Abstract
Microtermolides A (1) and B (2) were isolated from a Streptomyces sp. strain associated with fungus-growing termites. The structures of 1 and 2 were determined by 1D- and 2D-NMR spectroscopy and high-resolution mass spectrometry. Structural elucidation of 1 led to the re-examination of the structure originally proposed for vinylamycin (3). Based on a comparison of predicted and experimental 1H and 13C NMR chemical shifts, we propose that vinylamycin’s structure be revised from 3 to 4.
Actinomycetes, especially those belonging to the genus Streptomyces, are a rich source of biologically active natural products.1 A high rediscovery rate of known compounds from these bacteria in recent years has prompted the exploration of new environments.2 Actinomycetes associated with insects3 represent one such underexplored source.4 We have recently begun a collaborative effort to systematically explore the natural products from bacterial symbionts of eusocial insects.
In addition to exploring new niches for bacteria, we sought to further minimize rediscovery of known compounds in our large-scale survey. Various methods of early stage dereplication have been proposed.5 Recently, one of our laboratories has developed a rapid method to identify novel compounds from actinomycetes using HPLC-HRMS-based metabolomics.6 The method relies on careful processing of bacterial extracts and principal component analysis (PCA) of preprocessed LC-MS traces (Supporting Information). PCA was employed to rapidly identify unique producers from groups of actinomycetes obtained from a similar ecological niche.
Using this dereplication strategy, we have begun to investigate natural products produced by insect-associated actinomycete bacteria. In this report, we describe two new compounds likely produced by hybrid nonribosomal-polyketide (NRPS-PKS) pathways: microtermolides A (1) and B (2).
For this study, 30 termite-associated Streptomyces spp. were analyzed using LC-MS-based metabolomics. From inspection of the PCA of these 30 strains, Streptomyces sp. MspM5 was identified as having unique chemistry. The compound responsible for the variance observed in the PCA (Supporting Information Figure S1) was an [M + H]+ ion at m/z 452.2756, consistent with a molecular formula of C23H37N3O6, and a UV absorption with λmax = 221 and 262 nm. After a mass based search in Antibase, this peak could not be attributed to any known natural products and was therefore selected for further study.
Production cultures of Streptomyces sp. MspM5 were grown on solid ISP-2 medium for 7 days at 30 °C. The solid medium containing the mycelium was cut into small squares and soaked in EtOAc overnight. The EtOAc extract was dried in vacuo, redissolved in 80% MeOH/H2O, and passed through a C18 column to remove nonpolar components. The eluent from this column, which contained the compounds of interest based on LC-MS analysis, was diluted 2-fold with H2O to give a final MeOH concentration of 40%. This solution was passed through another C18 column and washed with additional 40% MeOH/H2O solution. The compounds of interest were then eluted from this column with 100% MeOH. This fraction was subjected to reversed-phase (C18) HPLC and silica gel chromatography (Supporting Information) to give pure microtermolides A (1) and B (2).
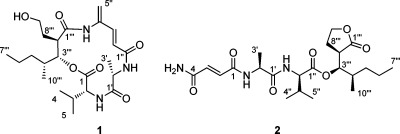
Microtermolide A (1) was isolated as a clear glass with the molecular formula C23H37N3O6, requiring seven sites of unsaturation. From the HMBC spectrum it was possible to identify four well-resolved carbonyl resonances as well as four additional olefinic carbon resonances, which together contribute six degrees of unsaturation. An analysis of the HSQC, HMBC, and COSY data obtained for 1 revealed the presence of the amino acids valine and alanine. A 1H resonance at δH 4.42 and attached to a carbon resonance at δC 59.2, typical for the α-position of an amino acid, showed a COSY correlation to a 1H resonance at δH 2.07 (Table 1). This resonance in turn showed COSY correlations to a pair of methyl resonances at δH 0.95 and δH 0.96, indicating the presence of a valine residue. A methyl resonance at δH 1.44 showed a COSY correlation to a 1H resonance at δH 4.40, which was in turn attached to a carbon resonance at δC 52.6, indicating the presence of an alanine residue. HMBC correlations from these resonances at δH 1.44 and δH 4.40 to a carbonyl carbon at δC 175.2 (C-1′) and from δH 4.42 to this same carbonyl carbon indicated that alanine was directly attached to valine in 1.
