The noncatalytic RabGAP protein TBC1D14 regulates the Rab11- and ULK1-positive recycling endosomes required for autophagosome formation upon starvation
Abstract
Autophagy is a bulk degradation process characterized by the formation of double membrane vesicles called autophagosomes. The exact molecular mechanism of autophagosome formation and the origin of the autophagosomal membrane remain unclear. We screened 38 human Tre-2/Bub2/Cdc16 domain–containing Rab guanosine triphosphatase–activating proteins (GAPs) and identified 11 negative regulators of starvation-induced autophagy. One of these putative RabGAPs, TBC1D14, colocalizes and interacts with the autophagy kinase ULK1. Overexpressed TBC1D14 tubulates ULK1-positive recycling endosomes (REs), impairing their function and inhibiting autophagosome formation. TBC1D14 binds activated Rab11 but is not a GAP for Rab11, and loss of Rab11 prevents TBC1D14-induced tubulation of REs. Furthermore, Rab11 is required for autophagosome formation. ULK1 and Atg9 are found on Rab11- and transferrin (Tfn) receptor (TfnR)–positive recycling endosomes. Amino acid starvation causes TBC1D14 to relocalize from REs to the Golgi complex, whereas TfnR and Tfn localize to forming autophagosomes, which are ULK1 and LC3 positive. Thus, TBC1D14- and Rab11-dependent vesicular transport from REs contributes to and regulates starvation-induced autophagy.
Introduction
Macroautophagy (here referred to as autophagy) is an evolutionarily conserved degradative process discovered in yeast as a response to starvation but now recognized in multicellular organisms as a response to a wide range of pathological conditions (Mizushima et al., 2008). In mammalian cells, the autophagosome, a double-membrane vesicle, arises from a phagophore or isolation membrane; a subset of Atg (autophagy related) proteins localize here and work together to expand it. During expansion, the autophagosome encloses portions of cytoplasm that are targeted for degradation by fusion with endosomes and lysosomes, completing the process of autophagy.
Among the set of phagophore-localized early autophagy proteins is the ULK1/2 kinase complex (Chan and Tooze, 2009). The activity of ULK1/2 is regulated by mammalian target of rapamycin, a key nutrient sensor that, under normal growth conditions, inhibits autophagy by repressing ULK1 activity (Jung et al., 2010). The ULK1/2 complex, together with a class III phosphatidylinositol-3-kinase complex that generates the autophagy-specific phosphatidylinositol-3-phosphate pool, is the key initiator of phagophore formation and expansion (Mizushima et al., 2011). After the action of these Atg complexes, two ubiquitin-like conjugates, Atg5-12-16 and phosphatidylethanolamine-conjugated LC3 (LC3-II), are sequentially recruited to the phagophore (Itakura and Mizushima, 2010). LC3-II associates with autophagosome membranes and is used as a marker for autophagosomes.
Many questions remain about autophagosome initiation and formation, the origin of the membrane, and phagophore expansion. The membrane trafficking events involved in autophagosome biogenesis are complex, poorly understood, and likely influenced by the conditions used to induce autophagy (Longatti and Tooze, 2009).
Intracellular membrane trafficking events are regulated by the Rab family of small monomeric GTPases (Stenmark, 2009). In their GTP-bound form, Rab proteins recruit effectors to regulate vesicle trafficking. The Rab proteins involved in starvation-induced autophagosome formation remain relatively unexplored. Rab1a has been shown to be involved in antibacterial autophagosome formation, and Rab1a and Rab5 are involved in the removal of aggregated proteins (Winslow et al., 2010; Huang et al., 2011). Several Rab GTPases have also been implicated in autophagosome–lysosome fusion during starvation-induced autophagy: The best characterized is Rab7 (Gutierrez et al., 2004; Jäger et al., 2004); Rab33b has been shown to be required for autophagosome maturation, and its activity has been shown to be regulated by OATL1 (TBC1D25; Itoh et al., 2011); and in K562 cells, Rab11 is required for autophagosome–multivesicular body fusion (Fader et al., 2008).
Rab GTPase activity is controlled by guanine nucleotide exchange factors and GTPase-activating proteins (GAPs). At least 38 putative RabGAPs have been identified in humans through the presence of a Tre-2/Bub2/Cdc16 (TBC) domain (Fuchs et al., 2007), which contains two conserved catalytic residues, an arginine and a glutamine, that promote GTP hydrolysis (Barr and Lambright, 2010). Overexpression of a RabGAP can inactivate its target Rab and disrupt the trafficking step mediated by the Rab GTPase. We undertook an overexpression screen testing 38 TBC domain proteins for inhibition of LC3-II formation to identify Rab proteins involved in starvation-induced autophagosome formation. Our candidate RabGAPs were further screened by testing their ability to colocalize with ULK1, the rationale being to identify Rabs acting in early formation events. Reflecting the complexity of autophagosome formation, we identified 11 RabGAPs that inhibit autophagy when overexpressed. One of these, TBC1D14, binds ULK1 and Rab11 and disrupts recycling endosome (RE) traffic. We show that REs contain two early acting autophagy proteins, ULK1 and mAtg9, and that TBC1D14 and Rab11, in a starvation-dependent mechanism, control the transport of membrane from ULK1- and transferrin (Tfn)-positive REs to forming LC3-positive autophagosomes.
Results
A RabGAP overexpression screen identifies negative regulators of autophagy
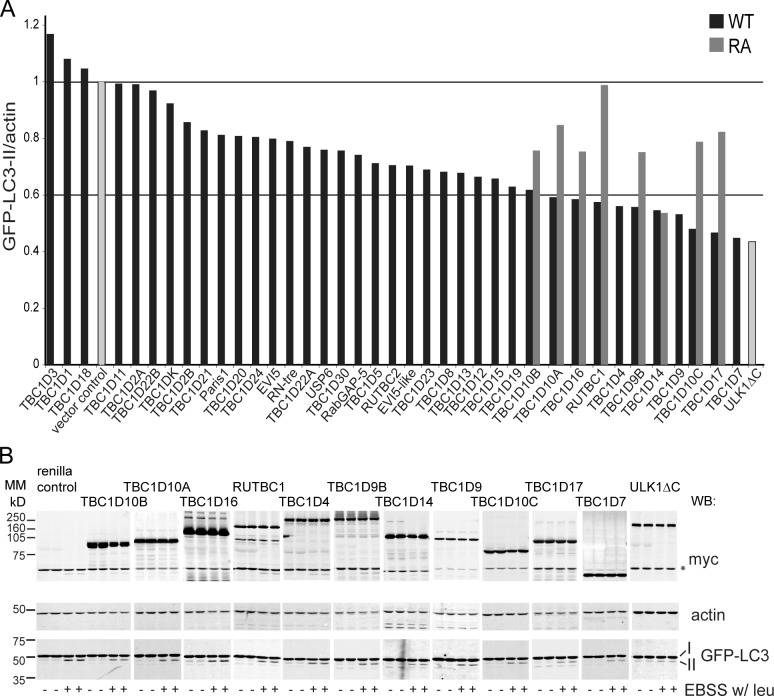
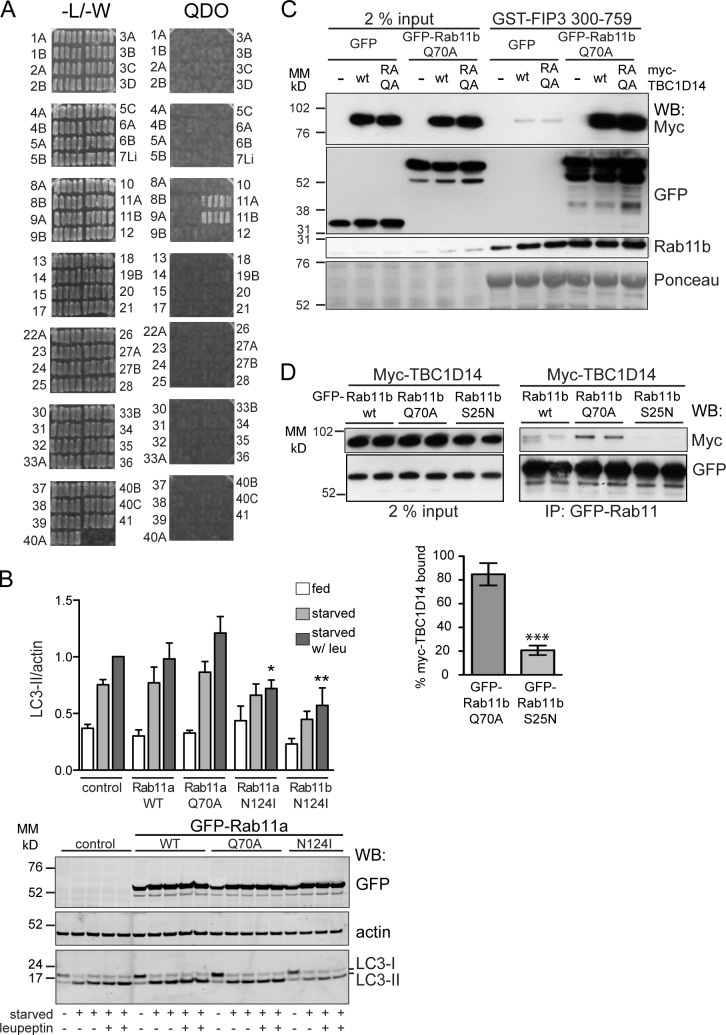
We screened a library of 38 TBC domain–containing proteins, predicted to be RabGAPs, for their ability to inhibit autophagosome formation when overexpressed in HEK293A cells stably expressing GFP-LC3 (2GL9 cells). To measure autophagosome formation, we assayed GFP–LC3-II accumulation after a 2-h starvation period (Fig. 1, A and B). We included the lysosomal protease inhibitor leupeptin in the Earle’s balanced saline solution (EBSS) starvation medium, which prevented GFP–LC3-II degradation in the lysosome, thus excluding the detection of RabGAPs that are involved in autophagosome maturation. We considered a 40% or more reduction in GFP–LC3-II as a strong inhibition: 11 RabGAPs were chosen based on this cutoff. To test whether GAP activity, and therefore Rab inactivation, was necessary for inhibition of autophagosome formation, we tested whether mutation of the catalytic arginine to alanine (RA mutant) within the TBC domain would abolish this inhibition. An arginine fitting the consensus site was identified in silico for 8 of the 11 RabGAPs, and these mutants were analyzed using GFP-LC3 lipidation. Indeed, we found that seven out of the eight RA mutants tested did not inhibit autophagosome formation to the same extent as the wild-type (WT) protein when overexpressed (Fig. 1 A).
