Abstract
Microwave synthesis was utilized to rapidly build Py-Im polyamides in high yields and purity using Boc-protection chemistry on Kaiser oxime resin. A representative polyamide targeting the 5′-WGWWCW-3′ (W = A or T) subset of the consensus Androgen and Glucocorticoid Response Elements was synthesized in 56% yield after 20 linear steps and HPLC purification. It was confirmed by Mosher amide derivatization of the polyamide that a chiral α-amino acid does not racemize after several additional coupling steps.
Defined arrangements of DNA-binding transcription factors drive gene expression programs.1 Aberrant gene expression caused by mutated or overexpressed transcription factors may lead to diseased states, such as cancer. Methods to disrupt specific protein–DNA interfaces in living systems may enable new chemical strategies for the design of therapeutics to control such transcriptional aberrations.
To this end, N-methylpyrrole-N-methylimidazole (Py-Im) polyamides have been developed to bind and recognize all four base pairs of the DNA minor groove.2 Oligomers constructed from cofacial pairings of aromatic amino acids Py/Py recognize A·T and T·A;3 Py/Im recognizes C·G; Im/Py recognizes G·C;4 3-hydroxypyrrole/Py (Hp/Py) recognizes T·A; and Py/Hp recognizes A·T.5 Cyclic (and by extension hairpin) Py-Im polyamides bind and widen the minor groove of DNA, distorting the DNA structure sufficiently to perturb binding of individual transcription factors.6 Polyamides can modulate hypoxia inducible factor-1α (HIF-1α), transforming growth factor-β1 (TGF-β1), androgen receptor (AR), glucocorticoid receptor (GR), and nuclear factor kappa B (NF-κB)-driven gene expression in cell culture.7
Early hairpin Py-Im polyamide solid phase syntheses contained β-alanine at the C-terminus due to the use of PAM resin.8 Nuclear uptake of these polyamides was improved by the deletion of the β-alanine moiety and hence use of other resins such as Kaiser oxime,9 or the safety-catch hydrazine resin was desirable.10 We have typically used the Kaiser oxime resin to synthesize biologically active Py-Im polyamides on solid phase.
Syntheses of eight-ring Py-Im polyamides, however, have been plagued by low yields (typically 1%) and lengthy durations (4–5 days).7a,7c−7e,9 Much research has explored impovements to Py-Im polyamide synthesis.11 Use of preassembled trimers and tetramers (synthesized in solution phase) enables Py-Im polyamide yields of ∼10% on Kaiser oxime resin;12 however, this necessitates a significant amount of solution phase chemistry to ensure generality for a variety of polyamide core sequences. The recent solution-phase total synthesis of a Py-Im polyamide core targeted to the androgen response element (ARE) offered gram scale quantities of unpurified material in 42% yield. Milligram quantities were purified by HPLC.11i While solution phase methods11b,11f,11i are an excellent option for large-scale preparation of known, useful polyamides, a faster method offering high, clean yields would enable a diverse library of polyamide cores to be rapidly synthesized and studied in cell culture and animal models. Advances in solid phase chemistry have employed HOAt ester (1-hydroxy-7-azabenzotriazole) activation to generate an achiral polyamide on Boc-β-Ala-PAM resin targeting a 5′-WGWWCW-3′ sequence in 20% yield.11dIn situ acid chloride activation methods enable as high as a 29% yield for synthesis of an eight-ring polyamide on a hydrazine solid support.10 Both methods reduce coupling times to 20 min.10,11d Acid chloride activation methods necessitate the use of triphosgene, a toxic chemical, and have not demonstrated retention of chirality during the incorporation of chiral α-amino acid monomers, a potential concern.10
Microwave synthesizers have gained popularity in solution phase and solid phase organic synthesis, enabling rapid, precise equilibration to programmed temperatures. Numerous examples demonstrate selective product formation, side-product minimization, high yields, and fast reaction times through use of a microwave synthesizer.13 Their utility has extended to the synthesis of peptides14 and Py-Im polyamides using Fmoc-protected coupling chemistry (Fmoc-β-Ala-CLEAR Resin).11h For the synthesis of peptides, microwave assisted couplings are known in many instances to preserve the stereochemistry encoded in chiral α-amino acids.15 Application of Kaiser oxime resin has precedent for use with Boc-protection chemistry.16 Because of the higher yields, faster coupling times, and cleaner products enabled by microwave synthesizers for the solid phase synthesis of peptides,14 we have developed microwave synthesis methods to build Py-Im polyamides on Kaiser oxime resin using Boc-protected amide coupling chemistry.
