Abstract
Natural product-derived bengamides possess potent antiproliferative activity and target human methionine aminopeptidases for their cellular effects. Using bengamides as a template, several derivatives were designed and synthesized as inhibitors of methionine aminopeptidases of Mycobacterium tuberculosis, and initial antitubercular activity were observed. Here, we present three new X-ray structures of the tubercular enzyme MtMetAP1c in complex with the inhibitors in the Mn(II) form and in the Ni(II) form. All amide moieties of the bengamide derivatives bind to the unique shallow cavity and interact with a flat surface created by His-212 of MtMetAP1c in the Mn(II) form. However, the active site metal has significant influence on the binding mode, because the amide takes a different conformation in the Ni(II) form. The interactions of these inhibitors at the active site provide the structural basis for further modification of these bengamide inhibitors for improved potency and selectivity.
Keywords: Metalloenzyme, Drug discovery, Tuberculosis, Antibiotic, X-ray structure
1. Introduction
Tuberculosis is a deadly disease caused by mycobacterial infection, and Mycobacterium tuberculosis is the major tuberculosis pathogen in human. Now, multidrug-resistant and extensively drug-resistant tuberculosis is happening at an alarming rate [1]. To overcome the drug resistance, new antibiotics with a novel mechanism of action are urgently needed. Methionine aminopeptidase (MetAP) is ubiquitous and carries out N-terminal methionine excision from majority of newly synthesized proteins [2]. The importance of this cotranslational modification is underscored by lethality of gene deletion in bacteria, such as Escherichia coli [3] and Salmonella typhimurium [4]. Therefore, MetAP is a potential target to develop novel antibacterial drugs [5].
M. tuberculosis has two MetAP genes (annotated as mapA and mapB in H37Rv genome). The protein from the mapB gene of M. tuberculosis, named MtMetAP1c, was purified, and its structures in apoform and in complex with methionine were reported [6]. The structural analysis revealed a SH3 binding motif in its N-terminus, and potential interaction with ribosome through this motif to facilitate cotranslational methionine excision was proposed [6]. We recently further characterized this enzyme for metal activation and inhibition and described three X-ray structures with small molecule inhibitors bound [7]. The other MetAP (from the mapA gene) of M. tuberculosis, named MtMetAP1a, is shorter at the N-terminus and has no such SH3 binding motif. No structural information for MtMetAP1a has been reported. Both MetAPs of M. tuberculosis were active as enzymes when purified [7–9], and their mRNA transcripts were analyzed and showed different levels in log phase and stationary phase [8]. MtMetAP1a gene (mapA) expressed more in log phase, while MtMetAP1c gene (mapB) showed higher level in stationary phase. It was concluded that the two MetAPs may perform an important function in different growth phases of M. tuberculosis [8]. The special characteristics of mycobacterial life cycle may require more than one MetAP enzyme to carry out the important cotranslational modification. Based on comparison of mycobacterial genomes, it was predicted that both MtMetAP1a and MtMetAP1c are essential for M. tuberculosis survival in vivo and pathogenicity [10].
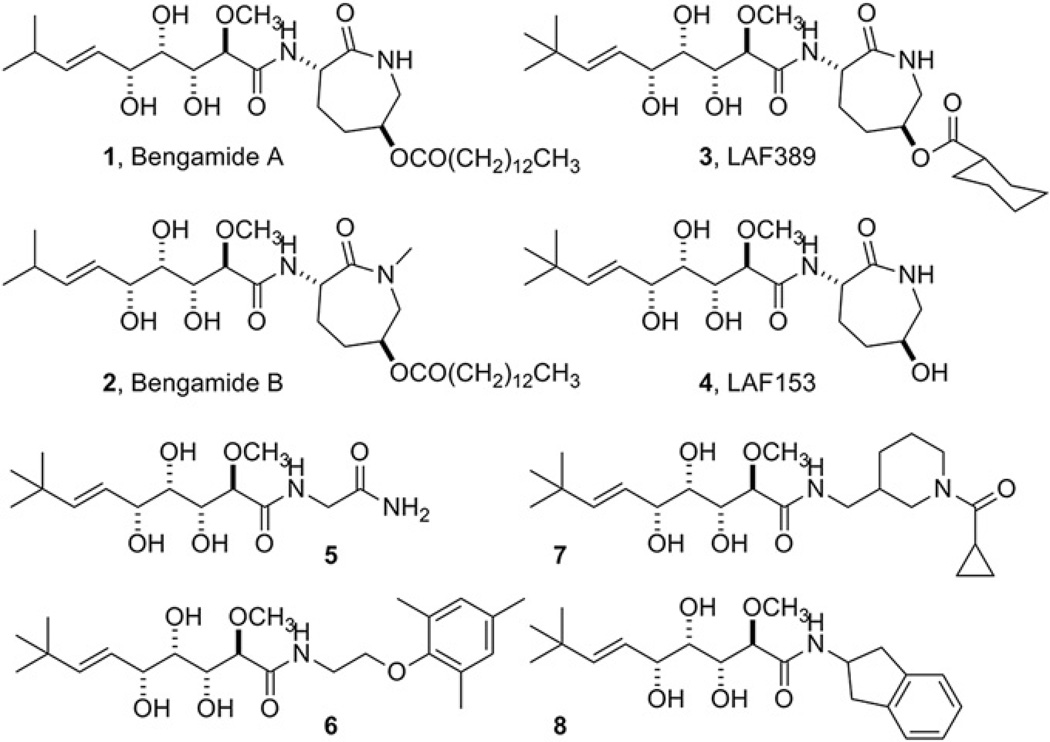
Eukaryotic cells usually have two MetAPs. Deletion of either of the two MetAP genes in Saccharomyces cerevisiae rendered a slow growth phenotype, and lethality was observed only when both genes were deleted [11]. Bengamides are natural products that were isolated from marine sponge [12]. Bengamides A and B (1 and 2 in Fig. 1) showed nanomolar potency again cancer cell lines [13,14], and bengamides arrest cells at the G1 and G2/M phases of the cell cycle [13,15]. A clinical trial was carried out for anticancer therapy, using the synthetic derivative LAF389 [16] (3). Human MetAP1 and MetAP2 were identified as the cellular targets of bengamides by a proteomic approach [17], and bengamides showed no selectivity in inhibition between the two human MetAP enzymes [17].