Figure 1.
A screen of 38 human RabGAPs reveals that 11 inhibit autophagy. (A) An overexpression screen for RabGAPs, which are autophagy inhibitors. 38 myc-TBC domain–containing proteins were expressed in 2GL9 cells for 24 h, and then, cells were fed or starved in EBSS with leupeptin (leu) for 2 h. GFP–LC3-II was quantified by the Odyssey system with antibodies against LC3, actin, and myc. Black bars show levels of GFP–LC3-II after starvation normalized to the renilla vector control. Gray bars show the effect of overexpression of eight GAP activity–deficient RA mutants. The data shown are from single representative experiments. All myc-TBC transfections were repeated (n ≥ 2) and, if inhibitory or if performed with the RA mutant, repeated three or more times (n ≥ 3). ULK1ΔC, an inhibitory C-terminal truncation mutant (Chan et al., 2009) was included in each experiment. No catalytic arginine could be identified in TBC1D4, TBC1D9, or TBC1D7. Horizontal lines illustrate the value of the vector control (set to 1) and 60% value of the control. (B) Representative Western blots for myc, GFP-LC3, and actin from the 11 myc-TBC proteins in a screen that inhibited autophagy by 40% or more. The asterisk indicates a nonspecific band. MM, molecular mass; WB, Western blot.
TBC1D14 and ULK1 interact and colocalize on TBC1D14-induced tubules
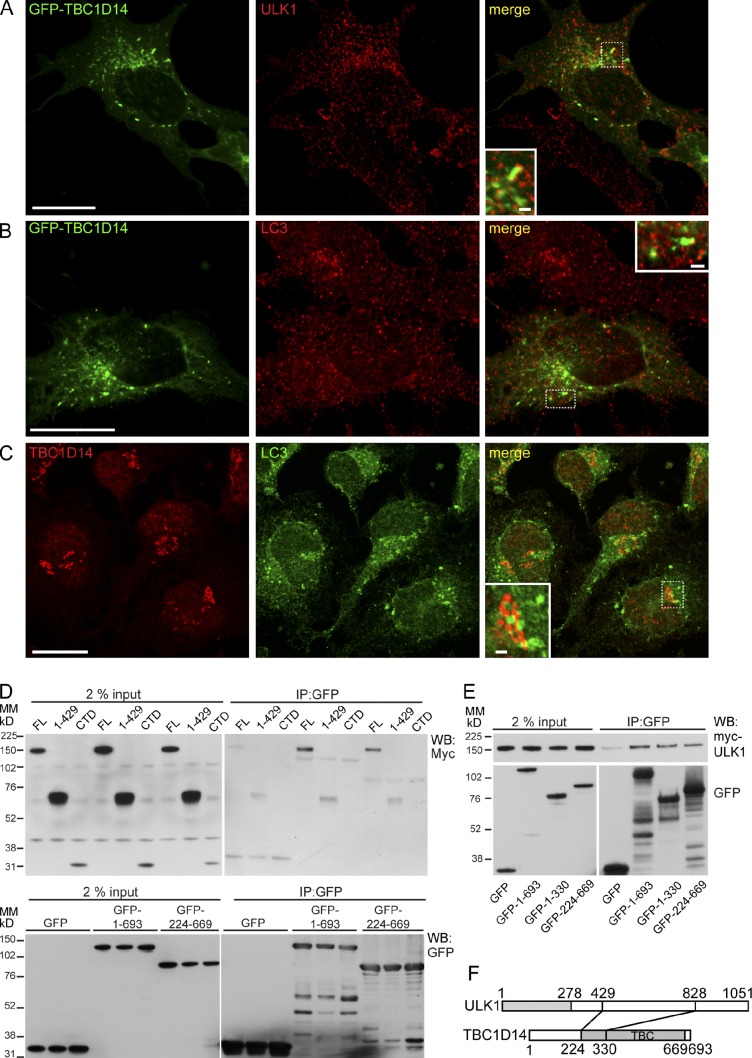
Next, we asked whether any of the 11 TBC domain proteins colocalize with autophagosome markers, as this would support the direct involvement of the RabGAP in autophagosome formation. We chose to examine colocalization of the overexpressed RabGAPs with endogenous ULK1 (Fig. S1), which is present on early autophagosomes. We found that of the RabGAPs tested, overexpressed TBC1D14 colocalizes extensively with ULK1: GFP-TBC1D14WT, RA, and RAQA mutants form tubular structures in the cytosol and recruit ULK1 to subdomains of these tubules (Fig. 2 A, Fig. S1, and not depicted). Approximately 35.3 (± 4.3)% of WT TBC1D14 colocalizes with endogenous ULK1. A small number of TBC1D14 tubules also contain low levels of LC3 (Fig. 2 B). Endogenous TBC1D14 also shows partial colocalization with LC3 (Fig. 2 C).
Figure 2.
TBC1D14 interacts with ULK1. (A) HEK293A cells transfected with GFP-TBC1D14 were fixed and stained 24 h later with the anti-ULK1 antibody. (B) HEK293A cells transfected with GFP-TBC1D14 were starved, fixed, and stained 24 h later with the anti-LC3 antibody. (C) HEK293A cells were starved for 2 h and then stained with anti-TBC1D14 and anti-LC3 antibodies. (A–C) Insets in merge show the colocalization of ULK1 and GFP-TBC1D14 on tubules. (D) GFP or GFP-tagged constructs encoding the full-length (FL; aa 1–693) or TBC domain (aa 224–669) of TBC1D14 were cotransfected into cells with myc-tagged ULK1 full-length (aa 1–1,051), aa 1–429, or C-terminal domain (CTD). Lysates were incubated with GFP-Trap resin, and bound proteins were detected by Western blotting. (E) GFP-tagged TBC1D14 as in D, plus GFP-tagged TBC1D14 aa 1–330, was cotransfected with myc-ULK1 (full length). (F) TBC1D14 (aa 224–330) is predicted to bind aa 429–828 of ULK1. Bars: (A–C, main images) 20 µm; (A–C, insets) 2 µm. IP, immunoprecipitation; MM, molecular mass; WB, Western blot.
A yeast two-hybrid assay revealed an interaction between TBC1D14 and ULK1 (Fig. S2 B). In HEK293A cells, we could detect an interaction between the TBC domain of TBC1D14 and HA-ULK1 by coimmunoprecipitation (Fig. S2, C and D). We were unable to coprecipitate the endogenous proteins possibly because the interaction is weak or the available antibodies have low affinity. TBC1D14 binds to full-length ULK1 but not the N-terminal 1–429 aa or the C-terminal domain (828–1,051 aa), suggesting the binding site is in the spacer domain (429–828 aa) of ULK1 (Fig. 2 D). The interaction between ULK1 and TBC1D14 was mapped to residues 224–330 in the TBC domain of TBC1D14 (Fig. 2, E and F; and Fig. S2 D).
TBC1D14 is a negative regulator of starvation-induced autophagy
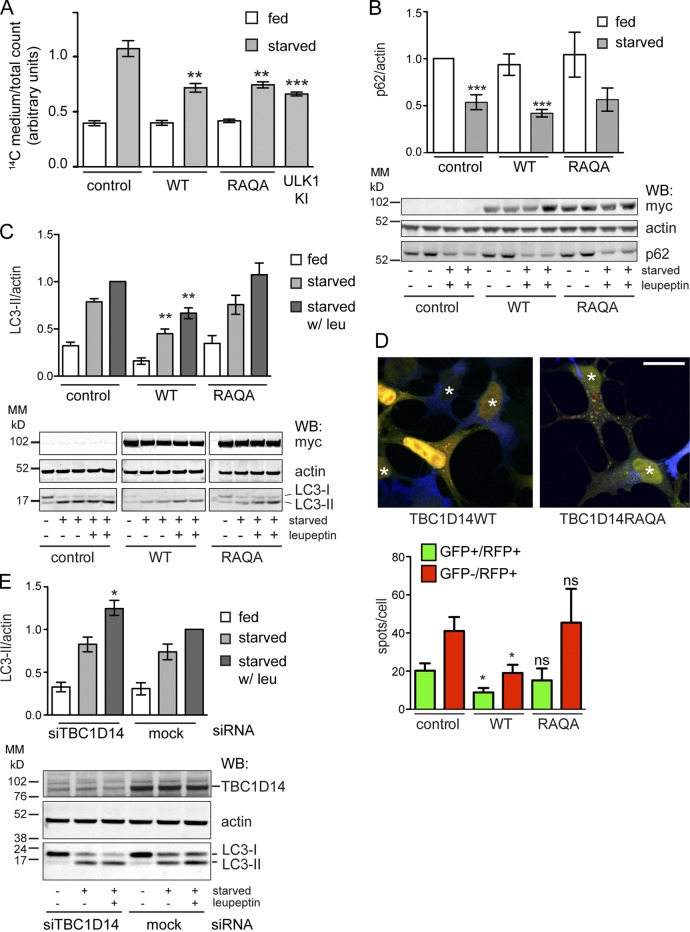
Nonselective long-lived protein degradation (LLPD) of [14C]valine-labeled proteins is a measure of autophagic flux. As shown in Fig. 3 A, we show that overexpression of TBC1D14WT inhibits starvation-induced LLPD and autophagic flux to a similar extent as the kinase mutant of ULK1, K46I (Chan et al., 2009). As the single mutant R472A (TBC1D14RA) also inhibited GFP-LC3 lipidation (Fig. 1), we mutated the second catalytic residue, glutamine 508, to alanine creating the TBC1D14R472A/Q508A (RAQA) mutant. Surprisingly, expression of TBC1D14RAQA inhibited LLPD to the same extent as WT (Fig. 3 A). We asked whether the degradation of p62, a well-characterized selective autophagic cargo (Pankiv et al., 2007), was inhibited by overexpression of WT or the RAQA mutant of TBC1D14 but saw no effect on loss of p62 (Fig. 3 B). However, although WT TBC1D14 inhibited the formation of endogenous LC3-II in starvation and in the presence of leupeptin, the TBC1D14RAQA mutant did not (Fig. 3 C). The level of autophagic inhibition was also assayed using tandem fluorescent-tagged (mRFP-GFP) LC3-expressing cells (Kimura et al., 2007). As seen in Fig. 3 D, TBC1D14WT significantly inhibited formation, but not maturation, of yellow (mRFP-GFP) LC3-positive autophagosomes, whereas the RAQA mutant did not (Fig. 3 D).