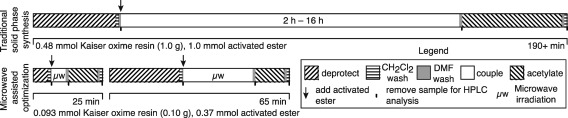
The solid phase synthesis of a Py-Im polyamide is iterative and divided into distinct phases. Following resin loading, a sequence of steps is repeated to add each monomer: deprotection of the terminal amine of the resin-bound polyamide, monomer activation, monomer coupling to resin-bound polyamide, and capping of unreacted amine groups. Between each step, the resin is washed with one or more solvents. Once the polyamide is complete, it is cleaved from the resin. A full coupling cycle—deprotection, washing, coupling, washing, capping, washing—is depicted in Figure 1. Typical coupling cycle times on Kaiser oxime resin utilizing room temperature manual solid phase synthesis methods have been significantly reduced by using a microwave synthesizer. Within each coupling cycle, we have used the microwave synthesizer to reduce the times necessary for coupling reactions; we have also reduced reaction times for the deprotection of Boc-protected amines and acetylation of unreacted amines.
Figure 1.
Overview of coupling cycle times for one full iteration of solid phase synthesis on Kaiser oxime resin. Room temperature solid phase synthesis cycles are significantly slower (≥190 min) than microwave optimized conditions for N-methylpyrrole nucleophiles (25 min total) and N-methylimidazole nucleophiles (median 65 min total). Boxes are scaled to the time required for each step within a coupling cycle.
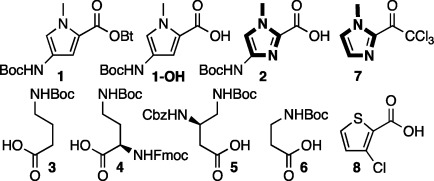
Py-Im polyamides are synthesized from a toolbox of monomers that include Boc-Py-OBt (1), Boc-Py-OH (1- OH), Boc-Im-OH (2), Boc-γ-Abu-OH (3), Fmoc-d-Dab(Boc)-OH (4), Z-β-Dab(Boc)-OH (5), Boc-β-Ala-OH (6), Im-CCl3 (7), and Ct-OH (8) (Figure 2). Monomers 3–5 function to covalently link two strands of aromatic heterocycles to form either a hairpin polyamide or a cyclic polyamide.2,11j,17 Monomer 6 functions as a surrogate for 1 in applications that need more conformational flexibility of the finished polyamide.18 Monomers 7 and 8 are capping monomers, used at the N-terminus of hairpin polyamides.19
Figure 2.
Amino acid monomers for synthesis of Py-Im polyamides.
The Kaiser oxime resin can be loaded with Boc-Py-OBt by treatment with 4 equiv each of 1 (0.4 M final concentration) and DIEA in DMF (80 °C, 3 h, microwave).
Reduced TFA deprotection times were found for resin-loaded Boc-protected Py (5 min), Im (25 min), and a representative Boc-protected aliphatic amine (5 min) using 80:1:19 TFA/triethylsilane/CH2Cl2 (SI Figure 2). For deprotection of Im, it was critical to utilize protein sequencing grade, redistilled TFA.
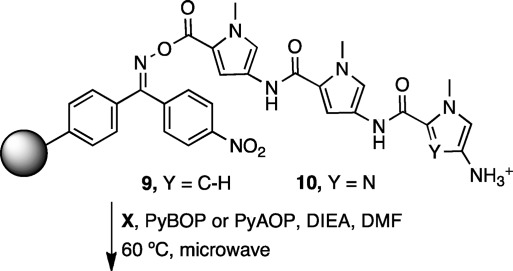
Two oligomer cores were studied for optimization of coupling times on Kaiser oxime resin (Figure 3), one terminating with a Py nucleophile (9) and the other with an Im nucleophile (10). Aliphatic amine nucleophiles have equivalent coupling times to the Py nucleophile. As revealed in Table 1, 5 min coupling times are sufficient for most monomers to 9. Typically, 25 min at 60 °C is required for coupling electrophilic X to 10. For couplings from 10 to certain monomers (especially 1), significantly extended reaction times were required to achieve acceptable yields. In these cases, PyAOP was used, which forms more reactive −OAt esters (Table 2).