Fig. 1.
Chemical structures of natural bengamides (1 and 2) and their synthetic derivatives (3 and 4). Compounds 5–9 are some of the newly designed and synthesized bengamide derivatives, used in the X-ray structural studies.
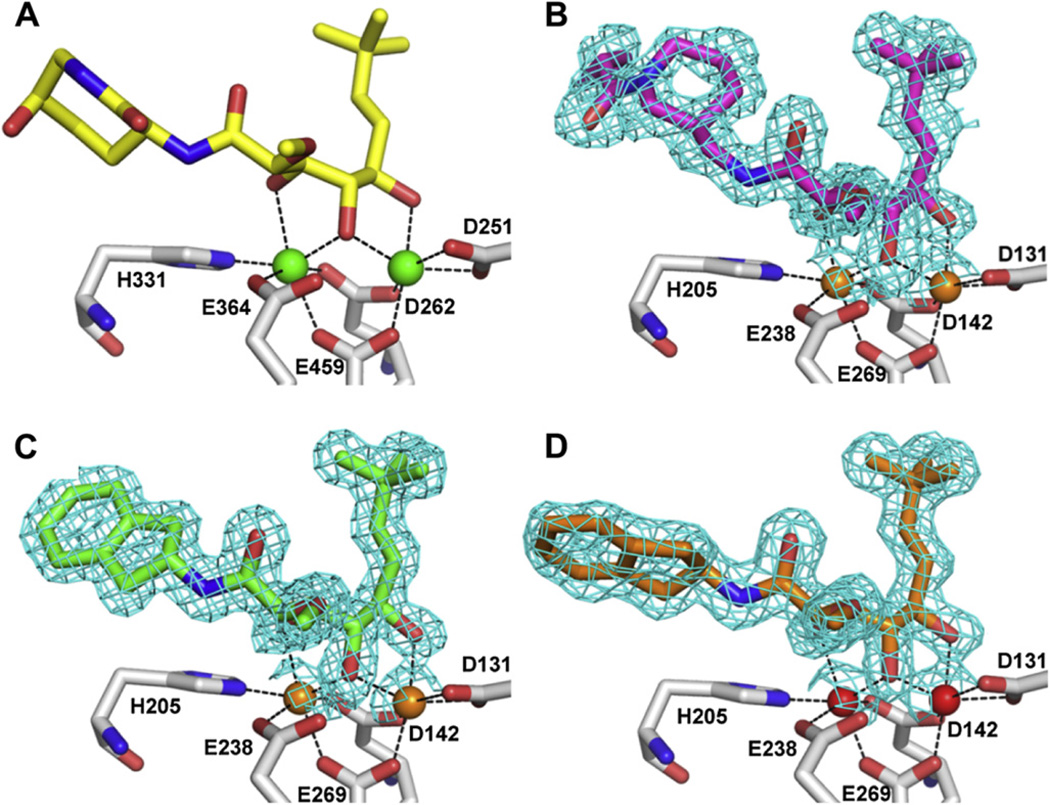
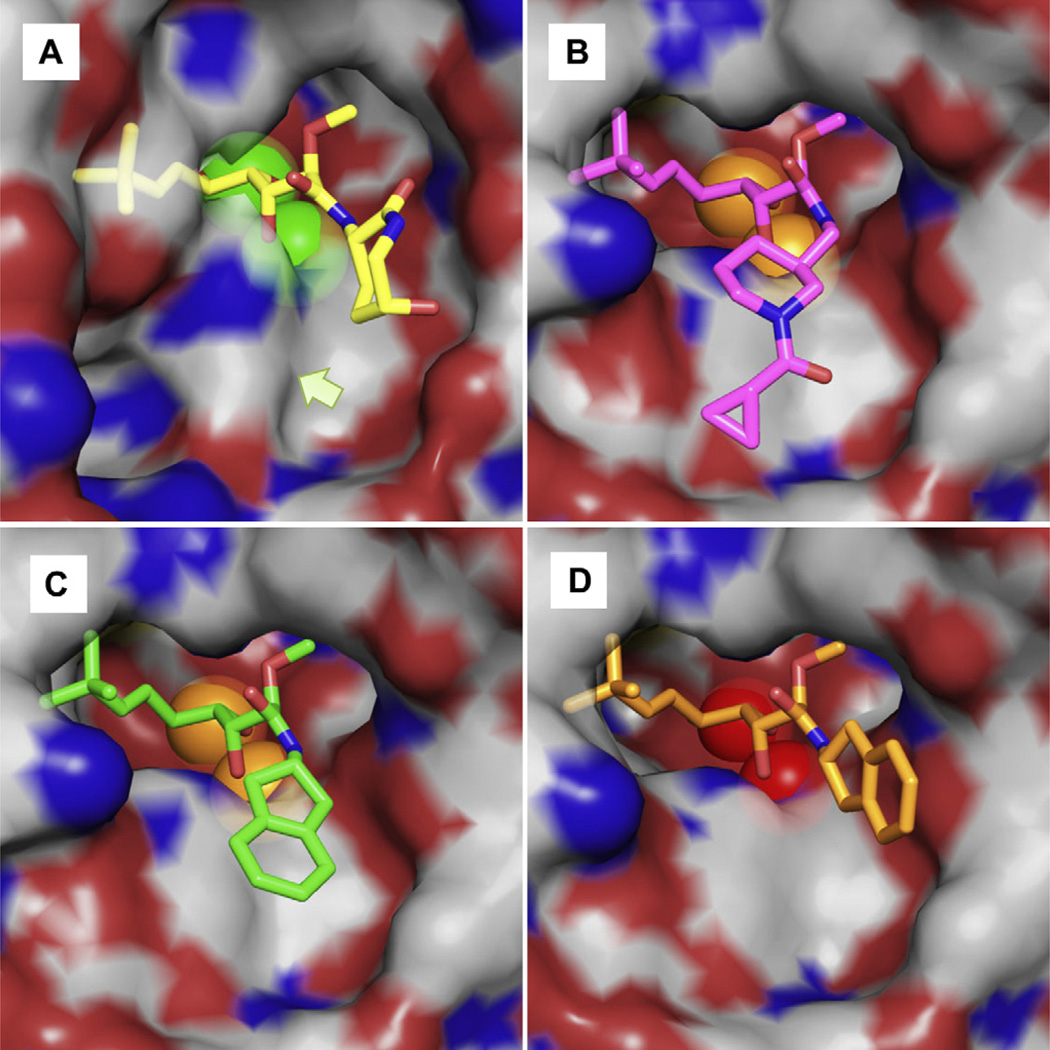
The unique bound conformation of bengamides at the active site was illustrated by the X-ray structure of a bengamide derivative LAF153 (4) in complex with human MetAP2 [17] (pdb code 1QZY) (Fig. 2A). In the dimetalated structure, the triol moiety of LAF153 coordinates with the two Co(II) ions to form two octahedral geometries, which is reminiscent of the binding of a bestatin-derived transition state inhibitor [18]. The spatial arrangement of three hydroxyl groups may uniquely satisfy the coordination requirement and possibly confer the high affinity. On one side of the triol moiety, a t-butylalkene substituent occupies the site reserved for the terminal methionine in a peptide substrate, and on the other side, a caprolactam ring beyond the amide bond interacts with residues towards the opening of the active site pocket (Fig. 3A). This unique binding mode of bengamides, coupled with their potent inhibition of MetAP enzymes and cancer cells, makes them an excellent template to develop potent MetAP inhibitors for mycobacterial and other MetAP enzymes as therapeutics.
Fig. 2.
Coordination of bengamide derivatives 7 and 8 with the metal ions at the dinuclear catalytic site, in comparison with LAF153. A. LAF153 with two Co(II) ions in human MetAP2. B. 7 with two Mn(II) ions in MtMetAP1c. C. 8 with two Mn(II) ions in MtMetAP1c. D. 8 with two Ni(II) ions in MtMetAP1c. Inhibitors are shown as sticks, and metal ions are shown as spheres. Coordination between the metal ions and the heteroatoms of the inhibitors or protein residues is shown as dashed lines. Only metal coordinating protein residues are shown. For coloring carbon atoms, LAF153 is yellow, 7 is magenta, 8 is green or orange, and protein residues are grey. For coloring non-carbon atoms, oxygen is red, and nitrogen is blue. For coloring the metal ions, Co(II) is green, Mn(II) is orange, and Ni(II) is red. The electron density (2Fo – Fc map) around the inhibitors is shown as cyan meshes at 1.0 σ level. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Binding of the bengamide derivatives 7 and 8 at the active site pocket, in comparison with LAF153. A. LAF153 with human MetAP2 in the Co(II) form. B. 7 with MtMetAP1c in the Mn(II) form. C. 8 with MtMetAP1c in the Mn(II) form. D. 8 with MtMetAP1c in the Ni(II) form. The inhibitors (sticks) and the metal ions (spheres) are colored in the same scheme as in Fig. 2. The semi-transparent surface formed by protein residues is colored grey for carbon, red for oxygen, and blue for nitrogen. The arrow in A indicates the position of the bump created by rotation of His-339 in Human MetAP2 by 80°. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Results and discussion
2.1. Design of bengamide derivatives for selective inhibition of mycobacterial MetAPs
Considering the importance of proper methionine removal, it is not surprising that MetAP shows stringent specificity for the N-terminal methionine [19,20]. No natural amino acid residues other than methionine are accepted at this position, and the formyl group in formylmethionine must be removed before methionine can be processed [21]. MetAP also shows high selectivity for the penultimate residue and has a strong preference for a small, uncharged amino acid (Gly, Ala, Ser, Thr, Pro, Val, or Cys) as the penultimate residue [22,23]. Therefore, different MetAPs use homologous residues to form the active site as a shallow and mostly hydrophobic pocket to fit the terminal methionine and the penultimate small residue. All MetAPs are homologous in the catalytic domain, and most protein residues inside the pocket are conserved [24], explaining the specificity for the 1st and 2nd amino acid residues in peptide substrates [25]. All MetAPs use the same five conserved residues for metal coordination. These present a challenge to develop selective MetAP inhibitors.