Figure 3.
TBC1D14 is a negative regulator of autophagy. (A) HEK293A cells were transfected in triplicate with vector, myc-TBC1D14WT (WT), kinase-inactive myc-ULK1K46I (ULK1 KI), or myc-TBC1D14R472AQ508A (RAQA). Cells were labeled for 18 h with [14C]valine, chased for 24 h, and then fed or starved for 2 h. LLPD was calculated as described (WT: **, P = 0.0025; RAQA: **, P = 0.0013; ULK1 kinase inactive: ***, P < 0.0001 by two-way unpaired t test compared with starved control values [n = 2]). (B) HEK293A cells were transfected and treated as in A, and p62 levels were detected by Western blotting and normalized to actin. ***, P < 0.001 by one-way analysis of variance (ANOVA) followed by Bonferroni posttest analysis comparing to the vector control. (C) HEK293A cells were transfected with control renilla vector and TBC1D14 constructs as in A and fed, starved, or starved with leupeptin (leu) for 2 h. A representative Western blot for myc, LC3, and actin is shown. Samples are separated for clarity. n = 3; **, P < 0.01 by one-way ANOVA followed by Bonferroni posttest. (D) HEK293A cells stably expressing mRFP-GFP-LC3 were transfected as in A and starved for 2 h. mRFP- and GFP-LC3 spots per cell were counted and analyzed using Imaris. Asterisks indicate myc-TBC1D14–transfected cells. Bar, 20 µm. n = 3; *, P < 0.05; result of two-way unpaired t test. (E) HEK293A cells mock or transfected with siRNA targeting TBC1D14 (catalog no. J-014032-12) for 24 h were starved with or without leupeptin for 2 h, and endogenous LC3 was measured as in B (n = 5). *, P < 0.05 by one-way ANOVA followed by Bonferroni posttest analysis and mock EBSS with leupeptin versus siTBC1D14 EBSS with leupeptin. A representative blot for TBC1D14, actin, and LC3 is shown. Error bars indicate SEM. MM, molecular mass; WB, Western blot.
LLPD measures nonselective autophagic degradation. As we show that TBC1D14WT and RAQA inhibit Tfn recycling (Fig. 6 B), we speculate that TBC1D14 may be involved in multiple membrane trafficking events, which could explain the lack of inhibition caused by the RAQA mutant on LC3 lipidation and/or trafficking. Conversely, p62 is a selective autophagic substrate, and the lack of effect of TBC1D14 overexpression on p62 suggests that p62 recruitment to LC3-II is independent of TBC1D14 function. In the presence of TBC1D14, oligomeric p62 may be efficiently recruited to the phagophore (Itakura and Mizushima, 2011), although this remains to be tested.
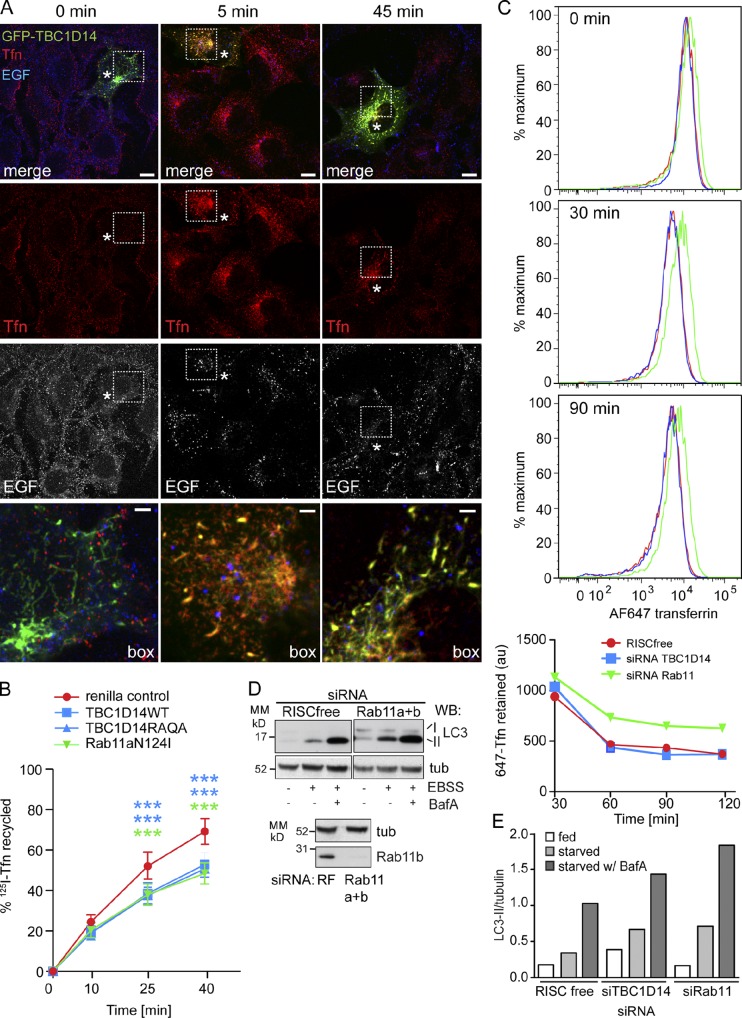
Figure 6.
Overexpression of TBC1D14 and Rab11N124I delays Tfn recycling, but only Rab11 is required for Tfn recycling. (A) HEK293A cells were transfected with GFP-TBC1D14, and 24 h later, Alexa Fluor–transferrin (Tfn) and Alexa Fluor–EGF were applied to the cells for 30 s before fixation (0 min). Alternatively, Tfn and EGF were internalized for 15 min at 37°C and followed by a 5- or 45-min chase. Asterisks indicate GFP-TBC1D14–transfected cells. Boxes are enlarged in bottom images. (B) HEK293A cells were transfected with renilla control, myc-TBC1D14, myc-TBC1D14RAQA, or GFP-Rab11aN124I. After 24 h, cells were serum starved for 2 h and fed 125I-Tfn for 20 min at 37°C and then chased in serum-free medium for 10, 25, or 40 min. The supernatant and cells containing recycled 125I-Tfn were collected, and 125I-Tfn was counted. Error bars indicate SEM (n = 3); two-way ANOVA followed by Bonferroni posttests; Renilla vector control at 45 and 60 min versus TBC1D14WT: ***, P < 0.001; versus TBC1D14RAQA: ***, P < 0.001; versus Rab11aNI: ***, P < 0.001. (C) Alexa Fluor 647–Tfn was internalized for 30 min into cells treated with RISCfree, siRNA against TBC1D14, or Rab11a and b and chased for 30, 60, and 90 min. At each time point, the cells were fixed, and the amount of Alexa Fluor 647 retained was analyzed by FACS. Representative FACS plots are shown with quantification of the geometric mean. (D and E) Cells were treated with siRNAs as in C and starved for 2 h in ESBB or EBSS plus Bafilomycin A (BafA). LC3 lipidation was quantified normalized to tubulin. A representative experiment is shown for LC3, tubulin (tub), and Rab11b. Bars: (A, main images) 10 µm; (A, insets) 2 µm. MM, molecular mass; WB, Western blot.
If TBC1D14 negatively regulates autophagy, loss of TBC1D14 should increase autophagy. Therefore, we depleted TBC1D14 with two different siRNAs and measured endogenous LC3 lipidation induced by starvation (Fig. 3 E and Fig. S2 E). In cells in which TBC1D14 levels are substantially reduced, LC3 lipidation and turnover during starvation increase significantly. TBC1D14-depleted cells showed little increase in LC3 lipidation in fed conditions, suggesting that the inhibitory role of TBC1D14 is confined to starvation-induced autophagy. This is supported by the lack of effect of TBC1D14 overexpression on the distribution of endosome and lysosome markers (Fig. S3), which when perturbed can inhibit both basal- and starvation-induced autophagic flux.
TBC1D14 binds to Rab11, which acts as a positive regulator of autophagy
To our knowledge, the target Rab GTPase for TBC1D14 remains unknown. Therefore, to identify a target Rab, we performed a yeast two-hybrid screen using TBC1D14RA as bait against a library of GTP-locked human Rab GTPases (Fuchs et al., 2005). TBC1D14 binds specifically to both Rab11a and 11b but not to any of the other 55 Rabs tested (Fig. 4 A). We then performed a GAP assay screen using a panel of 47 Rabs loaded with [32P]GTP but were unable to detect GAP activity with either recombinant GST-TBC1D14 (Fig. S4) or GFP-TBC1D14 immunoprecipitated from HEK293A cells (not depicted). Although TBC1D14 has a predicted TBC domain that contains the catalytic arginine and glutamine residues characteristic of a RabGAP (Fig. S2 A), structural analysis of the TBC domain has shown the overall spacing and position of these residues is at variance with canonical GAPs, such as Gyp1 (Tempel et al., 2008), suggesting it may not be functional.
Figure 4.
TBC1D14 binds Rab11, and inactive Rab11 inhibits autophagy. (A) Yeast two-hybrid screen to identify Rab-binding partners of TBC1D14. Yeast were transfected with the active (QA) Rab constructs (prey) and TBC1D14R472A (bait) and selected on plates lacking leucine and tryptophan (−L/−W) and medium lacking leucine, tryptophan, histidine, and adenine (QDO). Five colonies for each Rab were streaked on selective media. (B) HEK293A cells were transfected with GFP-tagged Rab11 constructs, 11aWT, 11aQ70A, 11aN124I, and 11bN124I and, 24 h later, fed or starved for 2 h. Western blot of a representative experiment with anti-GFP, anti-LC3, and antiactin and quantification of LC3-II/actin. n ≥ 3; one-way ANOVA followed by Bonferroni posttest analysis comparing to the vector control are starved with leupeptin (leu) versus Rab11aN124I starved with leupeptin: *, P < 0.05; starved with leupeptin versus Rab11bN124I starved with leupeptin: **, P < 0.01. (C) GST-FIP3 (aa 300–759) immobilized on glutathione–Sepharose beads was incubated with HEK293A lysate from cells cotransfected with either GFP or GFP-Rab11bQ70A and empty vector (EV), myc-TBC1D14, or myc-TBC1D14RAQA. Bound proteins were detected by Western blotting with anti-GFP (Rab11bQ70A) or myc (TBC1D14) antibodies. Endogenous Rab11b was detected with the anti-Rab11b antibody. Ponceau staining is the loading control. (D) HEK293A cells were transfected with myc-TBC1D14 and GFP-Rab11b constructs (WT, Q70A, and S25N). Quantification of pull-down of myc-TBC1D14 with GFP-Rab11bQ70A versus S25N (n = 3). ***, P < 0.0001 as result of two-way unpaired t test. Error bars indicate SEM. IP, immunoprecipitation; MM, molecular mass; WB, Western blot.