Figure 3.
Nucleophiles utilized to determine optimum coupling conditions.
Table 1. Coupling Conditions Using HOBt-Ester Chemistrya.
| resin | X |
X equiv |
DIEA equiv |
PyBOP equiv |
[X] in DMF |
time (min) |
yieldb (%) |
|---|---|---|---|---|---|---|---|
| 9 | 1 | 4 | 2 | – | 0.3 M | 5 | 99 |
| 2 | 4 | 6 | 4 | 0.3 M | 10 | 99 | |
| 3 | 4 | 6 | 4 | 0.3 M | 5 | 99 | |
| 4 | 4 | 6 | 4 | 0.3 M | 10 | 98 | |
| 5 | 4 | 6 | 4 | 0.3 M | 5 | 99 | |
| 6 | 4 | 6 | 4 | 0.3 M | 5 | 99 | |
| 7 | 4 | 2 | – | 0.3 M | 10 | 98 | |
| 8 | 4 | 6 | 4 | 0.3 M | 5 | 95 | |
| 10 | 1 | 6 | 3 | – | 0.45 M | 240 | 91c |
| 2 | 4 | 6 | 4 | 0.3 M | 30 | 96 | |
| 3 | 4 | 6 | 4 | 0.3 M | 25 | 99 | |
| 4 | 4 | 6 | 4 | 0.3 M | 25 | 98 | |
| 5 | 4 | 6 | 4 | 0.3 M | 25 | 98 | |
| 6 | 4 | 6 | 4 | 0.3 M | 5 | 97 | |
| 7 | 4 | 2 | – | 0.3 M | 60 | 78 | |
| 8 | 4 | 6 | 4 | 0.3 M | 20 | 93 |
Except as noted, all reactions were run at 60 °C (microwave).
Yield approximated by integration of analytical HPLC signal as described in the Supporting Information.
Coupling run at 80 °C (microwave).
Table 2. Coupling Conditions Using HOAt-Ester Chemistrya.
| resin | X |
X equiv |
DIEA equiv |
PyBOP equiv |
[X] in DMF |
time (min) |
yieldb (%) |
|---|---|---|---|---|---|---|---|
| 10 | 1-OH | 4 | 6 | 4 | 0.3 M | 60 | 96c |
| 2 | 4 | 6 | 4 | 0.3 M | 5 | 98 | |
| 6 | 4 | 6 | 4 | 0.3 M | 5 | 98 | |
| 8 | 4 | 6 | 4 | 0.3 M | 5 | 96 |
Except as noted, all reactions were run at 60 °C (microwave).
Yield approximated by integration of analytical HPLC signal as described in the Supporting Information.
Coupling run at 80 °C (microwave).
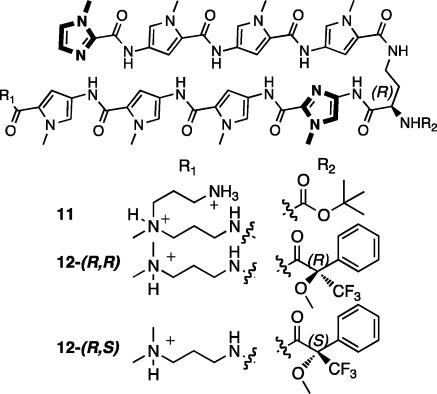
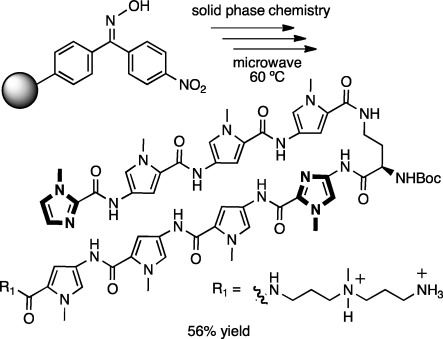
The core of a biologically relevant polyamide (11) targeting the 5′-WGWWCW-3′ subset of the consensus androgen and glucocorticoid response element sequences has been synthesized stepwise by solid phase synthesis in a microwave synthesizer (Figure 4, 100 mg, 93 μmol scale) using the loading methods, monomers, deprotection conditions, and coupling times described above.
