Abstract
Here, we present a photodegradable microparticle system that can be employed to entrap and deliver bioactive proteins to cells during culture. By using a photosensitive delivery system, experimenters can achieve a wide variety of spatiotemporally regulated release profiles with a single microparticle formulation, thereby enabling one to probe many questions as to how protein presentation can be manipulated to regulate cell function. Photodegradable microparticles were synthesized via inverse suspension polymerization with a mean diameter of 22 μm, and degradation was demonstrated upon exposure to several irradiation conditions. The protein-loaded depots were incorporated into cell cultures and release of bioactive protein was quantified during the photodegradation process. This phototriggered release allowed for the delivery of TGF-β1 to stimulate PE25 cells and for the delivery of fluorescently labeled Annexin V to assay apoptotic 3T3 fibroblasts during culture. By incorporating these photoresponsive protein delivery depots into cell culture, new types of experiments are now possible to test hypotheses about how individual or multiple soluble factors might affect cell function when presented in a uniform, temporally varying, or gradient manner.
Introduction
Signaling proteins influence a myriad of critical cell functions, including differentiation, migration, and cell fate decisions, and many of these effects are pleiotropic depending on the dose and persistence of the signal.1-4 Thus, spatiotemporal control over protein presentation is critical to study and understand the role that these biomacromolecules play in dynamic cellular processes. Toward this end, a prevalent method to protect, target, and locally deliver proteins and other therapeutics is to load such factors in polymeric microspheres.5,6 Such delivery vehicles enable the release of high doses of protein at specific locales, as well as controlled release over a desired time course.7,8
Microsphere systems, typically formed from hydrolytically degradable polymers with pre-determined release profiles, have been used in numerous controlled release applications, including in vitro delivery of factors that influence the differentiation of embryoid bodies9 or in vivo delivery of osteogenic factors to encourage robust bone growth.10 Corresponding to the increase in the discovery of biological factors that direct stem cell differentiation, treat a range of diseases, and encourage proper tissue morphogenesis, there has been a focus on developing advanced materials that offer precise control over the delivery of such molecules. To date, full spatiotemporal control over the release and presentation of these factors during cell culture has been limited and few systems allow experimenters to direct release in real time. As a result of the lack of more sophisticated protein delivery vehicles, it has become increasingly difficult and time consuming to determine appropriate doses and release profiles of biomacromolecules for specific applications. Further, advanced understanding of wound healing and developmental processes underscore the importance of the proper presentation of multiple cues, including proteins and co-factors or morphogen pairs, which is exceedingly difficult with current methods. Finally, few material systems allow the experimenter to introduce spatially heterogeneous gradients at any point in time that could be used to investigate how morphogens act during development and to fashion complex tissue structures ex vivo.
To circumvent these limitations and complement existing microsphere technologies, a unique delivery vehicle based on photolabile networks is presented that offers the experimenter control of entrapped biomolecule delivery in real time and in a manner that is compatible with 2D and 3D cell culture. Specifically, photodegradable, poly(ethylene glycol) (PEG) based hydrogel microspheres are fabricated that entrap and, subsequently, deliver proteins of interest on demand by exposure to selected wavelengths of light. Such delivery systems should prove beneficial for testing hypotheses related to how temporal and spatial protein presentation affects local cell function and have applied benefits for the controlled expansion and differentiation of stem cells.
The microsphere formulation includes PEGdiPDA (poly(ethylene glycol) di-photodegradable-acrylate)11 to render photodegradable, protein-loaded microspheres, on account of the o-nitrobenzyl ether moieties in the PEGdiPDA structure. Nitrobenzyl ethers (NBEs) undergo an irreversible cleavage upon irradiation, causing the network to degrade in response to specific wavelengths of light (Figure 1). Similar macromers have been employed to form photoactive monolithic materials for applications ranging from cell culture12-15 to drug delivery.16-19 However, none of these approaches have combined microsphere processing techniques with the ability to deliver bioactive proteins to cells during culture with full spatiotemporal control.
Figure 1.
