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Wiedemann N., Mick D.U., Guiard B., Rehling P., Pfanner N. 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16:2271–2276 10.1016/j.cub.2006.10.025 [DOI] [PubMed] [Google Scholar]
- van der Laan M., Meinecke M., Dudek J., Hutu D.P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., Rehling P. 2007. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9:1152–1159 10.1038/ncb1635 [DOI] [PubMed] [Google Scholar]
- Vögtle F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 139:428–439 10.1016/j.cell.2009.07.045 [DOI] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell. 21:694–707 10.1016/j.devcel.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Wachter C., Schatz G., Glick B.S. 1992. Role of ATP in the intramitochondrial sorting of cytochrome c1 and the adenine nucleotide translocator. EMBO J. 11:4787–4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., van der Laan M., Hutu D.P., Rehling P., Pfanner N. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179:1115–1122 10.1083/jcb.200709087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Esaki M., Kanamori T., Tamura Y., Nishikawa S., Endo T. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 111:519–528 10.1016/S0092-8674(02)01053-X [DOI] [PubMed] [Google Scholar]
- Yamano K., Yatsukawa Y., Esaki M., Hobbs A.E., Jensen R.E., Endo T. 2008. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283:3799–3807 10.1074/jbc.M708339200 [DOI] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S.B., Tomilin N., Castro J., Shupliakov O., Lendahl U., Nistér M. 2009. The novel conserved mitochondrial inner-membrane protein MTGM regulates mitochondrial morphology and cell proliferation. J. Cell Sci. 122:2252–2262 10.1242/jcs.038513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.