Abstract
In this paper, we describe the expression and characterization of recombinant human cystathionine β-synthase (CBS) in Escherichia coli. We have used a glutathione-S-transferase (GST) fusion protein vector and incorporated a cleavage site with a long hinge region which allows for the independent folding of CBS and its fusion partner. In addition, our construct has the added benefit of yielding a purified CBS which only contains one extra glycine amino acid residue at the N-terminus. In our two-step purification procedure we are able to obtain a highly pure enzyme in sufficient quantities for crystallography and other physical chemical methods. We have investigated the biochemical and catalytic properties of purified full-length human CBS and of two truncation mutants lacking the Cterminal domain or both the N-terminal heme-binding and the C-terminal regulatory regions. Specifically, we have determined the pH optima of the different CBS forms and their kinetic and spectral properties. The full-length and the C-terminally truncated enzyme had a broad pH 8.5 optimum while the pH optimum of the N- and C- terminally truncated enzyme was sharp and shifted to pH 9. Furthermore, we have shown unequivocally that CBS binds one mole of heme per subunit by determining both the heme and the iron content of the enzyme. The activity of the enzyme was unaffected by the redox status of the heme iron. Finally, we show that CBS is stimulated by S-adenosyl-L-methionine but not its analogs.
Keywords: Homocystinuria, S-adenosylmethionine, Pyridoxal-5′ phosphate, Heme
Cystathionine β-synthase (CBS)2 (L-Serine hydrolyase (adding homocysteine), EC 4.2.1.22) is central to sulfur amino acid metabolism in eukaryotes, and complete CBS deficiency is the principal cause of homocystinuria in humans. The CBS-catalyzed condensation of serine and homocysteine generates cystathionine which is then cleaved with cystathionine γ-lyase (CGL) to yield cysteine [1].
Cysteine synthase (O-acetylserine sulfhydrase, OASS; CS), the CBS counterpart in prokaryotes, plants and enteric protozoa, catalyzes the formation of cysteine from O-acetylserine and hydrogen sulfide (H2S) with the concomitant release of acetic acid. Unlike mammals, these organisms are able to assimilate inorganic sulfur into organic sulfur and are therefore essential players in the sulfur cycle in nature. CS and CBS are affiliated with the large β-family of pyridoxal-5′-phosphate (PLP) dependent enzymes and have a high degree of sequence and structural conservation within the catalytic core [2,3], (see also www.uchsc.edu/sm/cbs/cbsdata/cbsprotein.htm). An evolutionary relationship between these enzymes is also illustrated by the fact that some CBS enzymes have retained CS activity [4]. CS and CBS, together with other enzymes such as β-cyanoalanine synthase, the β-subunit of tryptophan synthase and cysteine lyase, catalyze primarily β-replacement reactions [5,6]. Despite catalyzing mostly one reaction type, CS and CBS can utilize a wide range of substrates [7].
CBS is the only PLP dependent enzyme which contains heme [7]. The heme is co-ordinated by Cys 52 and His 65 residues located in the N-terminal region of the enzyme. The function of this ligand is yet to be determined; however, several laboratories have shown that it is not directly involved in the catalytic process [8–11], thus contradicting previous reports that have shown catalytic involvement [12,13]. CBS from lower eukaryotes such as Saccharomyces cerevisiae do not contain heme [8,9]. Thus the absence of this cofactor in CBS from lower eukaryotes suggests that the function of heme in this enzyme is unique to higher organisms.
Human CBS activity is regulated by S-adenosyl-L-methionine (AdoMet) [14]. Interestingly, all CS and some CBS enzymes from lower eukaryotes are not responsive to AdoMet suggesting that the requirement by CBS of a heme moiety and regulation by AdoMet may have emerged at the same time evolutionarily and may be interrelated.
The purification of CBS from mammalian tissues is complicated by its tendency for aggregation and its susceptibility to proteolysis [15]. Previously, an Escherichia coli expression system using β-galactosidase (β-gal) was used for the purification of CBS [16]. This system had several disadvantages. There was variable undesirable proteolytic cleavage within E. coli between β-gal and CBS. The proteolytic cleavage produced free β-gal and CBS prematurely making it impossible to use an affinity chromatography step to obtain purified CBS. Because of these complications, we decided to purify CBS using an alternate expression system. The expression of CBS with a Glutathione-S-Transferase (GST) affinity tag was first described by Warren Kruger’s laboratory [17]. The disadvantage of that system was that the cleaved purified CBS still retained 11 non-CBS residues at its N-terminus [18].We have previously expressed human CBS with a GST tag, which upon purification and removal of the fusion partner yielded CBS with 23 extra residues at the N-terminus [19]. Subsequently, we demonstrated that N-terminal elongation altered the affinity of the enzyme for Hcy. This enzyme had ~9-fold lower Km for Hcy [20] compared to the WT enzyme [21] and was used to obtain the first CBS structure [22]. Here we describe a method to purify CBS using a GST fusion protein which yields CBS as close to normal sequence as possible. Using our construct we are able to obtain a purified enzyme with only one extra small amino acid residue, glycine, at the Nterminus of the purified CBS protein.
The new method gives us several other benefits. First, the vector we chose has a long recognition sequence for the precision protease cleavage site which provides a convenient long hinge region between GST and CBS allowing independent folding of the two enzymes. Second, this system gives the added benefit of a shorter fusion partner which enables us to rapidly purify large amounts of CBS in an easy, two-step procedure. In this paper we describe the expression and purification of the full-length wild type CBS as well as CBS which does not contain the C-terminal regulatory region (1–413) and CBS lacking both the N- and C-terminal regions (71–400). The 71–400 CBS does not contain heme and is highly conserved with the CS family of enzymes. We have further characterized all three forms of CBS with respect to their biochemical and physical properties.