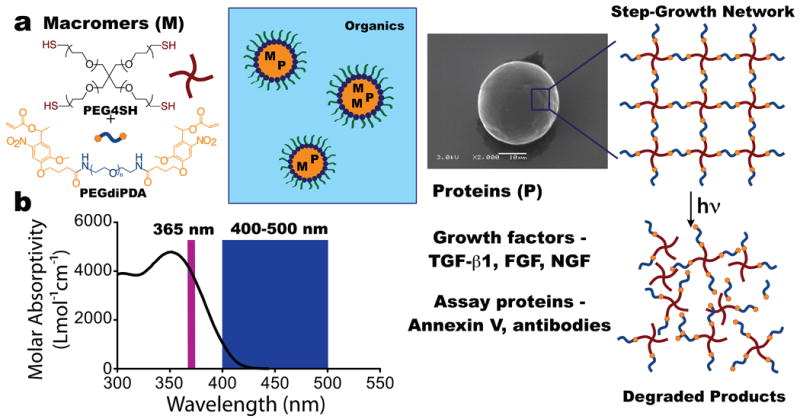
Photodegradable microparticle fabrication. a. Photodegradable particles were synthesized by reacting PEG4SH with PEGdiPDA via base-catalyzed Michael addition in an inverse-phase, suspension polymerization. The aqueous phase, consisting of macromers, the base catalyst triethanolamine, and the target protein, was suspended in an organic phase of hexanes and stabilized by surfactants. Upon completion of the polymerization, the particles were purified via centrifugation resulting in spherical particles, as imaged by SEM. The reaction of the PEG4SH with the PEGdiPDA forms a step-growth network, and owing to the presence of nitrobenzyl ether (NBE) moieties in the PEGdiPDA, the network degrades in response to light. b. The NBE moiety absorbs light strongly at 365 nm with a tail out past 405 nm. This allows both single photon irradiation at 365 nm or 400-500 nm to be used to degrade the particles, as well as two-photon irradiation using a wavelength of 740 nm.
The photodegradable microspheres described herein degrade upon single photon or multiphoton irradiation, which induces swelling and, ultimately, complete erosion and particle dissolution. During swelling, the entrapped protein diffuses into the surrounding environment and upon dissolution the total payload is released. In this system, the experimenter retains full control over the spatial and temporal presentation of the protein release by directing the irradiation. We demonstrate that biologically relevant proteins, namely TGF-β1 and Annexin V, can be entrapped within the microspheres and released on demand to direct or detect cell function. In total, we describe an innovative method to generate preloaded depots of protein agents, which can be employed to release bioactive proteins in the presence of cells.
Materials and Methods
Microparticle preparation
Poly (ethylene glycol) di-photodegradable-acrylate (PEGdiPDA; Mn ∼ 4,070 Da) was synthesized as previously described.11,12 Poly (ethylene glycol) tetrathiol (PEG4SH; Mn ∼ 5,000 Da) was synthesized as previously described.20 Photodegradable microparticles were prepared via inverse suspension polymerization, in which PEGdiPDA was copolymerized with PEG4SH via base-catalyzed Michael addition in an aqueous phase that was suspended in an organic phase. Briefly, the organic phase was comprised of 5 ml of hexane containing 150 mg of a 3:1 ratio by weight of sorbitan monooleate (Span 80, Sigma-Aldrich) and poly (ethylene glycol)-sorbitan monooleate (Tween 80, Sigma-Aldrich).21 The volume of the aqueous phase was 0.25 mL comprised of 300 mM triethanolamine (Sigma-Aldrich) at pH 8.0 with 6.2 wt % of PEGdiPDA, 3.8 wt % PEG4SH, and protein. Bovine serum albumin labeled with Alexa Fluor 488 or Alexa Fluor 594 (BSA-488 or BSA-594; Invitrogen) were entrapped at 0.8 mg/ml, TGF-β1 (Peprotech) was entrapped at 0.4 μg/ml, and the fluorescently labeled Annexin-V (Invitrogen) was entrapped at 20 v/v % Annexin-V conjugate solution. All of the components of the aqueous phase except for the PEG4SH solution were combined in a 1.7 ml microcentrifuge tube while the organic phase was added to a 20 ml scintillation vial with a stir bar. To initiate polymerization, the PEG4SH was added to the aqueous phase, which was subsequently vortexed for 10 s and quickly added to the organic phase. Mixing on a stir plate formed and maintained the inverse suspension between the two phases and the polymerization was allowed to proceed overnight.
Upon completion of the polymerization, the suspension was centrifuged (Eppendorf Centrifuge Model 5702) at 1000 rcf for 10 minutes and the supernatant was decanted. The microparticles were washed twice with hexanes and recovered with the same centrifugation conditions and once in 2-propanol and centrifuged at 2000 rcf for 10 minutes. The particles were then suspended in 1× PBS and washed three times by centrifuging (Eppendorf Centrifuge Model 5418) at 16,873 rcf for 15 minutes. The recovered particles were stored in PBS at 4°C and a portion was imaged on a low vacuum scanning electron microscope (LVSEM, JSM-6480LV).
Absorbance of PEGdiPDA
The molar absorptivity of the nitrobenzyl ether (NBE) moiety was calculated by measuring the absorbance of solutions of NBE in a water:DMSO (80:20 v/v) blend at concentrations of 110, 82.5, 55, and 27.5 μM. The absorbance was measured on a UV-visible spectrophotometer (NanoDrop Spectrophotometer ND-1000) for each solution and the molar absorptivity was calculated from these absorbance profiles.