Table 1. NMR Data for Microtermolide A (1) in CD3OD.
| no. | δH,a mult (J in Hz) | δCb | HMBC |
|---|---|---|---|
| 1 | 170.4 | ||
| 2 | 4.42, m | 59.2 | 1, 3, 4, 5, 1′ |
| 3 | 2.07, dq (13.7, 6.8) | 33.4 | 1, 2, 4, 5 |
| 4 | 0.96, d (6.5) | 19.7 | 2, 3, 5 |
| 5 | 0.95, d (7.0) | 18.5 | 2, 3, 4 |
| 1′ | 175.2 | ||
| 2′ | 4.40, m | 52.6 | 3′ |
| 3′ | 1.44, d (7.0) | 18.5 | 1′, 2′ |
| 1″ | 169.2 | ||
| 2″ | 6.24, d (15.3) | 118.2 | 1″, 3″, 4″ |
| 3″ | 7.11, d (14.7) | 141.6 | 1″, 4″, 5″ |
| 4″ | 138.6 | ||
| 5″ | 5.59, s | 119.3 | 3″, 4″ |
| 5.52, s | 3″, 4″ | ||
| 1‴ | 174.3 | ||
| 2‴ | 3.00, td (10.3, 4.1) | 46.9 | 1″, 3″, 8″ |
| 3‴ | 5.48, dd (10.3, 2.1) | 78.0 | 1, 1‴, 2‴, 4‴, 5‴, 10‴ |
| 4‴ | 1.88, m | 34.9 | 5‴, 10‴ |
| 5‴ | 1.34, m | 37.3 | 3‴, 4‴, 6‴, 10‴ |
| 1.16, m | 3‴, 4‴, 6‴, 10‴ | ||
| 6‴ | 1.42, m | 21.2 | 4‴, 5‴, 7‴ |
| 7‴ | 0.90, t (7.0) | 14.2 | 5‴, 6‴ |
| 8‴ | 1.80, m | 33.4 | 1‴, 2‴, 3‴, 9‴ |
| 9‴ | 3.66, m | 59.9 | 2‴, 8‴ |
| 3.56, m | 2‴, 8‴ | ||
| 10‴ | 1.06, d (7.0) | 13.4 | 3‴, 4‴, 5‴ |
600 MHz.
150 MHz.
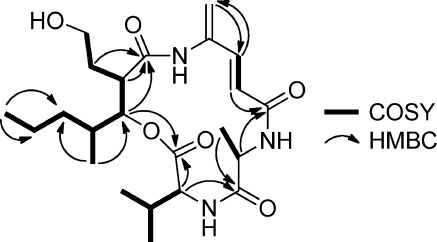
A COSY correlation between δH 6.24 and δH 7.11 and HMBC correlations from an olefinic methylene at δH 5.59 and δH 5.52 to δC 141.6 and from δH 7.11 to δC 119.3 showed that the two carbon–carbon double bonds in 1 were conjugated. The downfield nature of C-4″ (δC 138.6) and the lack of HMBC correlations from δH 5.59, δH 5.52, and δH 7.11 to an additional carbon resonance suggested that C-4″ was attached to a nitrogen atom. HMBC correlations from δH 7.11 and δH 6.24 to a carbonyl carbon resonance at δC 169.2 and from δH 4.40 (Ala-Hα) to this same carbonyl carbon indicated that this fragment was linked to the alanine residue.