We hypothesized that the inhibitory effect of TBC1D14 on autophagy may be through Rab11 and investigated the effect of Rab11 and its nucleotide-binding mutants on starvation-induced autophagy (Fig. 4 B). Overexpression of WT Rab11a did not alter LC3-II levels, whereas constitutively active Rab11aQ70A slightly increased LC3 lipidation. Importantly, overexpression of the dominant-negative (DN) Rab11a or 11bN124I mutants, which are defective for GTP binding, strongly inhibits LC3 lipidation with and without leupeptin, suggesting Rab11 is required for autophagosome formation.
Regulation of Rab activity can also be detected by binding of the activated Rab protein to its effectors. Family of Rab11–interacting proteins are effectors of Rab11 (Junutula et al., 2004; Horgan and McCaffrey, 2009), so we asked whether TBC1D14 alters the binding of FIP3 to endogenous Rab11 or activated Rab11Q70A. The binding of endogenous Rab11 or constitutively active Rab11Q70A to FIP3 was not affected by overexpression of TBC1D14WT or the RAQA mutant (Fig. 4 C), supporting the data that TBC1D14 has no GAP activity. However, small amounts of TBC1D14 were detected bound to GST-FIP3, and substantially higher amounts of TBC1D14 were bound to GST-FIP3 in the presence of activated Rab11bQ70A (Fig. 4 C). We confirmed that TBC1D14 binds preferentially to Rab11bQ70A (Fig. 4 D). Our results suggest that TBC1D14 is an effector of activated Rab11. We tested whether the inhibition of autophagy we observed by overexpression of TBC1D14 was seen with another Rab11 effector, FIP3, but saw no effect of overexpression of FIP3 on LC3-II formation (Fig. S4 B). Interestingly, FIP3 tubulates the Tfn-positive RE, and Atg13, a component of the ULK1 complex, is recruited to these tubules (Fig. S4 C).
Colocalization of TBC1D14 and Rab11
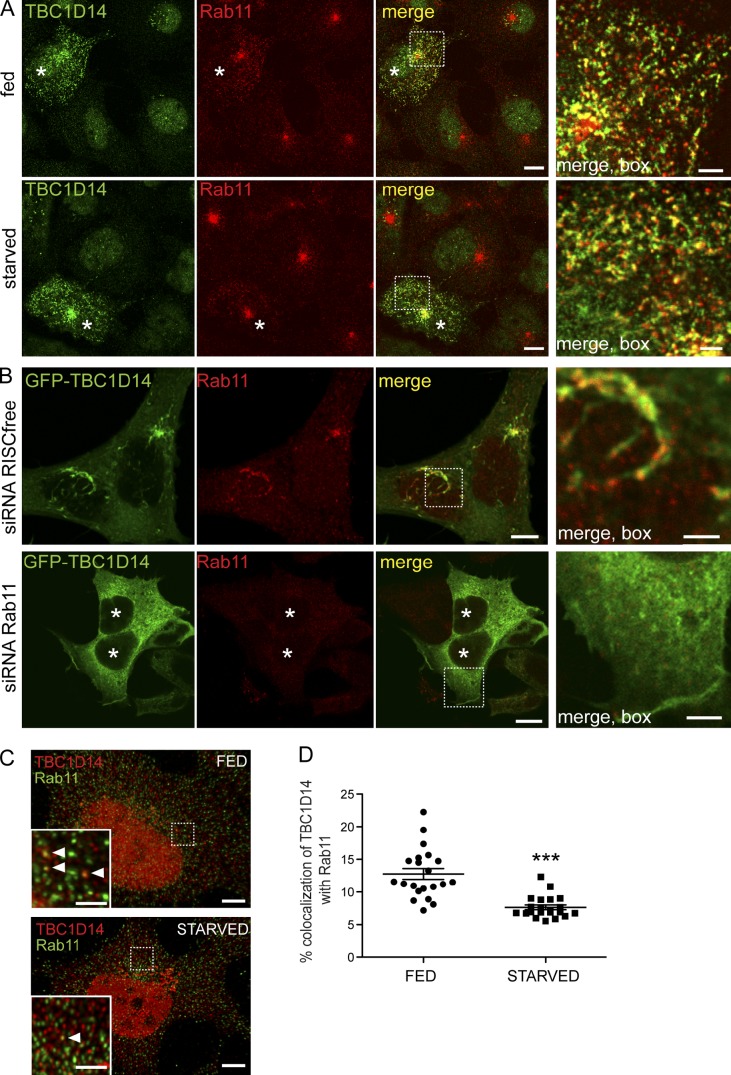
Rab11 localizes to the tubules formed in GFP-TBC1D14–expressing cells; this localization did not appear to be significantly affected by starvation (Fig. 5 A). However, the tubules formed by GFP-TBC1D14 were dependent on Rab11, as siRNA depletion of Rab11a and b compared with mock-treated cells resulted in a loss of the tubules and a diffuse localization of GFP-TBC1D14 (Fig. 5 B). Importantly, in fed cells, between 10 and 15% of TBC1D14 was found on Rab11-positive endosomes (Fig. 5, C and D).
Figure 5.
Rab11 is required for TBC1D14-induced tubulation. (A) HEK293A cells were transfected with myc-TBC1D14 and, 24 h later, fed or starved, fixed, and stained with antibodies against TBC1D14 or Rab11. (B) HEK293A cells were transfected with the indicated siRNA and transfected 24 h later with GFP-TBC1D14. After 72 h, cells were fixed and labeled with an anti-Rab11 antibody. (C) Endogenous TBC1D14 and Rab11 partially colocalize in HEK293A cells and were fed or starved for 2 h in EBSS, anti-TBC1D14, and Rab11. Multiple z stacks were deconvolved using Huygens software. TBC1D14 can be found on Rab11-positive endosomes (arrowheads in insets). (A–C) The insets show areas of colocalization between TBC1D14 and Rab11 (A), GFP-TBC1D14 and Rab11 (B), and TBC1D14 and Rab11 (C). Asterisks indicate transfected cells. (D) Colocalization between Rab11 and TBC1D14 decreases upon starvation. Images were acquired as in C. Colocalization quantified using Imaris software (***, P < 0.0001 by two-way unpaired t test). Error bars indicate SEM (n = 21 cells for each condition). Bars: (A and B, main images) 10 µm; (C, main images) 5 µm; (A–C, insets) 2 µm.
We found that endogenous TBC1D14 colocalizes with GM130, a cis/medial-Golgi marker, and TGN46, a TGN marker (Fig. S2 F). After amino acid starvation, the Golgi/TGN population significantly increased compared with fed conditions (Fig. S2, G and H), suggesting that TBC1D14 cycles between the Golgi and an endosomal pool, redistributing to the Golgi upon starvation. Consistent with the relocalization of TBC1D14 to the TGN during starvation, the colocalization of endogenous TBC1D14 and Rab11 decreased by almost 50% after 2 h of starvation (Fig. 5, C and D). These results show that the localization of TBC1D14 is sensitive to amino acid levels and that the Rab11-dependent tubulation and recruitment of ULK1 after overexpression of TBC1D14 correlates with the disruption of autophagy.
TBC1D14 perturbs recycling endosomal traffic
Rab11 regulates traffic through the RE and the endocytic recycling compartment (Stenmark, 2009). Tfn and EGF are both internalized into early endosomes, and although Tfn is efficiently recycled out of the cell via the endocytic recycling compartment, EGF is primarily targeted to the lysosome for degradation. If TBC1D14 expression alters Rab11 localization, the function of REs may be altered. To test this, we looked at endocytosis and recycling of fluorescent ligands, Tfn and EGF. In TBC1D14-transfected cells, internalized Tfn is delivered normally to TBC1D14-induced tubular structures after a 15-min uptake and a 5-min chase; however, recycling back to the plasma membrane is significantly delayed compared with untransfected cells in the same field (Fig. 6 A, red). Importantly, EGF uptake and degradation were unaffected, and no EGF could be observed in TBC1D14 tubules (Fig. 6 A).
To quantify the effect on Tfn, we assayed 125I-Tfn recycling after transfection with TBC1D14WT, TBC1D14RAQA, or Rab11aN124I. 125I-Tfn was internalized for 20 min and then chased for 10, 25, or 40 min. Endocytosis of 125I-Tfn was not affected by Rab11 or TBC1D14 overexpression. As shown in Fig. 6 B, Rab11aN124I significantly inhibited 125I-Tfn recycling, and overexpression of the TBC1D14WT or RAQA mutant inhibited Tfn recycling to the same extent as Rab11N124I. This result shows that RE function is inhibited by the TBC1D14WT or RAQA mutant, most likely as a consequence of tubulation of REs.
As ULK1 has been implicated in neuronal outgrowth and endocytosis (Tomoda et al., 2004; Zhou et al., 2007), and TBC1D14 sequestration of ULK1 on tubules may impact ULK1 function in endocytosis, we asked whether Tfn recycling was inhibited by overexpression of the DN mutant ULK1K46I: expression of ULK1K46I has no effect on 125I-Tfn uptake or recycling (Fig. S5 A). Overexpression of ULK1K46I also had no effect on the recycling of Tfn (Fig. S5 C). Thus, the decrease in autophagy we find after overexpression of TBC1D14 cannot be attributed to an inhibition of endocytic pathways as a result of loss of ULK1 function.