Microparticle size characterization with image analysis
Microparticles loaded with BSA-488 were used to characterize the size distribution of the particles. Particles were suspended in PBS and sealed between a glass slide and a cover slip in a rubber gasket, and imaged on an epifluorescent microscope (Nikon Eclipse TE2000-S). ImageJ (NIH) was used to threshold the images and the Analyze Particles plug-in was employed to determine the diameter of each microsphere. A total of 3130 particles were analyzed to determine the particle diameter distribution.
Degradation of microparticles
BSA-488 loaded microparticles were suspended in PBS in a sealed rubber gasket and exposed to 365 nm (I0 = 13.5 ± 0.5 mW/cm2; EXFO Omnicure 1000) or 400-500 nm (I0 = 20.0 ± 0.5 mW/cm2; EXFO Novacure) irradiation to induce degradation and erosion. To quantify the degradation induced changes in material properties, a time series of images was captured with an epifluorescent microscope. The images were analyzed with ImageJ by bounding each particle with a manually drawn circle to determine the particle diameter at each timepoint during irradiation. The diameters were used to calculate the ratio of the actual volume relative to the initial volume (V/V0) as a function of time for each particle, and data for the respective irradiation condition was plotted as an average of three particles.
To demonstrate focused irradiation induced degradation and erosion, BSA-488 and BSA-594 loaded microparticles were suspended in PBS in a sealed rubber gasket and placed on the stage of an overhead confocal laser-scanning microscope (Zeiss 710 NLO LSM). Particles were exposed to 405 nm (single photon; P = 1 mW) or 740 nm (two-photon; P = 100 mW) irradiation to degrade and, ultimately, erode the particles. Degradation and erosion were monitored by direct imaging on the LSM.
Quantification of BSA-488 release
To quantify the release profile of entrapped BSA-488 from the particles, BSA-488 loaded microspheres were exposed to flood irradiation (λ = 400-500 nm; I0 = 20.0 ± 0.5 mW/cm2) for 0 min to 15 min. Samples were collected at each time point and centrifuged to separate the soluble protein in the supernatant from intact particles in solution. The fluorescence of the supernatant was measured on a plate reader (BioTek Synergy H1 Hybrid Reader) to determine the relative amount of BSA-488 in the supernatant for each sample.
Diffusion in fibrin gels
Fibrin gels were formed by combining 50 μl of fibrin (20 mg/ml), 1 μl of thrombin (0.5 U/ml), and 150 μl PBS with BSA-488 and BSA-594 loaded particles (2 mg of particles/ml). The solution was allowed to gel at 37°C for 10 minutes in a sealed rubber gasket. The gels, with encapsulated particles, were imaged while the particles were degraded using an LSM (Zeiss 710 NLO LSM). Fluorescence intensity of the diffusing BSA-488 was quantified using the Image Processing Toolbox in MATLAB (MathWorks).
Cell Culture
All cell culture reagents were purchased from Invitrogen except where otherwise noted. PE25 cells, a cell line that produces luciferase in response to TGF-β1 exposure in a dose-dependent manner22 were cultured in low glucose DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 0.2% fungizone. PE25 cells were passaged every 2-3 days and maintained at less than 80% confluency. Passage 4-6 PE25 cells were used for TGF-β1 bioactivity assays. 3T3 fibroblasts were cultured in high glucose DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 0.2% fungizone. 3T3 cells were passaged every 2-3 days and maintained at less than 70% confluency. P5 3T3 cells were used for the apoptosis assays.
TGF-β1 delivery
For the TGF-β1 bioactivity assays, PE25 cells were plated on 24-well culture plates at 80,000 cells/well and allowed to adhere overnight. The following day, media with soluble TGF-β1 (2 ng/ml), media with TGF-β1 loaded particles (10 mg of particles/ml of media, which equates to 4 ng/ml TGF-β1 with complete release of the protein), media with blank particles (10 mg of protein-free particles/ml of media), and media were placed on the plated cells. Half of the wells were irradiated to degrade the particles (λ = 365 nm; I0 = 13.5 ± 0.5 mW/cm2) for 5 minutes to ensure complete erosion, while a duplicate set of conditions was not exposed to light. The solutions were left on the PE25 cells in an incubator for 16 hours. The following day, 200 μl of Glo-Lysis Buffer (Promega) was added to each well to lyse the cells and release any luciferase that had been produced. After 15 minutes, 50 μl of the lysis solution was combined with 50 μl of luciferin substrate in triplicate. The solutions were immediately quantified for luminescense on a plate reader (BioTek Synergy H1 Hybrid Reader).