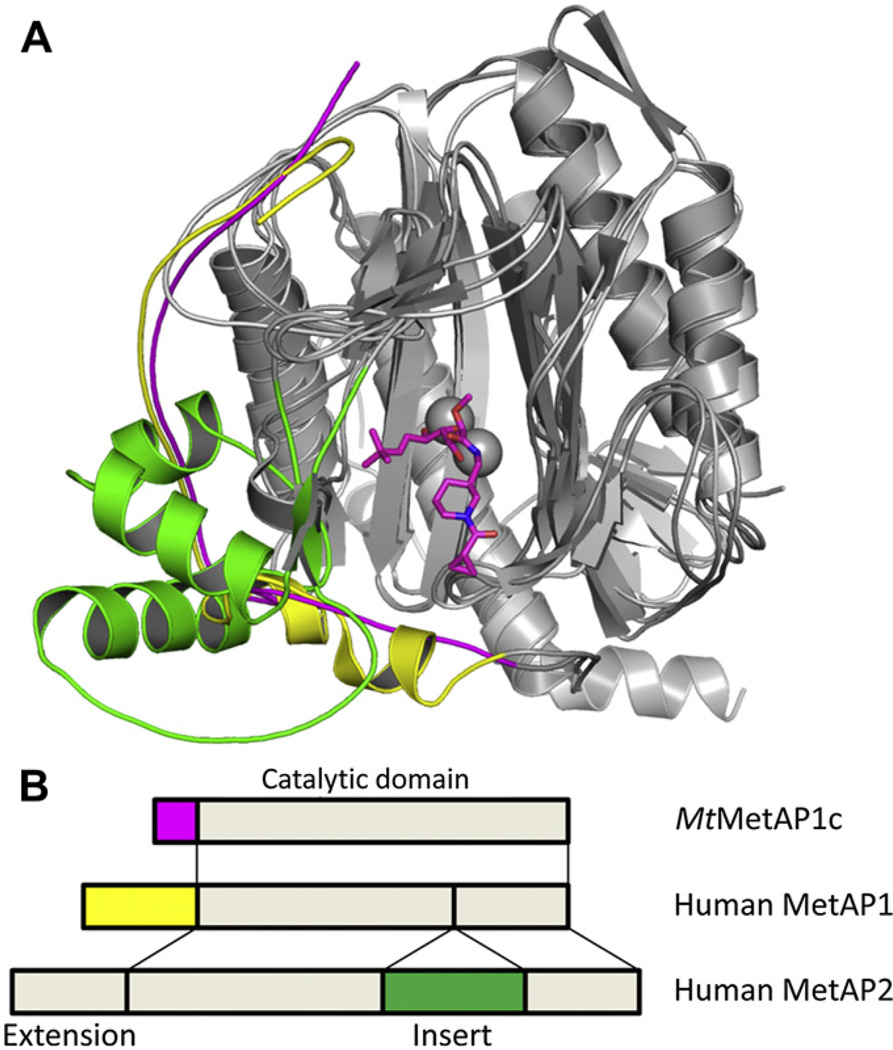
Because of the important roles played by human MetAP1 and MetAP2 in cell proliferation and angiogenesis [26,27], it is necessary to develop inhibitors of mycobacterial MetAPs with high inhibitory potency on the target mycobacterial enzyme and no potency on the human counterparts. MetAP is classified into two subtypes. Type 1 MetAP includes all bacterial and mycobacterial MetAPs (both MtMetAP1a and MtMetAP1c) and human MetAP1, while type 2 MetAP comprises archaeal MetAPs and human MetAP2. Type 2 MetAPs have a unique insert in the catalytic domain. Many MetAPs, including human MetAPs and MtMetAP1c, have extension at the N-terminus. These additional structural elements (N-terminal extension and insert) contribute to formation of the binding surface outside the active site and provide the structural diversity (Fig. 4). Consequently, to design selective MetAP inhibitors, exploration of the structural variation around the active site will offer the best chance for success. Therefore, our strategy is to explore the structural differences around the active site for selective inhibitors of mycobacterial MetAPs.
Fig. 4.
The differences among the mycobacterial MetAP (MtMetAP1c) and human Met-APs (MetAP1 and MetAP2) in forming part of the active site pocket. Both MtMetAP1c and human MetAP1 use the residues from the N-terminal extension, and human MetAP2 uses those from the insert. A. Overlay of the cartoon structures of the three MetAP enzymes, with the difference highlighted in magenta (MtMetAP1c), yellow (human MetAP1) and green (human MetAP2). The inhibitor 7 is shown as sticks at the active site. B. Schematic drawing of the domain structures for the three MetAP enzymes, with the corresponding regions highlighted in the same colors. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The caprolactam moiety of bengamides was believed to be essential for their potency on human MetAPs, and synthetic effort for anticancer therapy has been directed towards modification of this moiety by introducing different functional groups on the ring while keeping this seven-membered ring structure [13,14,28,29]. Recently, a few bengamide derivatives with the ring opened showed potent inhibitory activity on MDA-MB-435 human breast cancer cells [30], suggesting that the seven-membered ring structure may not be essential for their antiproliferative activity. To minimize potency on human MetAPs and increase potency on mycobacterial MetAPs, we keep the triol moiety and the t-butylalkene substituent as the bengamide core structure and replace this caprolactam ring with different amide moieties. We have synthesized several bengamide derivatives, including 5–8 in Fig. 1, and tested their enzyme inhibition of MtMetAP1c and MtMetAP1a and their growth inhibition of replicating and non-replicating M. tuberculosis [31]. We described two X-ray structures of MtMetAP1c in complex with 5 or 6 in the Mn(II) form initially [31], and here we report additional three X-ray structures of MtMetAP1c in complex with 7 or 8. Inhibitors 7 and 8, as well as 6, have significantly bulkier amide moieties than 5 and have very different binding interactions for the amide moieties at the active site of MtMetAP1c. Analysis of the binding and inhibition of MtMetAP1c with these bengamide derivatives provides the structural basis for further improvement of these MetAP inhibitors for potent and selective inhibition of mycobacterial MetAPs, as well as for designing effective inhibitors for other MetAP enzymes.