An oxygenated methine resonance at δH 5.48 and attached to a carbon resonance at δC 78.0 showed HMBC correlations to carbonyl carbons at δC 170.4 and δC 174.3. An HMBC correlation from δH 4.42 (Val-Hα) to the same carbonyl carbon at δC 170.4 indicated that C-3‴ was linked to C-1 through an ester bond and suggested that 1 is a depsipeptide. A series of COSY correlations (Figure 1) revealed a fragment likely derived from a polyketide pathway and suggests that 1 is a putative hybrid polyketide-nonribosomal peptide. Finally, in order to satisfy the molecular formula obtained for 1, C-4″ must be linked to C-1‴ through an amide bond to form a cyclic depsipeptide.
Figure 1.
Key COSY and HMBC correlations for microtermolide A (1).
The absolute configurations of the alanine and valine residues in 1 were determined by Marfey’s method.7 A small sample of 1 (0.1 mg) was hydrolyzed with 6 N HCl at 110 °C for 6 h and derivatized with Marfey’s reagent. A comparison by LC-MS of the Marfey’s derivatives derived from 1 with Marfey’s derivatives prepared from the d- or l-amino acids showed that 1 contains l-alanine and d-valine residues (Supporting Information).
The relative configuration of the polyketide-derived fragment was determined by a NOESY experiment combined with molecular modeling. The presence of three stereogenic centers in the polyketide-derived fragment of 1 leaves four possibilities for the relative configuration. A NOESY correlation was observed between H-2‴ (δH 3.00) and H-10‴ (δH 1.06), but not between H-2‴ (δH 3.00) and either of the valine methyls (δH 0.95 and 0.96). Since the distance between H-2‴ and the valine methyl protons in the energy-minimized conformation is smaller than the distance between H-2‴ and H-10‴ for both the 2‴R*,3‴S*,4‴S* and 2‴R*,3‴S*,4‴R* isomers (2.98 vs 3.99 Å and 2.73 vs 4.32 Å, respectively) these two possibilities were eliminated. An additional NOESY correlation was observed between H-3‴ (δH 5.48) and H-5a‴ (δH 1.34), while only a weak correlation was observed between H-3‴ (δH 5.48) and H-10‴ (δH 1.06). Since the distance between H-3‴ and H-10‴ in the energy-minimized conformation of the 2‴R*,3‴R*,4‴S* isomer is smaller than the distance between H-3‴ and H-5a‴ (2.32 vs 3.78 Å) this possibility was eliminated. On the other hand, all of the observed NOESY correlations for the 2‴R*,3‴R*,4‴R* isomer are consistent with its calculated energy-minimized conformation. Therefore, the relative configuration of the polyketide fragment in 1 has tentatively been assigned as 2‴R*,3‴R*,4‴R*. The absolute configuration of this fragment has not been determined, so the stereochemistry shown in 1 represents a mix of relative and absolute assignments.
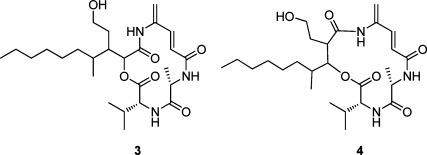
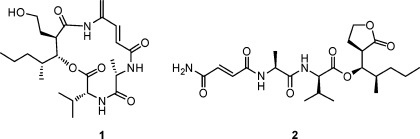
A literature search revealed that 1 is closely related to the known antibiotic vinylamycin.8 The reported structure of vinylamycin has the ester oxygen attached to C-2‴, while in 1 it is attached to C-3‴. The similarity in their structures, aside from the placement of this oxygen atom, prompted us to re-examine the structure of vinylamycin. The reported chemical shifts for C-3‴ (δC 44.97) and H-3‴ (δH 2.97) in vinylamycin do not closely match the values predicted by the ACD/NMR Predictor software. Placing this resonance α to the carbonyl carbon at C-1‴ (Figure 2) explains the downfield shift of this resonance and matches much more closely with the predicted values (Table 2). Thus we believe that the structure of vinylamycin should be revised to place the ester at the β-position, which is more in line with typical biosynthesis of mixed polyketide-nonribosomal peptides.9
Figure 2.