Fluorescently labeled Annexin V delivery
Particles were synthesized that were loaded with AlexaFluor-594 Annexin V (Invitrogen) at 5 μl of Annexin V solution per 250 μl of particle solution. 3T3 cells were plated on a 6-well plate at 100,000 cells/well and allowed to adhere overnight. The following day, half of the wells were treated with (+)camptothecin (Sigma) at 10 μM for 6 hours to induce apoptosis. After the treatment, the media was removed and substituted with 400 μl of Annexin V binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2 at a pH 7.4) containing soluble Annexin V (3 μl per 400 μl buffer) or Annexin V loaded particles (12.5 mg of particles/ml). A set of wells with Annexin V loaded particles was irradiated to release Annexin V (λ = 365 nm; I0 = 13.5 ± 0.5 mW/cm2) for 5 minutes. After 15 minutes, the samples were imaged on an LSM (Zeiss 710 LSM NLO).
Results and Discussion
Synthesis and characterization of microparticles
Photodegradable microparticles were fabricated by reacting PEGdiPDA (Mn ∼ 4,000 Da) with poly(ethylene glycol) tetrathiol (PEG4SH; Mn ∼ 5,000 Da) via base-catalyzed Michael addition in an inverse-phase, microsuspension polymerization (Figure 1a). The polymerization was carried out with the protein of interest included in the aqueous, macromer solution, which was suspended in an organic phase of hexanes with surfactants.21 This approach allowed the target protein to be entrapped within the particles upon gelation. Subsequently, the particles were purified via centrifugation, resulting in smooth, protein-loaded hydrogel microspheres (Figure 1a).
As a representative protein, fluorescently labeled bovine serum albumin (BSA-488) was incorporated into the macromer solution (Figure 2a) during polymerization and entrapped homogeneously within the microsphere network. BSA-488 loaded particles were employed to characterize the size distribution of the particles via image analysis (n = 3130 particles). The microspheres were synthesized with diameters on the order of 10 μm or greater, and more than 80% of the particles had a diameter less than 50 μm (Figure 2b, inset). The distribution had a first moment (Dn) = 22 μm, a second moment (Dw) = 42 μm, and a polydispersity index (PDI) = 1.9 (Figure 2b). This size distribution is appropriate for the delivery of a substantial local dose of protein with rapid light-triggered degradation.
Figure 2.
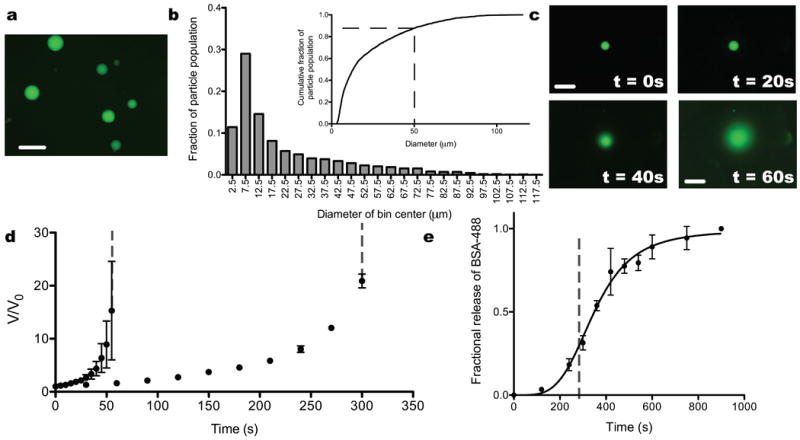
Size and degradation characteristics of photodegradable microparticles. a. BSA-488 was entrapped within photodegradable microspheres. b. Image analysis was used to quantify the size distribution of the particles synthesized by this method (n = 3130). The particles were formed with a number average diameter of 22 μm and a diameter average diameter of 42 μm, which resulted in a polydispersity index of 1.9. Over 80% of the particles had a diameter less than 50 μm. c. Photodegradable particles swell and, ultimately, erode in response to flood irradiation (λ = 365 nm; I0 = 13.5 ± 0.5 mW/cm2) over the time course of a minute. d. The swelling was quantified with image analysis and plotted as normalized volume (V/V0) as a function of irradiation time. Particles were exposed to 365 nm (I0 = 13.5 ± 0.5 mW/cm2; circles) and 400-500 nm (I0 = 20.0 ± 0.5 mW/cm2; triangles) irradiation, and the particles eroded at 55 ± 5 s and 300 ± 30 s for the two conditions, respectively (indicated by the dashed gray lines). e. The release of BSA-488 as a function of irradiation time was quantified as the particles swelled and dissolved. Prior to dissolution (indicated by the dashed gray line), BSA-488 began diffusing out as the particles swelled, and after dissolution the majority of the payload was released into solution. Scale bars, 100 μm.