Loss of TBC1D14 causes an increase in autophagy, which could be a result of altered endocytosis. We assayed Alexa Fluor 647–Tfn recycling after siRNA depletion of TBC1D14 or Rab11 by FACS analysis (Fig. 6 C) and found siRNA depletion of Rab11 increased Tfn retention over a 2-h chase, whereas siRNA depletion of TBC1D14 had no effect compared with RNA-induced silencing complex (RISC) free–treated cells. Rab11 depletion increased autophagy as measured by increased LC3-II (Fig. 6, D and E). This is in contrast to the effects of DN Rab11 (Fig. 4 B), which are possibly caused by effector depletion.
TBC1D14 regulates Tfn receptor (TfnR)–positive REs, which are required for autophagosome formation
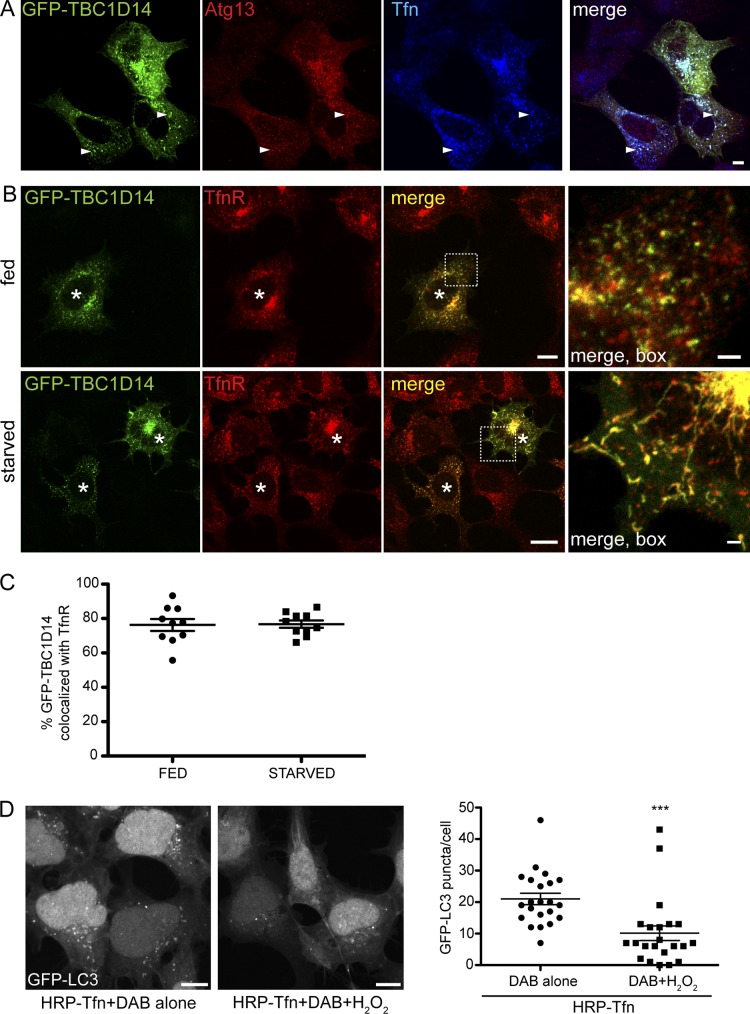
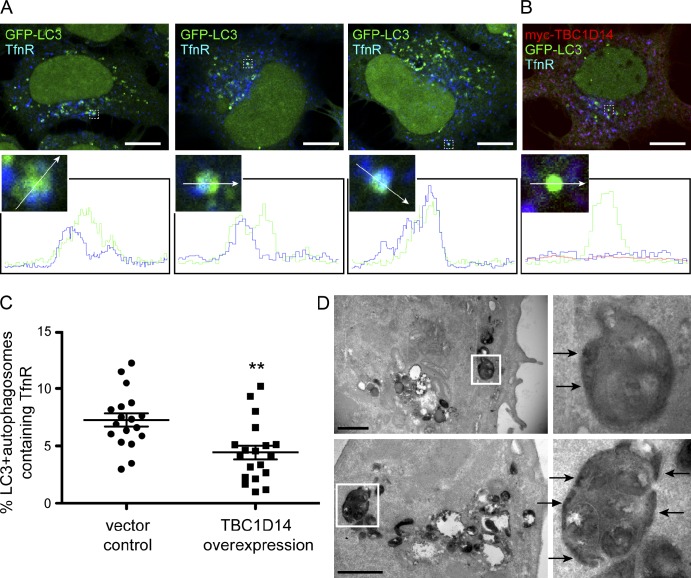
Our data support a model that the Tfn-positive RE is the target for TBC1D14-induced tubulation. Indeed, the Atg13 (Fig. 7 A)- and FIP200 (not depicted)-positive tubules induced by TBC1D14 contain the TfnR (Fig. 7 B). We quantified the extent of colocalization of overexpressed GFP-TBC1D14 and TfnR and found that the colocalization of TfnR-positive REs with GFP-TBC1D14 was independent of starvation (Fig. 7 C). Of note, tubulation and disruption of REs caused by TBC1D14 overexpression was selective to this compartment; other degradative endocytic routes seem to be unaffected (Fig. S3).
Figure 7.
TBC1D14 tubulates REs, which are required for autophagy. (A) HEK293A cells were transfected with GFP-TBC1D14, and 24 h later, Alexa Fluor 647–Tfn was internalized for 30 min. Cells were fixed and stained for Atg13. The arrows show regions of colocalization of Atg13 with Tfn on TBC1D14-induced tubules. (B) HEK293A cells were transfected with GFP-TBC1D14 and, 24 h later, fed or starved, fixed, and stained with an antibody against TfnR. The asterisks indicate transfected cells. (C) Colocalization of overexpressed GFP-TBC1D14 and TfnR was quantified using Imaris software (n = 10 cells fed and starved). (D) Ablation of Tfn-positive REs in 2GL9 cells loaded with HRP-labeled Tfn for 20 min at 37°C. Cells were washed, cooled to 4°C, and incubated for 60 min with DAB (control) or DAB and H2O2 to ablate Tfn-containing REs. Cells were incubated for 30 min at 37°C in fresh medium, starved for 2 h in EBSS, and fixed, and GFP-LC3 was imaged with a confocal microscope. (right) Quantification of GFP-LC3 puncta per cell. ***, P < 0.0001; result of two-way unpaired t test; n = 22 cells. Error bars indicate SEM. Bars, (A and B, main images; and D) 10 µm; (A and B, insets) 2 µm.
To directly demonstrate that REs are needed for the formation of autophagosomes, we used an established organelle ablation approach (Hopkins, 1983). After endosomal ablation, the cells were starved in EBSS. In controls without H2O2, the formation of GFP-LC3 puncta occurred as expected after starvation, but in the DAB/H2O2-ablated cells, the number of puncta was substantially reduced (Fig. 7 D). Cells treated with 0.003% H2O2 alone responded normally to starvation (unpublished data). Importantly, as phagophore formation can occur on or close to ER membranes (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009), ablation of RE function does not affect ER to Golgi transport (Ang et al., 2004). This result demonstrates that the function of REs and transport from the RE compartment are required for formation of autophagosomes.
REs contribute at an early step in autophagy
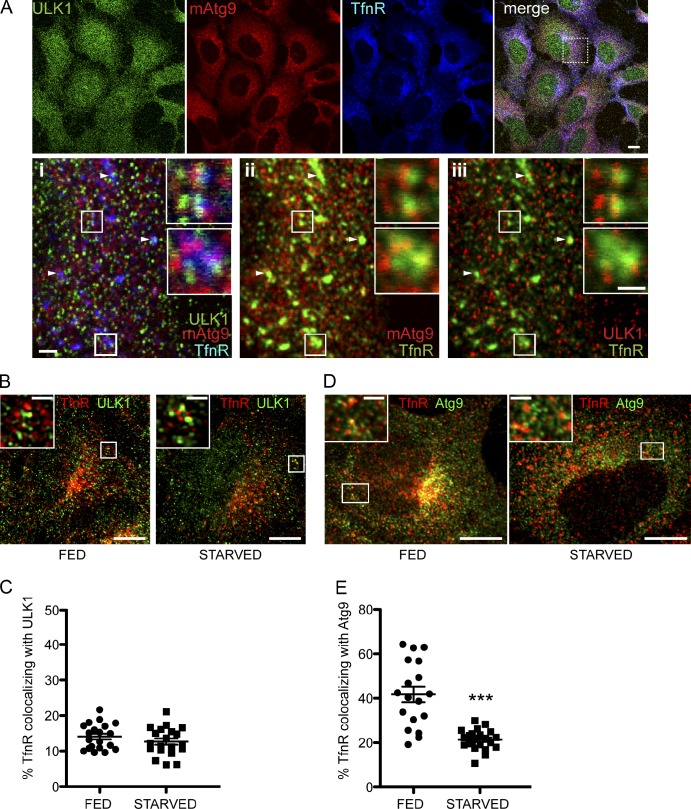
To address whether membranes derived from REs contribute to the early steps of autophagosome formation, we asked whether ULK1 and Atg9 are found on TfnR-positive REs. Mammalian Atg9 is involved in autophagosome formation (Young et al., 2006; Webber and Tooze, 2010; Orsi et al., 2012) and localizes to the Golgi and endosomes. Both endogenous ULK1 and mAtg9 are found partially colocalized with TfnR in fed cells (Fig. 8, A, B, and D). The colocalization of ULK1 and TfnR was unaffected by starvation (Fig. 8 C), but the population of TfnR that colocalized with mAtg9 significantly decreased after starvation (Fig. 8 E). These results suggest that ULK1 remains associated with the TfnR-positive RE compartment, whereas mAtg9 appears to follow an alternative route.
Figure 8.
REs are positive for ULK1 and mAtg9 and contribute membrane to autophagosomes. (A) HEK293A cells were starved for 2 h, fixed, and labeled with antibodies against ULK1, mAtg9, and TfnR. Inset i shows the triple-labeled merge. Insets ii and iii were pseudocolored: TfnR and mAtg9 are shown with ii, and ULK1 is shown in iii. The insets in i–iii indicate areas of colocalization of ULK1, mAtg9, and TfnR, mAtg9 and TfnR, and ULK1 and TfnR, respectively. Arrowheads indicate TfnR-, mAtg9-, and ULK1-positive puncta. Two enlarged images of REs are shown. (B) HEK293A cells were fed or starved for 2 h, fixed, and stained for ULK1 and TfnR. (C) Colocalization of TfnR with ULK1 was quantified from multiple z stacks and deconvolved using Huygens software using Imaris. No significant difference was detected (n = 20 cells). (D) mAtg9 is present on TfnR-positive REs in fed cells and redistributes upon starvation. HEK293A cells were fed or starved for 2 h in EBSS, fixed, and stained for mAtg9 and TfnR. (B and D) The insets indicate areas of colocalization of TfnR and ULK1 (B) and TfnR and Atg9 (D). (E) Quantification of TfnR with mAtg9 was performed as in C. ***, P < 0.0001; result of two-way unpaired t test; n = 18 cells. Error bars show SEM. Bars: (A, B, and D, main images) 10 µm; (A, i–iii; and A, i–iii, insets) 1 µm; (B and D, insets) 2 µm.