2.2. X-ray structure of MtMetAP1c in complex with 7 or 8 in the Mn(II) form
The three X-ray structures of MtMetAP1c in complex with 7 in the Mn(II) form or 8 in both the Mn(II) and Ni(II) forms were solved at high resolution (1.47–1.60 Å) (Fig. 2B–D and Fig. 3B–D), from crystals obtained individually by co-crystallization. As we have seen before for 5 and 6 [31], the bengamides 7 and 8 bound to MtMetAP1c in the sameway as LAF153 on human MetAP2 reported previously [17], for the part of the molecules including the triol moiety and the t-butylalkene chain. The triol moiety coordinated with the two active site metal ions and the t-butylalkene chain occupied the S1 site. However, due to the structural deviation from LAF153, each of the bengamide derivatives bound significantly differently from LAF153 at the amide moiety. For the 7 and 8 in the Mn(II) form, the amide moiety extended into the shallow cavity unique to MtMetAP1c (Fig. 3B and C). It is interesting to note that 6, as reported previously [31] and also in the Mn(II) form, bound to the enzyme with the amide moiety in the same general area (Fig. 5). The 5-membered and 6-membered ring structures in the inhibitors 6–8 directly interacted with His-212, which lay in parallel with the rings and formed a flat bottom surface for the cavity. The different amide moieties in 6–8 converged in binding to the same surface, which underscores the steric accommodation and favorable interactions in this cavity. This conclusion is consistent with the potent inhibition of 6, 7 and 8 on the Mn(II) form of MtMetAP1c (IC50 0.54, 0.96 and 0.76 µM, respectively). This histidine is conserved in all MetAPs (His-178 in E. coli MetAP, His-212 in MtMetAP1c, and His-339 in human MetAP2), because it is likely catalytically important [32]. However, although conserved, the His-339 in the human MetAP2 structure turned by 80°, so that it created a bump on the surface (Fig. 3A) and was spatially incompatible with this binding mode of these newly synthesized bengamide derivatives. Inhibitors 7 and 8 have not been tested directly on human MetAP2, but they showed no growth inhibition of human K562 cells at 333 µM [31], suggesting their weak or no inhibition of human MetAPs. It is interesting to note that inhibitor 5 has much shorter side chain and should tolerate the bump on the surface of human MetAP2 in binding. Although 5 has not been tested directly on human MetAP2, its observed inactivity on human K562 cells [31] could be due to several factors in the cellular assay, including its transport through the cell membrane.
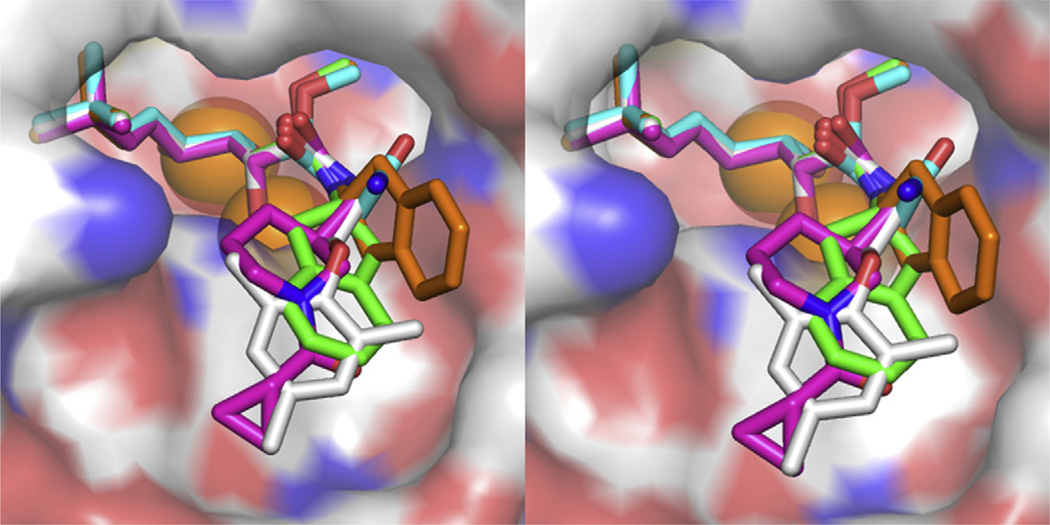
Fig. 5.
Stereo view of the superimposed bengamide derivatives 5–8 at the active site pocket of MtMetAP1c (four inhibitors and five structures). In addition to the color scheme used in the previous figures, 5 and 6 are colored cyan or grey for carbon, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. X-ray structure of MtMetAP1c in complex with 8 in the Ni(II) form
We obtained the structure of the same complex of MtMetAP1c with 8, but in the Ni(II) from (Fig. 2D and Fig. 3D). Instead of the C2 space group in the Mn(II) form, this Ni(II) structure was solved in a P63 space group. When the two structures were superimposed, the dihydroindene moiety displayed significantly different binding mode by turning almost 90° (Fig. 3C and D). This binding mode for the dihydroindene moiety is similar to that of the caprolactam moiety in LAF153 (Fig. 3A and D). For enzyme inhibition, 8 was highly potent against the Mn(II) form of MtMetAP1c, but it showed no significant inhibition at 250 µM against the Ni(II) form [31]. It is likely that this loss of binding interactions with His-212 in the Ni(II) form contributed significantly to its much reduced inhibitory activity on the Ni(II) form of the enzyme. With the metal ion being the only difference, it is interesting that 8 crystallized in two different space groups and the dihydroindene moiety oriented significantly differently. The presence of Ni(II) instead of Mn(II) at the metal site forced the dihydroindene moiety to take a different conformation and find different interactions.