Photodegradation of microparticles
Since the o-nitrobenzyl ether (NBE) moiety in the PEGdiPDA macromer is susceptible to cleavage with single photon or multiphoton excitation,23,24 a broad range of irradiation conditions can be used to erode the microspheres and release the entrapped payload on the order of milliseconds to minutes. This process works as the NBE moieties in the PEGdiPDA structure introduce a photolabile linker into the network backbone of the microspheres. NBE moieties absorb light strongly in the UV (peak at 365 nm) with a tail that extends into the visible (Figure 1b) and may undergo an irreversible cleavage upon absorption of light at these wavelengths, as well as absorption of two-photon irradiation centered at 740 nm. When a NBE is cleaved, the corresponding bond in the particle backbone is also cleaved. This process, which will be referred to as degradation, induces swelling in the particle as bonds are cleaved in the microsphere and the crosslinking density is decreased. Eventually, when a sufficient fraction of the bonds have been cleaved, erosion (i.e., mass loss) occurs and at these later stages of degradation, the microsphere is no longer a network, but soluble branched polymers that dissolve.
To demonstrate degradation and protein release in response to single photon irradiation, BSA-488 loaded microspheres were irradiated with collimated light (λ = 365 nm or 400-500 nm). Particles swelled initially, as bonds were cleaved throughout the network, as quantified by the increase in V/V0 with irradiation time (Figure 2c,d). Ultimately, the microspheres eroded completely when a sufficient number of bonds in the network were cleaved (pc = 0.42; the critical fraction of bonds that need to be cleaved to dissolve the network as determined by the Flory-Stockmayer equation) (Figure 2c,d). For 365 nm irradiation at an intensity of 13.5 ± 0.5 mW/cm2, the microspheres swelled prior to eroding into solution over the course of 55 ± 5 s. Whereas 400-500 nm irradiation at an intensity of 20.0 ± 0.5 mW/cm2 induced swelling and erosion over the course of 300 ± 30 s. The fractional release of entrapped BSA-488 from the microspheres followed the degradation-induced swelling profile at short times and for the first 30% of release, while the bulk of the payload was released after complete particle dissolution (Figure 2e). In this manner, collimated irradiation provides the user with temporal control over protein release within a culture system.
Selective release of proteins
Oftentimes the release of multiple factors within a single culture system is desirable, as cells respond in vivo to combinations of factors. For example, opposing gradients of transcriptional repressors, Hunchback and Knirps, direct proper development in Drosophila.25 Light responsive protein release affords the unique ability to deliver multiple factors selectively within a single system. To demonstrate this concept, photodegradable microspheres were loaded with BSA-594 (BSA labeled with Alexa Fluor 594) and combined with BSA-488 loaded particles. A mixture of BSA-488 and BSA-594 spheres were plated and imaged on a confocal LSM (Figure 3a). Focused irradiation (λ = 405 nm single photon or 740 nm multiphoton) was employed to erode individual particles in sequence to release each desired protein (Figure 3a). Initially, t = t1, the focused irradiation (λ = 740 nm; P = 100 mW) was used to selectively erode a BSA-594 loaded microsphere. At a subsequent point in time, t = t2, focused irradiation was employed to selective erode a microsphere containing a second entrapped protein, BSA-488. In this manner, different growth factors or cytokines could be delivered locally and in combination over short distances to specific locations during culture. This system should prove useful for studies aimed at the investigation of synergistic protein interactions or to elucidate how multiple and/or opposing gradients influence cell fate or function, such as chemotaxis or tissue morphogenesis.
Figure 3.