We hypothesized that the TfnR- and ULK1-positive RE contributes membrane directly to the autophagosome and tested whether we could find TfnR colocalizing with other autophagosome markers, such as GFP-LC3. Endogenous TfnR is found partially colocalizing with GFP-LC3–positive autophagosomes: in representative images (Fig. 9 A), TfnR is found at the edges of the GFP-LC3–positive structures, suggesting that TfnR and the RE membrane were incorporated into the forming autophagosome. In addition, a complete overlap of TfnR with GFP-LC3 can also be detected. Approximately 8% of the GFP-LC3–positive autophagosomal membranes colocalize with TfnR (Fig. 9 C). If this colocalization is dependent on Rab11, overexpression of TBC1D14, which disturbs Rab11–TfnR-positive REs, should decrease this colocalization. As shown in Fig. 9 (B and C), overexpression of TBC1D14 significantly decreases GFP-LC3 and TfnR colocalization.
Figure 9.
Autophagosomal membranes contain TfnR. (A and B) 2GL9 (A) or 2GL9 (B) cells transfected with myc-TBC1D14 were starved for 2 h in EBSS, fixed, and labeled with antibodies against TfnR (A) and TfnR and myc (B). Line scan profiles of GFP-LC3–positive autophagosomes were acquired using LSM software. Three representative mock-transfected cells are shown in A, and one myc-TBC1D14–expressing cell is shown in B. The enlarged areas show the colocalization between GFP-LC3 and TfnR. The arrows indicate the region that was analyzed by line scanning. Bars, 10 µm. (C) Colocalization of TfnR with GFP-LC3–positive autophagosomes is significantly reduced in myc-TBC1D14–overexpressing cells. Quantification was performed as in Fig. 7 C excluding the nuclear GFP-LC3 signal. **, P < 0.01 by two-way unpaired t test. Error bars show SEM. (D) Tfn-HRP is found in autophagosomes and appears in the intraluminal membrane space (arrows). Cells were serum starved for 1 h and incubated with Tfn-HRP for 2 h, fixed, incubated with DAB and H2O2, and embedded in Epon. Thin (70 nm) sections were imaged using a transmission electron microscope. Two representative images are shown with enlarged autophagosomes on the right. Bars, 1 µm.
Lastly, we used the Tfn-HRP-DAB reaction in fixed cells to ask whether we could visualize the contribution of the Tfn-positive RE compartment to autophagosomes by transmission EM. Both REs and autophagosomes contained the dense dark reaction product (Fig. 9 D). Importantly, the HRP reaction product was found concentrated in areas on the edge of the autophagosomes (Fig. 9 D, arrows). We therefore conclude that REs or at least the population of ULK1-positive REs is involved in autophagosome formation and contributes to autophagosome precursor membranes.
Discussion
Previous morphological data suggest a role for early endosomes and REs in autophagosome formation (Tooze et al., 1990; Liou et al., 1997). We show that REs contribute to autophagosome formation: ULK1 localizes to TfnR-positive REs, which are incorporated into forming autophagosomes upon starvation, and the delivery of this membrane requires Rab11. The TBC domain protein TBC1D14, when overexpressed, tubulates ULK1–TfnR–Rab11-positive REs, disrupting their function and inhibiting autophagy at an early step (Fig. 10). From our results, it appears that TBC1D14 has no RabGAP activity, a possibility supported by structural analysis between a canonical RabGAP, Gyp1, and TBC1D14 (Tempel et al., 2008). Nonetheless, TBC1D14 does interact with activated Rab11, and expression of the inactive Rab11N124I mutant inhibits autophagy. In all our assays, except LC3 lipidation, the RAQA and the RA mutant behaved as a WT protein, and the RAQA mutant still tubulates REs. The exact mechanism of membrane tubulation caused by the overexpressed TBC1D14 remains to be elucidated but requires Rab11.
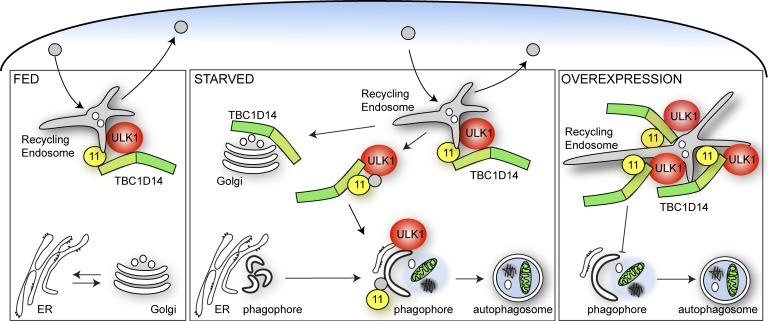
Figure 10.
Model for role of TBC1D14 and Rab11 mediating vesicular transport from the RE to the expanding phagophore. Under fed conditions, Rab11-positive REs function in recycling to the plasma membrane. Upon amino acid starvation, Rab11 mediates vesicle formation from the RE directed to forming autophagosomes. This process is negatively regulated by TBC1D14, which functions as an effector for Rab11. TBC1D14 dissociates from Rab11-positive REs in starvation and accumulates on the Golgi complex. Overexpression of TBC1D14 causes tubulation of REs, accumulation of the ULK1 complex and Rab11 on REs, and inhibition of vesicular transport from the RE.
We propose a population of ULK1 and mAtg9 resides on REs in normal conditions, and upon nutrient starvation, the ULK1–TfnR-positive REs contribute to the forming autophagosome. Our results show that overexpression of TBC1D14 tubulates REs and sequesters the ULK1 complex and TfnR, thus inhibiting autophagosome formation. Rab11 on REs is required for autophagosome formation, and we predict that Rab11 performs the function of targeting the REs to the growing autophagosome. Although we cannot exclude Rab11 inactivation as the principal mechanism of autophagy inhibition by overexpressed TBC1D14, we propose that TBC1D14 may function as a non-GAP effector for Rab11. This would be similar to the interaction between RUTCB1 and Rab9a: RUTCB1 binds Rab9a and alters Rab9-dependent processes but does not function as a Rab9a GAP (Nottingham et al., 2011). However, mAtg9 trafficking may also be disturbed, as its RE localization is decreased on TBC1D14 overexpression (unpublished data), and this could result in an inhibition of autophagy. During nutrient starvation, although ULK1 remains with the RE, mAtg9 moves away from REs and may be involved in transport of other membranes, such as the Golgi, to the growing autophagosomes (Geng et al., 2010; Ohashi and Munro, 2010; van der Vaart et al., 2010).
TBC1D14 was identified in an overexpression screen in which we tested 38 putative human RabGAPs for their effect on autophagy. Out of these, 11 RabGAPs inhibited autophagy by 40% or more, highlighting autophagosome formation as a complex, multistep process that may well require the activity of several Rab proteins. Within the group of 11 RabGAPs classified as having a substantial effect on LC3 lipidation, the Rab target of some of these has been shown to have a role in autophagy. Recently, 14 RabGAPs were identified as LC3-binding proteins (Popovic et al., 2012), and out of these 14, we identified seven as being inhibitors of GFP-LC3 lipidation, including TBC1D17, shown to bind Rab5 (Itoh et al., 2006). We also identified RUTBC1/SGSM2 (Yang et al., 2007), which binds Rab9a and was shown to be a GAP for Rab32 and Rab33b (Nottingham et al., 2011), both have been implicated in autophagy (Itoh et al., 2008; Hirota and Tanaka, 2009).
We have shown that TBC1D14 acts in the TfnR–Rab11 recycling pathway, and our data strongly support the hypothesis that TBC1D14 acts as a Rab11 effector. Overexpression of TBC1D14 may sequester Rab11 or downstream effectors, thus inhibiting vesicular transport from the RE and autophagosome formation. In support of this, we have shown that a DN mutant of Rab11 inhibits autophagy. Although we have not yet identified any other Rab targets for TBC1D14, TBC1D14 may be recruited by active Rab11 to act on another Rab immediately upstream as part of a Rab cascade. In yeast, the Rab Ypt32 recruits the RabGAP Gyp1 to inactivate the upstream Rab Ypt1 (Rivera-Molina and Novick, 2009). Intriguingly, in the same Rab cascade, Trs85, a subunit of the novel Ypt1 guanine nucleotide exchange factor TRAPPIII (transport protein particle III), is recruited to the yeast phagophore assembly site, allowing it and Ypt1 to function in autophagy (Lynch-Day et al., 2010). Rab11 is proposed to be the mammalian orthologue of Ypt32, whereas Ypt1 is thought to be the orthologue of Rab1. Thus, our future studies on the role of TBC1D14 in autophagy will encompass an examination of its effect on the TRAPPIII complex and the function of Rab11 and Rab1a.
Materials and methods
Constructs
HA-tagged ULK1 was generated by amplifying human ULK1 by PCR from a full-length IMAGE (Integrated Molecular Analysis of Genomes and their Expression) clone (ID 40080552) containing human ULK1 using 5′-GGCCGGATCCGCCACCATGGAGTACCCATACGACGTACCAGATTACGCTGGCGCCATGGAGCCCGGCCGCGGCGGC-3′ and 5′-GGCCGAATTCTCAGGCACAGATGCCAGTCAGC-3′ and inserted into pcDNA3.1 (Invitrogen). 38 human RabGAP constructs were myc-tagged and cloned into pcDNA3.1 as described previously (Fuchs et al., 2007). Mutant human myc-TBC1D14QA and myc-TBC1D14RAQA constructs were generated using a site-directed mutagenesis kit (QuikChange; Agilent Technologies) and either the WT or RA mutant vector DNA as a template for the primer 5′-CCAGATGTGGGTTATGTCGCGGGCATGTCCTTCATAGCAGC-3′. Other human RabGAPs that were mutated by QuikChange site-directed mutagenesis in pcDNA3.1 are as follows: TBC1D17R381A, TBC1D10CR141A, TBC1D14R472A, TBC1D9BR559A, RUTBC1R848A, TBC1D16R494A, TBC1D10AR160A, and TBC1D10BR134A.