Figure 4.
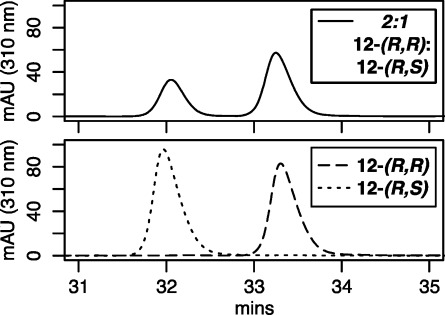
Polyamide 11 specifying the 5′-WGWWCW-3′ subset of the ARE and GRE consensus sequences. Polyamides 12-(R,S) and 12-(R,R) were used for Mosher amide analysis.
Polyamide 11 was cleaved from resin in the microwave using 3,3′-diamino-N-methyl-dipropylamine, precipitated using diethyl ether, and purified by HPLC, affording a 56% yield. Synthesis of polyamide 11 involves nine coupling cycles, including resin loading (which can be viewed as coupling Py to the resin), plus a protecting group exchange and a cleavage step. If the latter two steps and purification are assumed to be quantitative, our 56% overall yield would indicate an average yield of 94% per coupling cycle. However, the actual synthetic yield is likely to be higher (see discussion in the SI).
Because the chiral turn unit 4 used to synthesize polyamide 11 contains an acidic alpha proton adjacent to an amide carbonyl and the coupling reactions were run at elevated temperature in modestly basic conditions, we were concerned whether the stereochemical information encoded in the turn unit would survive several additional coupling steps. We synthesized Mosher amide derivatives2012-(R,S) and 12-(R,R) to approximate the enantiomeric excess of the chiral polyamide 11 (Figure 4). UPLC was utilized to separate the two diastereomers (Figure 5). By integrating the areas corresponding to each diastereomer UV/vis peak, we calculate a 99% de for both 12-(R,S) and 12-(R,R). This de compares favorably to the 98% de found for samples synthesized by nonmicrowave methods.
Figure 5.
Polyamides 12-(R,S) and 12-(R,R) are separable by UPLC. An overlay of three separate UPLC runs (λ = 310 nm) is shown. A 2:1 ratio of 12-(R,R):12-(R,S) was utilized to optimize separation and to assign retention times to diastereomers. Individual runs of 12-(R,R) and 12-(R,S) each revealed 99% de by integration of the area of the UV–vis signal (λ = 310 nm).
We anticipate that the simplicity and accessibility of these microwave-assisted solid phase synthesis methods will enable a broadening of Py-Im polyamide research into the biological community.
Acknowledgments
Research support was provided by the National Institutes of Health Research Grant GM27681. UPLC work was performed in the Center for Catalysis and Chemical Synthesis in the Warren and Katharine Schlinger Laboratory for Chemistry and Chemical Engineering. We thank Amanda Hargrove and Scott Virgil (California Institute of Technology) for helpful discussions.
Supporting Information Available
Supplementary data, including synthetic procedures and analytical characterization data, can be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Farnham P. J. Nat. Rev. Genet. 2009, 10, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Bioorg. Med. Chem. 2001, 9, 2215–2235. [DOI] [PubMed] [Google Scholar]
- Pelton J. G.; Wemmer D. E. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 5723–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade W. S.; Mrksich M.; Dervan P. B. J. Am. Chem. Soc. 1992, 114, 8783–8794. [Google Scholar]
- White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B. Nature 1998, 391, 468–471. [DOI] [PubMed] [Google Scholar]