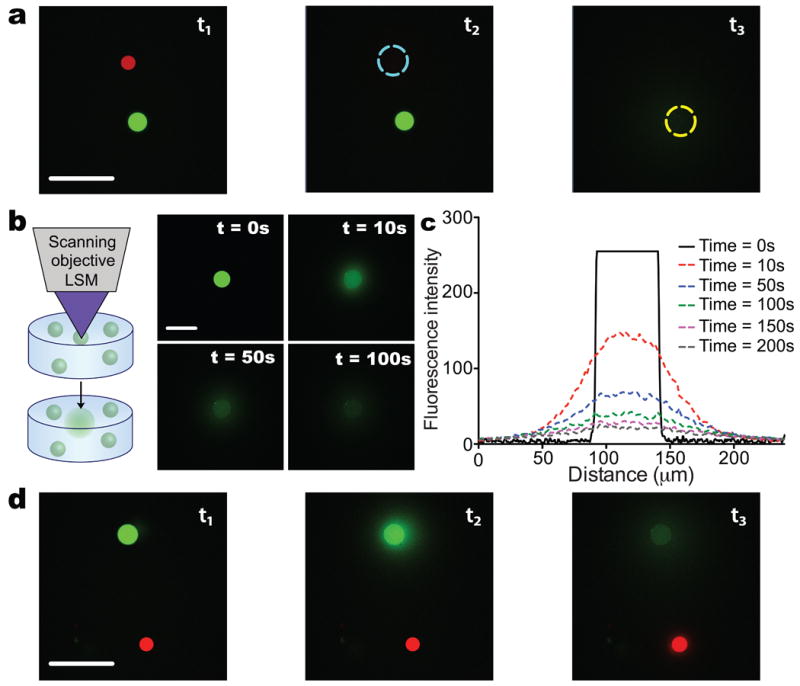
Spatially controlled degradation of photodegradable particles. a. BSA-488 loaded microparticles were combined with BSA-594 loaded particles in a single system and focused irradiation from an LSM (λ = 740 nm; two-photon) was used to erode individual particles selectively. Here, a BSA-594 loaded particle was dissolved (t1 to t2) followed by the erosion of a BSA-488 loaded particle (t2 to t3) without disrupting neighboring particles. b. Photodegradable particles can also be encapsulated within three-dimensional hydrogels and selectively photodegraded with focused light from an LSM (λ = 405 nm). Here, BSA-488 loaded particles were encapsulated within fibrin hydrogels and eroded with focused LSM irradiation after the image t = 0 s was taken. Images were captured after erosion to monitor diffusion of the protein through the fibrin gel. c. Profiles of the diffusing protein were quantified over the time course of imaging and demonstrate that the BSA-488 diffused radially at a detectable level to a distance of 50 μm from the edge of the original particle. d. Multiple protein loaded particle populations were encapsulated within a single fibrin gel and individual particles were eroded selectively as was demonstrated in 2D. Scale bars, 100 μm.
Release of proteins in 3D culture platforms
Advanced three-dimensional culture platforms are increasingly employed for the study of cell biology and pathophysiology ex vivo.26-28 Accompanying these advances is the need for methods to deliver proteins within these platforms in sophisticated manners, systematically introducing cues that recapitulate aspects of the native extracellular environment. Photoresponsive, pre-loaded depots of proteins were encapsulated within fibrin hydrogels (Figure 3b,c) to demonstrate how this system might be used to deliver factors during 3D culture. Focused irradiation (λ = 405 nm; P = 1 mW) from a confocal LSM was used to dissolve individual particles, allowing the entrapped payload to release and diffuse through the gel (Figure 3b). The released protein diffused, at a detectable level, ∼50 μm radially from the edge of the particle (Figure 3c). As was demonstrated in 2D, multiple proteins were released selectively within a single hydrogel to motivate combinatorial studies in 3D (Figure 3d). In this manner, signaling proteins of interest can be delivered locally within a 3D cell culture scaffold. This light-controlled release and diffusion can be tailored to cell binding and uptake levels to influence cells and their function over reasonable length scales.
Release of bioactive proteins to direct cell function
The microsphere formulation was designed to accommodate a broad range of proteins including growth factors, cytokines, antibodies, and extracellular matrix components. To demonstrate that bioactive proteins can be incorporated and released from the photodegradable particles in the presence of cells, we entrapped a common and potent growth factor, TGF-β1,29 within the microspheres. TGF-β1 loaded particles, as well as blank particles, were delivered to plated PE25 cells, a reporter cell line that produces luciferase in response to TGF-β1 exposure. The particles were dissolved with collimated irradiation (λ = 365 nm, I0 = 13.5 ± 0.5 mW/cm2) for 5 minutes to release the TGF-β1. PE25 cells that were exposed to TGF-β1 loaded particles significantly up-regulated luciferase production as compared to blank particles and media control (Figure 4a). This demonstrates that the majority of the TGF-β1 remains bioactive upon entrapment and subsequent release. Furthermore, viability, as measured by a membrane integrity assay, was greater than 90% for all conditions (data not shown) indicating that the irradiation conditions and microsphere degradation products do not adversely affect cell function.
Figure 4.