Myc-tagged mouse ULK1ΔC (aa 1–828), C-terminal domain (aa 829–1,051), aa 1–427, and ULK1K46I cloned into pRK5 have been described previously (Chan et al., 2007, 2009). GFP-tagged constructs were made in the pEGFP-C2 vector (Takara Bio Inc.). Human GFP-tagged TBC1D14 aa 1–330, aa 1–223, and aa 224–669 were generated by PCR amplification from human GFP-TBC1D14 (Fuchs et al., 2007), and point mutations were introduced using the QuikChange method. pGEX-4T1 human FIP3 300–759 and GFP-tagged FIP3 cloned in pEGFP, identified as KIAA0665, and originally called Arfophilin (Wilson et al., 2005) were gifts from R. Prekeris (University of Colorado, Denver, CO).
Cell culture, starvation, and transfections
The HEK293A cell line stably expressing GFP-LC3 is a stable cell line generated by transfection of the plasmid DNA encoding GFP-tagged rat LC3 in EGFP-C1 followed by G418 selection, single-cell cloning, and screening of stably cells expressing GFP-LC3 to obtain a single-cell clone called 2GL9 (Chan et al., 2007). All cells were cultured in DME (Invitrogen) with 10% FBS (Sigma-Aldrich). Where cells are indicated as fed, they were grown in this medium. Starvation medium consisted of EBSS and, where indicated, contained 0.25 mg/ml (0.5 mM) leupeptin (Roche). For biochemical analyses, cells were transfected using Lipofectamine 2000 (Invitrogen), and for immunofluorescence imaging, cells were transfected using FuGENE 6 (Roche), both according to the manufacturers’ instructions.
RNA interference
All siRNAs were purchased from Thermo Fisher Scientific, and cells were transfected using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. Knockdown was performed for a 72-h period with HEK293A cells being transfected twice, on day 1 and 2, or mock transfected. For ULK1 and TBC1D14, 50 nm siRNA was used for each transfection, and for Rab11a and 11b, 25 nm of each duplex was used together. Cells were then left in full medium for 24 h and treated as indicated.
TBC1D14 silencing was performed using siRNA duplexes (catalog no. J-014032-12 and J-014032-11) targeting the following sequences: 5′-UCACAGAAAUGAACCGUUU-3′ and 5′-GGAAAGCGCCACUCCGAGA-3′. ULK1 was silenced with an siRNA duplex (no. D-005049-04-0010) targeting the following sequence: 5′-UGUAGGUGUUUAAGAAUUG-3′. Rab11a was targeted with a siRNA duplex (no. D-004726-01) with the following sequence: 5′-GUAGGUGCCUUAUUGGUUU-3′. Rab11b was targeted with a siRNA duplex (no. D-004727-03-0005) with the following sequence: 5′-GACAGAAGCCCAACAAGCU-3′.
Antibodies
Antibodies used were rabbit anti-ULK1 (1:250; Santa Cruz Biotechnology, Inc.), mouse antimyc clone 9E10 (Cancer Research UK), rabbit anti–β-actin (Abcam), mouse antiactin clone AC40 (Sigma-Aldrich), mouse anti-LC3 clone 5F10 (nanoTools), rabbit anti-ATG13 (Sigma-Aldrich), mouse anti-HA (Covance), rabbit anti-GFP clone SGE5/6 (Cancer Research UK), mouse anti-GFP clone 3E1 (Cancer Research UK), mouse anti-GM130 (BD), sheep anti-TGN46 (AbD Serotec), mouse anti-Rab11 (BD), mouse anti-Rab11b (Cell Signaling Technology), mouse anti-TfnR clone 13E4 (Abcam), rabbit anti–cation-independent mannose-6-phosphate receptor (CI-MPR; Dittié et al., 1999), mouse anti-EEA1 (BD), mouse anti-LAMP2 (BD), hamster anti-mAtg9 (Webber and Tooze, 2010), and rabbit anti-Rab7 (Cell Signaling Technology). Rabbit anti-TBC1D14 was generated by injecting rabbits with a C-terminal peptide consisting of the aa 671–691 of TBC1D14 with the sequence CLTALQKDSREMEKGSPSLR.
Immunofluorescence
Cells were grown on poly-d-lysine (Sigma-Aldrich)–coated coverslips and transfected with FuGENE 6, and 24 h later, cells were starved. Cells were washed with PBS at 37°C once, fixed in 4% PFA in PBS at 37°C for 20 min, washed with PBS three times, quenched with 50 nM NH4Cl/PBS for 10 min, permeabilized with 0.2% Triton X-100 in PBS for 3 min, and blocked with 0.2% gelatin in PBS (PBS-G) for 20 min. Cells were incubated with primary antibodies diluted in PBS-G for 20 min, washed with PBS-G, incubated with secondary antibodies diluted in PBS-G for 20 min, washed with PBS-G, PBS, and sterile water, and then mounted on microscope slides with Mowiol 488 mounting medium. Confocal images with a slice thickness of 0.7 µm were obtained with a microscope (Axioplan 2; Carl Zeiss) using a 63× oil immersion objective, and images were processed using LSM 510 or LSM 710 software (Carl Zeiss). For deconvolution, five z stacks 0.33 µm apart were acquired with a microscope (LSM 710) as previously described in this paragraph. Image stacks were deconvolved using Huygens software (Scientific Volume Imaging). Colocalization was quantified using Imaris software (Bitplane) with all images processed identically within one experiment. Unspecific (TBC1D14) or irrelevant (GFP-LC3) nuclear labeling was excluded for quantification.
Immunoblotting
HEK293A cells were lysed in TNTE (20 mM Tris, pH 7.5, 150 mM NaCl, 0.3% vol/vol Triton X-100, and 5 mM EDTA) containing EDTA-free protease inhibitors cocktail (Complete; Roche) and phosphatase inhibitors (PhosSTOP; Roche) for 5 min on ice. Cell debris was pelleted at 2,500 g for 5 min at 4°C. Lysates were mixed with 5× SDS sample buffer, heated to 65°C for 15 min, and then analyzed on 4–12% Bis-Tris NuPAGE gels in MES buffer (Invitrogen).
2GL9 cells were lysed in SDS Laemmli sample buffer, heated to 65°C for 5–10 min, passed through a 27-gauge needle eight times to reduce viscosity before analysis on 10% Laemmli SDS-PAGE, and transferred to polyvinylidene difluoride membranes. After incubation with primary antibodies, signals were detected and quantified using secondary antibodies coupled to HRP and ECL or infrared chromophores and a two-channel scanning method (Odyssey; LI-COR Biosciences) as previously described (Chan et al., 2007).
Immunoprecipitation
Doubly transfected HEK293A cells were lysed in TNTE as described in the previous section. Cleared lysates were incubated with protein G–Sepharose beads (Sigma-Aldrich) coupled to either anti-HA (Covance) or anti-FLAG (mouse M2; Sigma-Aldrich) for ≥1 h at 4°C on a rotating wheel, the beads were washed three times with TNTE, and proteins were eluted using 3× SDS sample buffer, heated to 65°C for 15 min, and analyzed on 4–12% Bis-Tris NuPAGE gels in MOPS buffer (Invitrogen).
GFP-Trap
10-cm plates of HEK293A cells expressing the indicated GFP fusions were lysed in 400 µl lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.5% NP-40) and cleared by centrifugation at 16,000 g for 15 min at 4°C. The lysates were precleared against blocked agarose beads (Chromotek) and incubated with GFP-Trap resin (Chromotek) for 1 h at 4°C. Immobilized complexes were washed with wash buffer (10 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 0.5 mM EDTA), eluted from the beads in 30 µl of 2× Laemmli sample buffer for 5 min at 100°C, and analyzed by SDS-PAGE and immunoblotting.
GST fusion protein preparation
400-ml cultures of BL21 (DE3) Star Escherichia coli (Invitrogen) harboring pGEX-4T1 FIP3 300–759 were grown to OD600 = 0.6 at 37°C, and protein expression was induced for 4 h. Cells were harvested in PBS and lysed by sonication, and insoluble components were removed at 17,000 g. The cleared lysate was mixed with 600 µl glutathione–Sepharose 4B (GE Healthcare) for 1 h at 4°C to immobilize GST-FIP3 300–759, and the beads were washed with cold PBS and used in pull-down assays.
GST pull-down
10-cm dishes of HEK293A cells expressing the appropriate proteins were lysed in 400 µl lysis buffer (150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 20 mM Hepes, pH 7.4, and 0.1% Triton X-100 supplemented with Complete EDTA-free protease inhibitor cocktail and PhosSTOP phosphatase inhibitors) for 15 min on ice. Insoluble debris was pelleted at 16,000 g for 15 min. The lysates were mixed with 20 µl of a 50% glutathione–Sepharose bead slurry harboring immobilized GST-FIP3 300–759 at 4°C for 2 h. The beads were washed three times with lysis buffer, and bound proteins were eluted in 2× SDS Laemmli sample buffer and analyzed by Western blotting.
LLPD
Autophagy-dependent degradation of [14C]valine-labeled cellular proteins was measured in transfected HEK293A as previously described (Chan et al., 2007). In brief, HEK293A cells were transfected for 6 h and then labeled overnight with 0.2 µCi/ml [14C]valine in DME containing 10% dialyzed FCS and 65 µM valine and chased for 24 h with full medium containing 2 mM valine. The medium was collected, and the cells were lysed and harvested in 1% Triton X-100 in PBS. The extent of LLPD was expressed as the percentage of the TCA-soluble counts in the medium compared with the total TCA-soluble counts in the medium plus TCA-insoluble counts from the cells and normalized across experiments containing the same controls to control-treated starved cells.
HRP ablation of REs
REs in 2GL9 cells were loaded with HRP-labeled Tfn (a gift from C. Futter, Institute of Ophthalmology, University College London, London, England, UK) in full growth medium for 20 min at 37°C. Cells were then washed once with PBS, cooled to 4°C, and incubated for 60 min with 0.1 mg/ml DAB as a control or 0.1 mg/ml DAB and 0.003% H2O2 to ablate Tfn-containing RE (Hopkins, 1983; Ang et al., 2004; Henry and Sheff, 2008). The medium was removed, and the cells were washed four times with PBS plus 1% BSA at 4°C to quench the HRP, DAB, and H2O2, replaced with full medium at 37°C, incubated for 30 min to recover, and then starved for 2 h in EBSS and fixed. GFP-LC3 was imaged with a confocal microscope (LSM 510), and the number of GFP-LC3 puncta was counted from 22 cells per condition from two experiments.