The critical question is which metal is the native metal used by MetAP in cells, and which of the metalloforms is more physiologically relevant. We provided experimental evidence to show that E. coli MetAP uses Fe(II) as its cofactor in cells [33], and Wang et al. proposed Mn(II) as the physiological metal for human MetAP2 [34]. When MtMetAP1c was expressed in E. coli, it likely functioned as a Fe(II)-enzyme in E. coli cells [7]. However, neither it is known which metal MtMetAP1c uses in M. tuberculosis cells, nor the X-ray structure for any MetAP in the Fe(II)-form is known, due to the easy oxidation from Fe(II) to Fe(III). Therefore, the X-ray structures in the two different metalloforms provide two possible binding modes for bengamides on the Fe(II)-form of MtMetAP1c.
2.4. Comparison among the X-ray structures of MtMetAP1c in complex with different bengamide derivatives
When the five MtMetAP1c structures with 5, 6, 7, and 8 were overlaid (Fig. 5), the core bengamide structure (the triol moiety and the t-butylalkene substituent) is almost identical in binding, and the different amide groups, as designed, explore different interactions at the opening of the active site pocket. These diverse interactions provide the structural basis for exploring the shallow but unique binding pocket in MtMetAP1c for further inhibitor improvement for potency and selectivity. Clearly, the four Mn(II) form structures have their amide groups converging in the same area, and there are rooms for additional structural modifications on these groups for additional interactions in this pocket. The amide in the Ni(II) form takes a different position, and while its potency is much weaker, it provides a different mode of binding and can be potentially used in combination with other interactions for inhibitor design.
3. Conclusions
Bengamides are potential anticancer therapeutics, targeting human MetAPs [13,16,17]. Their unique binding mode on MetAP makes them an excellent template to develop MetAP inhibitors as therapeutics for tuberculosis and other diseases. Keeping the core structure that is responsible for interactions with the active site metals and S1 site residues and replacing the caprolactam moiety that is required for inhibition of human MetAPs, we have obtained initial bengamide derivatives with potent inhibition of tubercular enzymes MtMetAP1a and MtMetAP1c and modest antitubercular activity [31]. The three structures of MtMetAP1c in complex with two of the newly synthesized bengamides revealed different binding modes at the active site, and this information is valuable for further modification of these bengamide derivatives to enhance their potency on tubercular MetAPs and reduce their interactions with human MetAPs.
Metal ions play an important role in the MetAP catalysis and inhibition [24]. Among the activating metals, Fe(II) and Mn(II) are likely the physiologically relevant metal ions [33,34]. The four structures of the Mn(II) form of MtMetAP1c have the amide moieties converging in the same shallow concave immediately outside of the active site, revealing the important interaction with the His-212 and potential room for expansion to interact with other surface residues. Ni(II) is an excellent activator of MtMetAP1c [7]. Nevertheless, Ni(II) is not a common cofactor, and only seven nickel enzymes are known [35]. So far, no evidence indicates that Ni(II) is used by MetAP in cells. Nevertheless, the structure of MtMetAP1c in the Ni(II) form provides a possible binding mode, especially when the structural information for the Fe(II)-form of MetAP is lacking.
4. Experimental
Crystals of the enzyme–inhibitor complexes were obtained independently by a hanging-drop vapor-diffusion method at room temperature. Each of the inhibitors (7 and 8; 100 mM in DMSO)was added to concentrated metalated enzyme (10 mg/mL, 0.32 mM protein; 2 mM metal) in 50 mM Tris, pH 8.0, and 150 mM NaCl, and the molar ratio of inhibitor to MtMetAP1c was 5:1 or 10:1. The enzyme/inhibitor mixture was mixed with a reservoir buffer in a 1:1 ratio. The reservoir buffer was 100 mM Bis-Tris, pH 5.5, 1.4 M NH4SO4, and 15% glycerol for 7; and 100 mM Bis-Tris, pH 5.5, 1.3 M NH4SO4, and 15% glycerol for 8. Diffraction data were collected at the Advanced Photon Source, Argonne National Laboratory (beamlines 19BM and 19ID) and processed with HKL3000 [36]. The crystals belong to space group C2 or P63. In each case, one molecule is in the asymmetric unit. The structures were solved by molecular replacement with MolRep [37] in CCP4 [38] with CCP4i interface [39], using the previously published MtMetAP1c structure (PDB code 3IU7) [7] as the search model. The structure was refined with REFMAC5 [40] with iterative model building using WinCoot [41]. The refinement was monitored with 5% of the reflections set aside for Rfree factor analysis throughout the whole refinement process. Electron density was clear for all residues except a few residues at the N-terminus, and residues from the fourth (Arg-4) in the native protein to the end (Leu-285) were modeled. Comparison of structures and generation of structural drawings were carried out by using PyMOL [42]. Statistic parameters in data collection and structural refinement are shown in Table 1. Atomic coordinates and structure factors for the three structures were deposited in the Protein Data Bank.
Table 1.