- Chenoweth D. M.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 13175–13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Olenyuk B. Z.; Zhang G.-J.; Klco J. M.; Nickols N. G.; Kaelin W. G. Jr.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16768–16773. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lai Y.-M.; Fukuda N.; Ueno T.; Kishioka H.; Matsuda H.; Saito S.; Matsumoto K.; Ayame H.; Bando T.; Sugiyama H.; Mugishima H.; Kim S.-J. J. Pharmacol. Exp. Ther. 2005, 315, 571–575. [DOI] [PubMed] [Google Scholar]; c Nickols N. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Muzikar K. A.; Nickols N. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Raskatov J. A.; Meier J. L.; Puckett J. W.; Yang F.; Ramakrishnan P.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2011, 109, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird E. E.; Dervan P. B. J. Am. Chem. Soc. 1996, 118, 6141–6146. [Google Scholar]
- Edelson B. S.; Best T. P.; Olenyuk B.; Nickols N. G.; Doss R. M.; Foister S.; Heckel A.; Dervan P. B. Nucleic Acids Res. 2004, 32, 2802–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.; Gray S. J.; Dondi R.; Burley G. A. Org. Lett. 2009, 11, 3910–3913. [DOI] [PubMed] [Google Scholar]
- a König B.; Rödel M. Chem. Commun. 1998, 5, 605–606. [Google Scholar]; b Xiao J. H.; Yuan G.; Huang W. Q.; Chan A. S. C.; Lee K. L. D. J. Org. Chem. 2000, 65, 5506–5513. [DOI] [PubMed] [Google Scholar]; c Wurtz N. R.; Turner J. M.; Baird E. E.; Dervan P. B. Org. Lett. 2001, 3, 1201–1203. [DOI] [PubMed] [Google Scholar]; d Krutzik P. O.; Chamberlin A. R. Bioorg. Med. Chem. Lett. 2002, 12, 2129–2132. [DOI] [PubMed] [Google Scholar]; e Choi J. S.; Lee Y.; Kim E.; Jeong N.; Yu H.; Han H. Tetrahedron Lett. 2003, 44, 1607–1610. [Google Scholar]; f Sinyakov A. N.; Feshchenko M. V.; Ryabinin V. A. Russ. J. Bioorg. Chem. 2004, 30, 98–99. [Google Scholar]; g Harris D.; Stewart M.; Sielaff A.; Mulder K.; Brown T.; Mackay H.; Lee M. Heterocycl. Commun. 2007, 13, 17–23. [Google Scholar]; h Bando T.; Fujimoto J.; Minoshima M.; Shinohara K.-I.; Sasaki S.; Kashiwazaki G.; Mizumura M.; Sugiyama H. Bioorg. Med. Chem. 2007, 15, 6937–6942. [DOI] [PubMed] [Google Scholar]; i Chenoweth D. M.; Harki D. A.; Dervan P. B. J. Am. Chem. Soc. 2009, 131, 7175–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]; j Chenoweth D. M.; Harki D. A.; Phillips J. W.; Dervan P. B. J. Am. Chem. Soc. 2009, 131, 7182–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harki D. A.; Satyamurthy N.; Stout D. B.; Phelps M. E.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappe C. O. Angew. Chem., Int. Ed. 2004, 43, 6250–6284. [DOI] [PubMed] [Google Scholar]
- a Bacsa B.; Horváti K.; Bõsze S; Andreae F.; Kappe C. O. J. Org. Chem. 2008, 73, 7532–7542. [DOI] [PubMed] [Google Scholar]; b Olivos H. J.; Alluri P. G.; Reddy M. M.; Salony D.; Kodadek T. Org. Lett. 2002, 4, 4057–4059. [DOI] [PubMed] [Google Scholar]; c Murray J. K.; Gellman S. H. Org. Lett. 2005, 7, 1517–1520. [DOI] [PubMed] [Google Scholar]; d Baptiste B.; Douat-Casassus C.; Laxmi-Reddy K.; Godde F.; Huc I. J. Org. Chem. 2010, 75, 7175–7185. [DOI] [PubMed] [Google Scholar]
- Palasek S. A.; Cox Z. J.; Collins J. M. J. Pept. Sci. 2007, 13, 143–148. [DOI] [PubMed] [Google Scholar]
- DeGrado W. F.; Kaiser E. T. J. Org. Chem. 1980, 45, 1295–1300. [Google Scholar]
- Dose C.; Farkas M. E.; Chenoweth D. M.; Dervan P. B. J. Am. Chem. Soc. 2008, 130, 6859–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M.; Swalley S. E.; Baird E. E.; Dervan P. B. J. Am. Chem. Soc. 1998, 120, 6219–6226. [Google Scholar]
- Dervan P. B.; Poulin-Kerstien A. T.; Fechter E. J.; Edelson B. S. Top. Curr. Chem. 2005, 253, 1–31. [Google Scholar]
- Based on the original work in: Dale J. A.; Mosher H. S. J. Am. Chem. Soc. 1973, 95, 512–519. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.