Yeast two-hybrid
The yeast two-hybrid assay was performed as previously described (Haas et al., 2005).
Statistical analysis
Statistical analysis was performed using Prism (GraphPad Software) using the indicated tests. Data represent means of the indicated number of independent experiments. Error bars indicate SEM. P < 0.05 was considered to be significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Fluorescent Tfn and EGF trafficking
Transfected HEK293A cells plated on poly-d-lysine–coated coverslips were serum starved for 2 h and then incubated in 37°C warm serum-free DME containing 1 mg/ml Alexa Fluor 647–labeled Tfn and 1 mg/ml Alexa Fluor 555–labeled EGF for 15 min at 37°C, washed once with 37°C PBS, and then chased for the indicated time in serum-free medium in cell culture conditions at 37°C. Cells were then fixed with 4% PFA warmed to 37°C for 20 min and mounted on microscope slides in Mowiol 4–88.
125I-Tfn recycling
Transfected or siRNA-treated HEK293A cells plated in poly-d-lysine–coated wells were serum starved for 2 h and then incubated with 0.3 µg/ml 125I-Tfn (PerkinElmer) in binding medium (0.05% BSA in DME) for 10 min at 37°C. After each time point, the medium was collected and replaced by fresh medium for the subsequent time point. Cells were lysed in PBS containing 1% Triton X-100, and the radioactivity in the chase medium and the cell lysates was counted using a multipurpose scintillation counter (LS 6500; Beckman Coulter). Data presented include the stimulation period. The percentage of recycled 125I-Tfn was determined by dividing the cpm in the medium (adding all counts from previous time points if applicable) by the total cpm (lysate + medium).
FACS analysis
For flow cytometry analysis of Tfn recycling, cells were incubated with Alexa Fluor 647–Tfn in EBSS for 30 min, and then, the medium was removed and replaced with fresh EBSS for different lengths of time to chase out the internalized Tfn. The time presented in the figures represents the 30-min uptake plus the chase time. At the end of the experiment, cells were placed on ice, trypsinized, and fixed with 4% PFA. The amount of fluorescent Tfn was measured using a cell analyzer (LSRFortessa; BD) using excitation of 633 nm and emission of a 670/14-nm band pass filter.
Online supplemental material
Fig. S1 shows the colocalization of the 11 most inhibitory overexpressed myc-TBC proteins with endogenous ULK1. Fig. S2 shows the domain structure of TBC1D14 and its interaction with ULK1 by yeast two-hybrid and coimmunoprecipitation. Fig. S3 shows that TBC1D14 does not significantly colocalize with endocytic markers, including CI-MPR, EEA1, Lamp2, or Rab7. Fig. S4 shows that TBC1D14 has no GAP activity toward a panel of GTP-loaded mammalian Rab proteins. Fig. S5 shows that overexpression of ULK1K46I does not cause inhibition of Tfn recycling. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111079/DC1.
Supplementary Material
Acknowledgments
S.A. Tooze and A. Longatti thank Dr. G. Schiavo for his invaluable support and advice throughout the project and members of the Secretory Pathways Laboratory at the London Research Institute. We are grateful to Prof. Clare Futter for supplying Tfn-HRP and expert advice.
F.A. Barr and S. Yoshimura acknowledge support from the Deutsche Forschungsgemeinschaft and the Wellcome Trust. S.A. Tooze acknowledges Cancer Research UK for generous support.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- CI-MPR
- cation-independent mannose-6-phosphate receptor
- DN
- dominant negative
- EBSS
- Earle’s balanced saline solution
- GAP
- GTPase-activating protein
- LLPD
- long-lived protein degradation
- mRFP
- monomeric RFP
- RE
- recycling endosome
- RISC
- RNA-induced silencing complex
- TBC
- Tre-2/Bub2/Cdc16
- Tfn
- transferrin
- TfnR
- Tfn receptor
- WT
- wild type
References
- Ang A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., Mellman I. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F., Lambright D.G. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y.W., Tooze S.A. 2009. Evolution of Atg1 function and regulation. Autophagy. 5:758–765 [DOI] [PubMed] [Google Scholar]
- Chan E.Y., Kir S., Tooze S.A. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282:25464–25474 10.1074/jbc.M703663200 [DOI] [PubMed] [Google Scholar]
- Chan E.Y., Longatti A., McKnight N.C., Tooze S.A. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29:157–171 10.1128/MCB.01082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittié A.S., Klumperman J., Tooze S.A. 1999. Differential distribution of mannose-6-phosphate receptors and furin in immature secretory granules. J. Cell Sci. 112:3955–3966 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250 10.1111/j.1600-0854.2007.00677.x [DOI] [PubMed] [Google Scholar]
- Fuchs E., Short B., Barr F.A. 2005. Assay and properties of rab6 interaction with dynein-dynactin complexes. Methods Enzymol. 403:607–618 10.1016/S0076-6879(05)03053-3 [DOI] [PubMed] [Google Scholar]
- Fuchs E., Haas A.K., Spooner R.A., Yoshimura S., Lord J.M., Barr F.A. 2007. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J. Cell Biol. 177:1133–1143 10.1083/jcb.200612068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D.J. 2010. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2257–2269 10.1091/mbc.E09-11-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I. 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117:2687–2697 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- Haas A.K., Fuchs E., Kopajtich R., Barr F.A. 2005. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 7:887–893 10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- Henry L., Sheff D.R. 2008. Rab8 regulates basolateral secretory, but not recycling, traffic at the recycling endosome. Mol. Biol. Cell. 19:2059–2068 10.1091/mbc.E07-09-0902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Tanaka Y. 2009. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell. Mol. Life Sci. 66:2913–2932 10.1007/s00018-009-0080-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C.R. 1983. Intracellular routing of transferrin and transferrin receptors in epidermoid carcinoma A431 cells. Cell. 35:321–330 10.1016/0092-8674(83)90235-0 [DOI] [PubMed] [Google Scholar]
- Horgan C.P., McCaffrey M.W. 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37:1032–1036 10.1042/BST0371032 [DOI] [PubMed] [Google Scholar]
- Huang J., Birmingham C.L., Shahnazari S., Shiu J., Zheng Y.T., Smith A.C., Campellone K.G., Heo W.D., Gruenheid S., Meyer T., et al. 2011. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 7:17–26 10.4161/auto.7.1.13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Mizushima N. 2010. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 6:764–776 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Mizushima N. 2011. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192:17–27 10.1083/jcb.201009067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Satoh M., Kanno E., Fukuda M. 2006. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 11:1023–1037 10.1111/j.1365-2443.2006.00997.x [DOI] [PubMed] [Google Scholar]
- Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M. 2008. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 19:2916–2925 10.1091/mbc.E07-12-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Kanno E., Uemura T., Waguri S., Fukuda M. 2011. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 192:839–853 10.1083/jcb.201008107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.-L. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- Jung C.H., Ro S.-H., Cao J., Otto N.M., Kim D.-H. 2010. mTOR regulation of autophagy. FEBS Lett. 584:1287–1295 10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula J.R., Schonteich E., Wilson G.M., Peden A.A., Scheller R.H., Prekeris R. 2004. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 279:33430–33437 10.1074/jbc.M404633200 [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Liou W., Geuze H.J., Geelen M.J.H., Slot J.W. 1997. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 136:61–70 10.1083/jcb.136.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R., Brumell J.H., Ferro-Novick S., Klionsky D.J. 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA. 107:7811–7816 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Nottingham R.M., Ganley I.G., Barr F.A., Lambright D.G., Pfeffer S.R. 2011. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J. Biol. Chem. 286:33213–33222 10.1074/jbc.M111.261115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Munro S. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 21:3998–4008 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A., Razi M., Dooley H.C., Robinson D., Weston A.E., Collinson L.M., Tooze S.A. 2012. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol. Biol. Cell. 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Popovic D., Akutsu M., Novak I., Harper J.W., Behrends C., Dikic I. 2012. Rab GTPase-Activating Proteins in Autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32:1733–1744 10.1128/MCB.06717-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Molina F.E., Novick P.J. 2009. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. USA. 106:14408–14413 10.1073/pnas.0906536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Tempel W., Tong Y., Dimov S., Bochkarev A., Park H. 2008. First crystallographic models of human TBC domains in the context of a family-wide structural analysis. Proteins. 71:497–502 10.1002/prot.21885 [DOI] [PubMed] [Google Scholar]
- Tomoda T., Kim J.H., Zhan C., Hatten M.E. 2004. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 18:541–558 10.1101/gad.1151204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Hollinshead M., Ludwig T., Howell K., Hoflack B., Kern H. 1990. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J. Cell Biol. 111:329–345 10.1083/jcb.111.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A., Griffith J., Reggiori F. 2010. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2270–2284 10.1091/mbc.E09-04-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber J.L., Tooze S.A. 2010. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 29:27–40 10.1038/emboj.2009.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.M., Fielding A.B., Simon G.C., Yu X., Andrews P.D., Hames R.S., Frey A.M., Peden A.A., Gould G.W., Prekeris R. 2005. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol. Biol. Cell. 16:849–860 10.1091/mbc.E04-10-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow A.R., Chen C.-W., Corrochano S., Acevedo-Arozena A., Gordon D.E., Peden A.A., Lichtenberg M., Menzies F.M., Ravikumar B., Imarisio S., et al. 2010. α-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J. Cell Biol. 190:1023–1037 10.1083/jcb.201003122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Sasaki T., Minoshima S., Shimizu N. 2007. Identification of three novel proteins (SGSM1, 2, 3) which modulate small G protein (RAP and RAB)-mediated signaling pathway. Genomics. 90:249–260 10.1016/j.ygeno.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 5:1180–1185 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Young A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhou X., Babu J.R., da Silva S., Shu Q., Graef I.A., Oliver T., Tomoda T., Tani T., Wooten M.W., Wang F. 2007. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. USA. 104:5842–5847 10.1073/pnas.0701402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.