Original (3) and revised (4) structures of vinylamycin.
Table 2. Calculated and Experimental 13C and 1H NMR Chemical Shifts for Reported (3) and Revised (4) Vinylamycin.
| reported (3) |
revised (4) |
|||||
|---|---|---|---|---|---|---|
| position | calcda | exptl | Δδ | calcda | exptl | Δδ |
| C-1‴ | 171.7 | 171.6 | 0.1 | 175.6 | 171.6 | 4.0 |
| C-2‴ | 70.0 | 76.4 | –6.4 | 47.3 | 45.0 | 2.3 |
| C-3‴ | 29.8 | 45.0 | –15.2 | 78.2 | 76.4 | 1.8 |
| C-4‴ | 34.8 | 33.6 | 1.2 | 32.1 | 33.6 | –1.5 |
| H-2‴ | 4.94 | 5.22 | –0.28 | 2.92 | 2.97 | –0.05 |
| H-3‴ | 1.78 | 2.97 | –1.19 | 5.35 | 5.22 | 0.13 |
| H-4‴ | 1.52 | 1.77 | –0.25 | 1.84 | 1.77 | 0.07 |
Weighted average experimental values (ppm) calculated for DMSO-d6 using ACD/NMR predictor. Δδ represents the deviation in chemical shift between calculated and experimental values (δcalcd – δexptl)
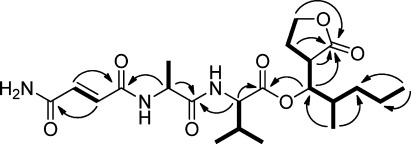
Microtermolide B (2) was isolated as a clear glass with the molecular formula C22H35N3O7 and differed from the molecular formula of 1 by the addition of an oxygen atom and the loss of CH2. The NMR data obtained for 2 were very similar to those of 1, with the major differences being in the region of C-2″ to C-5″, as well as C-8‴ to C-9‴. The 1H NMR chemical shifts of the oxygenated methylene at H-9‴ (δH 3.66 and 3.56) in 1 were shifted downfield in 2 to δH 4.31 and 4.18 (H-9‴), and both of these resonances showed HMBC correlations to a carbonyl carbon at δC 178.6 (C-1‴) suggesting that the hydroxyl group attached to C-9‴ forms a δ-lactone with C-1‴. The exocyclic methylene in 1 (C-5″) was not present in 2. Instead, the olefinic protons, which had shifted to δH 6.94 (H-2) and δH 6.90 (H-3), showed HMBC correlations to carbonyl carbons at δC 168.7 (C-1) and δC 166.2 (C-4). Thus, the C-5″ methylene found in 1 has been replaced with an oxygen atom in 2, satisfying the molecular formula of 2 and completing its structure (Figure 3).
Figure 3.
Key COSY and HMBC correlations in microtermolide B (2).
Microtermolide B (2) is a member of the relatively rare class of linear depsipeptides, which includes dolastatin 15, symplocin A, symplostatin 3, and gallinamide A, all of which were isolated from cyanobacteria.10 To our knowledge this is the first such example from a Streptomyces sp. It is unclear whether 1 is a precursor to 2 and whether such a transformation would be spontaneous or enzymatically catalyzed.
Marfey’s analysis of 2 was performed using the same procedure as that for 1. This analysis indicated that 2 contains the same configuration of amino acids, l-alanine and d-valine, as in 1. The polyketide fragment in 2 is expected to be less conformationally restricted than 1, precluding the use of NOESY or ROESY in determining the relative configuration of this fragment. However, since 1 and 2 are likely derived from the same biosynthetic pathway, it is likely that they have the same stereochemistry. Thus, the relative configuration in this fragment in 2 has also tentatively been assigned as 2‴R*,3‴R*,4‴R*.