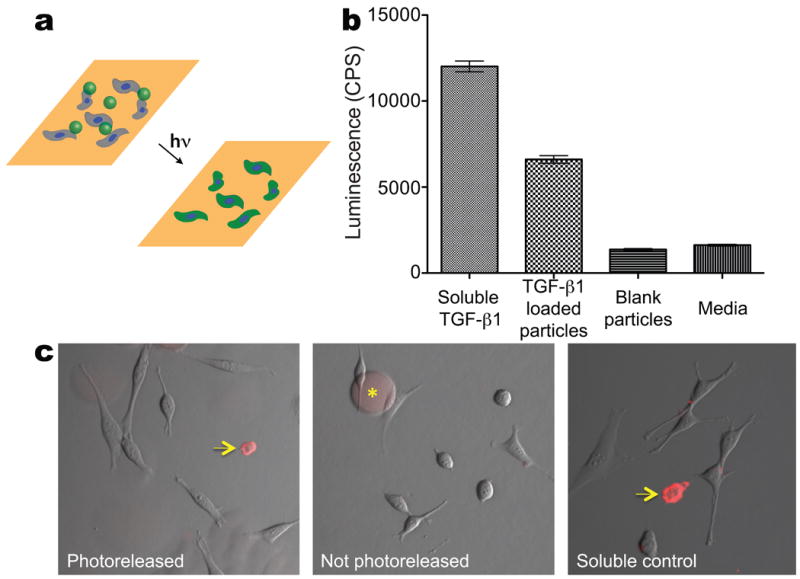
Release of bioactive proteins in the presence of cells. a. Protein-loaded, photodegradable microparticles were incorporated into cell culture with plated cells. Collimated irradiation (λ = 365 nm; I0 = 13.5 ± 0.5 mW/cm2) was used to release the entrapped protein to direct or detect cell function. b. TGF-β1 loaded particles were added to the media of plated PE25 cells, a TGF-β1 responsive reporter cell line, and compared to conditions: media with blank particles, media with soluble TGF-β1, and plain media. All samples were irradiated with the same dose used to erode particles and release the TGF-β1, and the response of the PE25 cells was compared between each condition. The TGF-β1 loaded particles had a significantly higher response than negative controls (blank particles and media alone), though not as strong a response as the positive control of soluble TGF-β1. This demonstrates that a significant fraction of the TGF-β1 remained bioactive upon entrapment and photorelease. c. Fluorescently-labeled Annexin V loaded particles were added to the media on plated 3T3 cells. (+)Camptothecin was used to induce apoptosis and selected samples were irradiated to release the Annexin V. The samples were imaged to visualize cells labeled as apoptotic. Samples exposed to soluble Annexin V and photoreleased Annexin V stained positively for apoptosis (indicated by arrows at regions of red staining on the cell membranes), while there was no staining in the sample that was exposed to particles that were not photoreleased (particles are still visible in this image, denoted by the asterix). Scale bar, 100 μm.
Release of bioactive proteins to assay cell function
A further difficulty of in vitro culture is assaying a specific cell's functions during culture, and this can be especially challenging when culturing cells in 3D. To illustrate how photodegradable microspheres can be employed as protein loaded depots for assaying cell function, fluorescently conjugated Annexin V was loaded into microspheres. Annexin V loaded particles were delivered to plated NIH 3T3 fibroblasts, and the protein was photoreleased to identify apoptotic cells (Figure 4c). Camptothecin was dosed to the cells prior to release to increase the rate of apoptosis in culture. Annexin V staining on the membranes of apoptotic cells was observed in the samples with photoreleased Annexin V and soluble Annexin V, whereas no membrane staining was observed in the sample in which the microspheres were not irradiated. To circumvent the challenge of assaying cell function during 3D culture, protein-loaded microspheres could be included in cell encapsulations so that the assay protein of interest can be delivered at a later time during culture.
Conclusion
The synthesis of photodegradable, PEG-based microspheres was demonstrated and these microspheres were employed to entrap and release soluble proteins. Cytocompatible irradiation conditions were determined to dissolve the particles with light, and the corresponding release of the entrapped payload was quantified during the degradation and erosion process. Multiple factors were loaded into batches of microspheres and focused irradiation was used to degrade individual particles selectively to release specific proteins of interest. TGF-β1 was loaded into the microspheres and was released with light to a reporter cell line to demonstrate that the entrapped and released protein remained bioactive. Similarly, Annexin V was loaded into particles to illustrate that protein-loaded depots could be incorporated into cell cultures to assay local cell function. By incorporating protein loaded, photoresponsive microspheres within cell aggregates, in media fed to plated cells, or in cell-laden scaffolds, the externally controlled and on-demand release of entrapped biological signals will allow experimenters to answer complex questions regarding the influence of sequential protein presentation on stem cell function or the response of cells to local gradients of chemokines or cytokines.
Acknowledgments
The authors would like to thank Josh McCall for assistance with TGF-β1 studies, Dan McKinnon for assistance with SEM imaging, and Emi Tokuda for assistance with Annexin V studies. The authors would like to thank the National Science Foundation (DMR 1006711D) and the Howard Hughes Medical Institute for funding this work. M.W. Tibbitt would like to thank the National Institutes of Health (T32 GM-065103) and the Teets Family Endowed Doctoral Fellowship for funding assistance. B.W. Han would like to thank the HHMI UROP (UC-Boulder) for funding assistance.