X-ray data collection and refinement statistics.
| Inhibitor | 7 | 8 | 8 |
|---|---|---|---|
| Inhibitor code | Y08 | Y10 | Y10 |
| PDB code | 3PKC | 3PKD | 3PKE |
| Metal ions | 2 Mn(II) | 2 Mn(II) | 2 Ni(II) |
| Cell Parameters | |||
| Space group | C2 | C2 | P63 |
| a (Å) | 57.7 | 57.5 | 105.5 |
| b (Å) | 79.9 | 79.8 | 105.5 |
| c (Å) | 65.1 | 64.6 | 50.1 |
| α (deg) | 90 | 90 | 90 |
| β (deg) | 91.8 | 91.6 | 90 |
| γ (deg) | 90 | 90 | 120 |
| X-ray Data Collection | |||
| Resolution range (Å)a | 50–1.47 (1.50–1.47) | 50–1.47 (1.50–1.47) | 50–1.60 (1.63–1.60) |
| Collected reflections | 219,806 | 201,889 | 261,003 |
| Unique reflections | 45,772 | 49,388 | 41,930 |
| Completeness (%)a | 91.2 (98.4) | 99.9 (98.6) | 99.5 (100) |
| I/σ (I)a | 29.5 (2.7) | 33.9 (4.8) | 14.0 (2.4) |
| Rmerge (%)a | 7.5 (48.1) | 5.0 (28.7) | 15.3 (81.3) |
| Refinement Statistics | |||
| R (%) | 18.4 | 15.9 | 17.3 |
| Rfree (%) | 21.6 | 19.2 | 20.8 |
| R.m.s.d. bonds (Å) | 0.029 | 0.028 | 0.028 |
| R.m.s.d. angles (°) | 2.39 | 2.39 | 2.21 |
| No. of solvent molecules | 167 | 240 | 179 |
| <B> protein (Å2) | 16.6 | 11.7 | 10.5 |
| <B> inhibitor (Å2) | 17.7 | 12.8 | 11.1 |
| <B> water (Å2) | 22.1 | 19.8 | 17.4 |
Values given in parentheses correspond to the outer shell of data.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 AI065898 and R56 AI065898 and by Indiana University School of Medicine (BRG) and Indiana University and Purdue University at Indianapolis (RSFG) (to Q.-Z.Y.). We thank the staffs at Structural Biology Center of the Advanced Photon Source, Argonne National Laboratory (beamlines 19BM and 19ID) for assistance with data collection.
Abbreviations
- MetAP
methionine aminopeptidase
- MtMetAP1c
M. tuberculosis methionine aminopeptidase type 1c
- IC50
concentration of 50% inhibition
Footnotes
Supplementary data
Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.ejmech.2011.11.017. These data include MOL files and InChiKeys of the most important compounds described in this article.
References
- 1.Fauci AS. Multidrug-resistant and extensively drug-resistant tuberculosis: the national institute of allergy and infectious diseases research agenda and recommendations for priority research. J. Infect. Dis. 2008;197:1493–1498. doi: 10.1086/587904. [DOI] [PubMed] [Google Scholar]
- 2.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol. Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang SY, McGary EC, Chang S. Methionine aminopeptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol. 1989;171:4071–4072. doi: 10.1128/jb.171.7.4071-4072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller CG, Kukral AM, Miller JL, Movva NR. pepM is an essential gene in Salmonella typhimurium. J. Bacteriol. 1989;171:5215–5217. doi: 10.1128/jb.171.9.5215-5217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughan MD, Sampson PB, Honek JF. Methionine in and out of proteins: targets for drug design. Curr. Med. Chem. 2002;9:385–409. doi: 10.2174/0929867023371102. [DOI] [PubMed] [Google Scholar]
- 6.Addlagatta A, Quillin ML, Omotoso O, Liu JO, Matthews BW. Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome. Biochemistry. 2005;44:7166–7174. doi: 10.1021/bi0501176. [DOI] [PubMed] [Google Scholar]
- 7.Lu JP, Chai SC, Ye QZ. Catalysis and inhibition of Mycobacterium tuberculosis methionine aminopeptidase. J. Med. Chem. 2010;53:1329–1337. doi: 10.1021/jm901624n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Chen S, Hu Z, Zhang L, Wang H. Expression and characterization of two functional methionine aminopeptidases from Mycobacterium tuberculosis H37Rv. Curr. Microbiol. 2009;59:520–525. doi: 10.1007/s00284-009-9470-3. [DOI] [PubMed] [Google Scholar]
- 9.Lu JP, Ye QZ. Expression and characterization of Mycobacterium tuberculosis methionine aminopeptidase type 1a. Bioorg. Med. Chem. Lett. 2010;20:2776–2779. doi: 10.1016/j.bmcl.2010.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro-Guimaraes ML, Pessolani MC. Comparative genomics of mycobacterial proteases. Microb. Pathog. 2007;43:173–178. doi: 10.1016/j.micpath.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Chang YH. Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine amino-peptidases. Proc. Natl. Acad. Sci. U S A. 1995;92:12357–12361. doi: 10.1073/pnas.92.26.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinoa E, Adamczeski M, Crews P, Bakus GJ. Bengamides, heterocyclic anthelmintics from a Jaspidae marine sponge. J. Org. Chem. 1986;51:4494–4497. [Google Scholar]
- 13.Kinder FR, Jr, Versace RW, Bair KW, Bontempo JM, Cesarz D, Chen S, Crews P, Czuchta AM, Jagoe CT, Mou Y, Nemzek R, Phillips PE, Tran LD, Wang RM, Weltchek S, Zabludoff S. Synthesis and antitumor activity of ester-modified analogues of bengamide B. J. Med. Chem. 2001;44:3692–3699. doi: 10.1021/jm010188c. [DOI] [PubMed] [Google Scholar]