Vinylamycin is reported to have moderate antimicrobial activity against a variety of gram-positive bacteria.8 Neither 1 nor 2 showed any antimicrobial activity against Bacillus subtilis 3610 at concentrations as high as 100 μg/mL. They also had no antifungal activity against Saccharomyces cerevisiae at concentrations as high as 100 μg/mL, but further assays to define their functional roles are continuing.
The discovery of 1 and 2 from termite-associated Streptomyces sp. provides further evidence that insect-associated actinomycetes are a promising yet underexplored source of novel natural products.
Acknowledgments
This work was supported by NIH RC4GM096347 (C.R.C. and J.C.) and an NSERC (Canada) postdoctoral fellowship (to J.L.K.).
Supporting Information Available
Experimental details, tables of NMR assignments, and NMR spectra for 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Nett M.; Ikeda H.; Moore B. S. Nat. Prod. Rep. 2009, 26, 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bérdy J. J. Antibiot. 2005, 1, 1–26. [DOI] [PubMed] [Google Scholar]
- a Thornburg C. C.; Zabriskie T. M.; McPhail K. L. J. Nat. Prod. 2010, 73, 489–499. [DOI] [PubMed] [Google Scholar]; b Pettit R. K. Mar. Biotechnol. 2011, 13, 1–11. [DOI] [PubMed] [Google Scholar]; c Rateb M. E.; Houssen W. E.; Harrison W. T. A.; Deng H.; Okoro C. K.; Asenjo J. A.; Andrews B. A.; Bull A. T.; Goodfellow M.; Ebel R.; Jaspars M. J. Nat. Prod. 2011, 74, 1965–1971. [DOI] [PubMed] [Google Scholar]
- Bode H. B. Angew. Chem., Int. Ed. 2009, 48, 6394–6396. [DOI] [PubMed] [Google Scholar]
- a Oh D.-C.; Poulsen M.; Currie C. R.; Clardy J. Nat. Chem. Biol. 2009, 5, 391–393. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Oh D.-C.; Scott J. J.; Currie C. R.; Clardy J. Org. Lett. 2009, 11, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Oh D.-C.; Poulsen M.; Currie C. R.; Clardy J. Org. Lett. 2011, 13, 752–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashidhara K. V.; Rosaiah J. N. Nat. Prod. Commun. 2007, 2, 193–202. [Google Scholar]
- Hou Y.; Braun D. R.; Michel C. R.; Klassen J. L.; Adnani N.; Wyche T. P.; Bugni T. S.. Anal. Chem.2012, 84, 4277–4283. [DOI] [PMC free article] [PubMed]
- Marfey P. Carlsberg Res. Commun. 1984, 49, 591–596. [Google Scholar]
- Igarashi M.; Shida T.; Sasaki Y.; Kinoshita N. J. Antibiot. 1999, 52, 873–879. [DOI] [PubMed] [Google Scholar]
- Koglin A.; Walsh C. T. Nat. Prod. Rep. 2009, 26, 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bai R.; Friedman S. J.; Pettit G. R.; Hamel E. Biochem. Pharmacol. 1992, 43, 2637–2645. [DOI] [PubMed] [Google Scholar]; b Molinski T. F.; Reynolds K. A.; Morinaka B. I. J. Nat. Prod. 2012, 75, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Luesch H.; Yoshida W. Y.; Moore R. E.; Paul V. J.; Mooberry S. L.; Corbett T. H. J. Nat. Prod. 2002, 65, 16–20. [DOI] [PubMed] [Google Scholar]; d Linington R. G.; Clark B. R.; Trimble E. E.; Almanza A.; Urena L.-D.; Kyle D. E.; Gerwick W. H. J. Nat. Prod. 2009, 72, 14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.