References
- 1.Dekanty A, Milán M. The interplay between morphogens and tissue growth. Embo Rep. 2011;12:1003–1010. doi: 10.1038/embor.2011.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 3.Plouet J, Schilling J, Gospodarowicz D. Isolation and characterization of a newly identified endothelial-cell mitogen produced by AtT-20 cells. Embo J. 1989;8:3801–3806. doi: 10.1002/j.1460-2075.1989.tb08557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang HY, Speicher DW, Sanger JW, Sanger JM, Discher DE. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freiberg S, Zhu XX. Polymer microspheres for controlled drug release. Int J Pharm. 2004;282:1–18. doi: 10.1016/j.ijpharm.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Peppas NA, Bures P, Leobandung W, Ichikawa H. Hydrogels in pharmaceutical formulations. Eur J Pharm Biopharm. 2000;50:27–46. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]
- 7.Timko BP, Whitehead K, Gao W, Kohane DS, Farokhzad O, Anderson D, Langer R. Advances in Drug Delivery. Annu Rev Mater Res. 2011;41:1–20. [Google Scholar]
- 8.Berkland C, King M, Cox A, Kim K, Pack DW. Precise control of PLG microsphere size provides enhanced control of drug release rate. J Control Release. 2002;82:137–147. doi: 10.1016/s0168-3659(02)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenedo RL, Bratt-Leal AM, Marklein RA, Seaman SA, Bowen NJ, McDonald JF, McDevitt TC. Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules. Biomaterials. 2009;30:2507–2515. doi: 10.1016/j.biomaterials.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito N, Okada T, Horiuchi H, Murakami N, Takahashi J, Nawata M, Ota H, Nozaki K, Takaoka K. A biodegradable polymer as a cytokine delivery system for inducing bone formation. Nat Biotechnol. 2001;19:332–335. doi: 10.1038/86715. [DOI] [PubMed] [Google Scholar]
- 11.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat Protoc. 2010;5:1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JA, Anseth KS. Tunable hydrogels for external manipulation of cellular microenvironments through controlled photodegradation. Adv Mater. 2010;22:61–66. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibbitt MW, Kloxin AM, Dyamenahalli KU, Anseth KS. Controlled two-photon photodegradation of PEG hydrogels to study and manipulate subcellular interactions on soft materials. Soft Matter. 2010;6:5100–5108. doi: 10.1039/C0SM00174K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz JS, Zhong S, Ricart BG, Pochan DJ, Hammer DA, Burdick JA. Modular synthesis of biodegradable diblock copolymers for designing functional polymersomes. J Am Chem Soc. 2010;132:3654–3655. doi: 10.1021/ja910606y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yesilyurt V, Ramireddy R, Thayumanavan S. Photoregulated release of noncovalent guests from dendritic amphiphilic nanocontainers. Angew Chem Int Ed. 2011;50:3038–3042. doi: 10.1002/anie.201006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinger D, Landfester K. Photo-sensitive PMMA microgels: light-triggered swelling and degradation. Soft Matter. 2011;7:1426–1440. [Google Scholar]
- 19.Peng K, Tomatsu I, van den Broek B, Cui C, Korobko AV, van Noort J, Meijer AH, Spaink HP, Kros A. Dextran based photodegradable hydrogels formed via a Michael addition. Soft Matter. 2011;7:4881–4887. [Google Scholar]
- 20.Fairbanks BD, Singh SP, Bowman CN, Anseth KS. Photodegradable, photoadaptable hydrogels via radical-mediated disulfide fragmentation reaction. Macromolecules. 2011;44:2444–2450. doi: 10.1021/ma200202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murthy N, Xu M, Schuck S, Kunisawa J, Shastri N, Fréchet JMJ. A macromolecular delivery vehicle for protein-based vaccines: acid-degradable protein-loaded microgels. P Natl Acad Sci. 2003;100:4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke DC, Brown ML, Erickson RA, Shi Y, Liu X. Transforming growth factor beta depletion is the primary determinant of Smad signaling kinetics. Mol Cell Biol. 2009;29:2443–2455. doi: 10.1128/MCB.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Zheng Q, Dakin K, Xu K, Martinez M, Li W. New caged coumarin fluorophores with extraordinary uncaging cross sections suitable for biological imaging applications. J Am Chem Soc. 2004;126:4653–4663. doi: 10.1021/ja036958m. [DOI] [PubMed] [Google Scholar]
- 24.Aujard I, Benbrahim C, Gouget M, Ruel O, Baudin JB, Neveu P, Jullien L. o-Nitrobenzyl photolabile protecting groups with red-shifted absorption: Syntheses and uncaging cross-sections for one- and two-photon excitation. Chem Eur J. 2006;12:6865–6879. doi: 10.1002/chem.200501393. [DOI] [PubMed] [Google Scholar]
- 25.Clyde DE, Corado MSG, Wu X, Paré A, Papatsenko D, Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 26.Abbott A. Cell culture: Biology's new dimension. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 27.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 28.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]