- 14.Thale Z, Kinder FR, Bair KW, Bontempo J, Czuchta AM, Versace RW, Phillips PE, Sanders ML, Wattanasin S, Crews P. Bengamides revisited: new structures and antitumor studies. J. Org. Chem. 2001;66:1733–1741. doi: 10.1021/jo001380+. [DOI] [PubMed] [Google Scholar]
- 15.Phillips PE, Bair KW, Bontempo J, Crews P, Czuchta AM, Kinder FR, Vattay A, Versace RW, Wang B, Wang J, Wood A, Zabludoff S. Bengamide E arrests cells at the G1/S restriction point and within the G2/M phase of the cell cycle. Proc. Am. Assoc. Cancer Res. 2000;41:59. [Google Scholar]
- 16.Dumez H, Gall H, Capdeville R, Dutreix C, van Oosterom AT, Giaccone G. A phase I and pharmacokinetic study of LAF389 administered to patients with advanced cancer. Anticancer Drugs. 2007;18:219–225. doi: 10.1097/CAD.0b013e328010ef5b. [DOI] [PubMed] [Google Scholar]
- 17.Towbin H, Bair KW, DeCaprio JA, Eck MJ, Kim S, Kinder FR, Morollo A, Mueller DR, Schindler P, Song HK, van Oostrum J, Versace RW, Voshol H, Wood J, Zabludoff S, Phillips PE. Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors. J. Biol. Chem. 2003;278:52964–52971. doi: 10.1074/jbc.M309039200. [DOI] [PubMed] [Google Scholar]
- 18.Lowther WT, Orville AM, Madden DT, Lim S, Rich DH, Matthews BW. Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis. Biochemistry. 1999;38:7678–7688. doi: 10.1021/bi990684r. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsunasawa S, Izu Y, Miyagi M, Kato I. Methionine aminopeptidase from the hyperthermophilic Archaeon Pyrococcus furiosus: molecular cloning and overexpression in Escherichia coli of the gene, and characteristics of the enzyme. J. Biochem. (Tokyo) 1997;122:843–850. doi: 10.1093/oxfordjournals.jbchem.a021831. [DOI] [PubMed] [Google Scholar]
- 21.Solbiati J, Chapman-Smith A, Miller JL, Miller CG, Cronan JE., Jr Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J. Mol. Biol. 1999;290:607–614. doi: 10.1006/jmbi.1999.2913. [DOI] [PubMed] [Google Scholar]
- 22.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol. Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Lowther WT, Matthews BW. Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta. 2000;1477:157–167. doi: 10.1016/s0167-4838(99)00271-x. [DOI] [PubMed] [Google Scholar]
- 25.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:7678. 5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Addlagatta A, Lu J, Matthews BW, Liu JO. Elucidation of the function of type 1 human methionine aminopeptidase during cell cycle progression. Proc. Natl. Acad. Sci. U S A. 2006;103:18148–18153. doi: 10.1073/pnas.0608389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sin N, Meng L, Wang MQ, Wen JJ, Bornmann WG, Crews CM. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. U S A. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Ma YM, Tai WY, Xie CM, Li YL, Li J, Nan FJ. Design, synthesis, and biological evaluation of caprolactam-modified bengamide analogues. Chem. Med. Chem. 2008;3:74–78. doi: 10.1002/cmdc.200700214. [DOI] [PubMed] [Google Scholar]
- 29.Kinder FR, Jr, Wattanasin S, Versace RW, Bair KW, Bontempo J, Green MA, Lu YJ, Marepalli HR, Phillips PE, Roche D, Tran LD, Wang R, Waykole L, Xu DD, Zabludoff S. Total syntheses of bengamides B and E. J. Org. Chem. 2001;66:2118–2122. doi: 10.1021/jo0017133. [DOI] [PubMed] [Google Scholar]
- 30.Tai WY, Zhang RT, Ma YM, Gu M, Liu G, Li J, Nan FJ. Design, synthesis, and biological evaluation of ring-opened bengamide analogues. Chem. Med. Chem. 2011;6:1555–1558. doi: 10.1002/cmdc.201100164. [DOI] [PubMed] [Google Scholar]
- 31.Lu JP, Yuan XH, Yuan H, Wang WL, Wan B, Franzblau SG, Ye QZ. Inhibition of Mycobacterium tuberculosis methionine aminopeptidases by bengamide derivatives. Chem. Med. Chem. 2011;6:1041–1048. doi: 10.1002/cmdc.201100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copik AJ, Swierczek SI, Lowther WT, D’Souza VM, Matthews BW, Holz RC. Kinetic and spectroscopic characterization of the H178A methionyl aminopeptidase from Escherichia coli. Biochemistry. 2003;42:6283–6292. doi: 10.1021/bi027327s. [DOI] [PubMed] [Google Scholar]
- 33.Chai SC, Wang WL, Ye QZ. FE(II) is the native cofactor for Escherichia coli methionine aminopeptidase. J. Biol. Chem. 2008;283:26879–26885. doi: 10.1074/jbc.M804345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Sheppard GS, Lou P, Kawai M, Park C, Egan DA, Schneider A, Bouska J, Lesniewski R, Henkin J. Physiologically relevant metal cofactor for methionine aminopeptidase-2 is manganese. Biochemistry. 2003;42:5035–5042. doi: 10.1021/bi020670c. [DOI] [PubMed] [Google Scholar]
- 35.Wattt RK, Ludden PW. Nickel-binding proteins. Cell Mol. Life Sci. 1999;56:604–625. doi: 10.1007/s000180050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 37.Vagin A, Teplyakov A. MOLREP: an Automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 38.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System on World Wide Web. 2002 http://www.pymol.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.