Materials and methods
Chemicals
Unless stated otherwise, all materials were purchased from Sigma. L-[U-14C] serine was obtained from Perkin Elmer Life Sciences.
Cloning of CBS wild type, CBS (1–413) and CBS (71–400)
In order to express the human CBS wild type protein, a construct pGEX-6P-1 HCBS WT was prepared. This construct enabled us to express CBS as a fusion protein with a glutathione-S-transferase (GST) tag, which could be later cleaved off with PreScission protease (Amersham Biosciences, Piscataway, NJ). First, the commercially available pGEX-6P-1 vector (Amersham Biosciences, Piscataway, NJ) was modified by destroying the internal Apa I site located at the nucleic acid position 3890 of the pGEX-6P-1 vector as described previously [11] and cut with ApaI/SalI restriction enzymes to create an ApaI–SalI cassette. Then, two primers (sense: 5′-ctctgagaccccccaggcagaagtggggcc-3′ and antisense: 5′-ccacttctgcctggggggtctcagagggcc-3′) encoding the last two C-terminal amino acids of the PreScission protease site and the first ten amino acids of the CBS enzyme were designed. Each primer (1.5 nmol) in 50 μl total volume was phosphorylated in a separate tube using 10 U of polynucleotide kinase (New England Biolabs Ipswich, MA). Following the phosphorylation, the primers were mixed and boiled in a water bath for 2 min. 4M sodium chloride was added immediately to a final concentration of 10 mM, and the mixture was allowed to slowly cool at room temperature. Under these conditions, the two primers hybridize while forming a short double-stranded insert with ApaI–ApaI overhangs.
The cDNA portion of the previously constructed pAX5− HCBS WT vector [16] encoding CBS amino acids 1–551 was cut out using ApaI and SalI restriction endonucleases. Finally, the pGEX-6P-1 vector cassette, the two hybridized primers and the fragment cut out of the pAX5− HCBS WT vector were ligated.
The pGEX-6P-1 HCBS 1–413 construct, yielding human CBS protein truncated at the C-terminus by 138 amino acid residues was prepared from the pGEX-6P-1 HCBS WT clone. The latter was digested with SphI and SalI, which released the portion coding for the C-terminal region spanning amino acids 337–551. Subsequently, we replaced the missing region with the corresponding cDNA portion cut out from a previously constructed pAX5− CBS 1–413 clone.
The construction of the pGEX-6P-1 HCBS 71–400 expression plasmid has been described previously [11].
All constructs were transformed into E. coli BL21 cells (Stratagene) and their authenticity was confirmed by DNA sequencing using a Thermo Sequenase Cy5.5 sequencing kit (Amersham Biosciences) and the Visible Genetics Long-Read Tower System-V3.1 DNA Sequencer according to the manufacturer’s instructions.
Expression and purification of wild type, 1–413 and 71–400 CBS
The purification of wild type human CBS (1–551) and two truncated forms, 1–413 CBS and 71–400 CBS, was performed as described previously [11,19] with some modifications. Protein expression was carried out at 30 °C, as the slower expression appears to have beneficial influence on the solubility and concomitantly, activity of the resulting fusion protein. Cells were lysed in a buffer containing 1× TBS, pH 8.0, supplemented with 5 mM DTT, 1% Triton X-100, 2 mg/ml lysozyme, 100 μM PLP, and Sigma protease inhibitor cocktail (final concentration of 0.9 mM 4-(2- aminoethyl) benzenesulfonyl fluoride, 4.3 mM EDTA, 0.085 mM bestatin, 0.125 mM Pepstatin, and 0.0105 mM E-64). The presence of 1% Triton X- 100 in the lysis buffer increased the yield of the wild type and 1–413 CBS proteins; in direct contrast, addition of this detergent led to inactivation of the 71–400 CBS mutant protein.
The soluble fraction of the cell lysate was incubated for 10 min at room temperature in the presence of 2 mM ATP and 10 mM MgSO4 to prevent non-specific interaction between the E. coli 70 kDa DNA K protein and the affinity resin.
The isolated GST–CBS fusion proteins were cleaved with PreScission Protease in 1× cleavage buffer (50 mM Tris–HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) at 5 °C for 12 h at a final concentration of 0.5 U/mg of protein and the GST tag was subsequently removed by two different methods. In the case of the wild type and 1–413 CBS proteins, the GST tag was removed on a Phenyl Sepharose column (Sigma). The column was equilibrated with 0.3M ammonium sulfate, 1 mM DTT, 50 μM PLP and 10 mM Tris–HCl, pH 7.4. Under these conditions, both wild type and 1–413 CBS proteins bind to the Phenyl Sepharose resin, but the GST does not and is eventually completely removed by washing the resin with ~20 column volumes of the equilibration buffer. CBS is then eluted with 20 mM Tris–HCl, pH 8.0. In the case of the 71–400 CBS mutant, the GST tag is removed by size exclusion chromatography as described previously [11].
Characterization of purified wild type, 1–413, and 71–400 CBS
Heme content
The extent of saturation of the enzyme preparations with the heme cofactor was determined by a previously described pyridine-hemochromogen method [23] using hemin hydrochloride as a standard.
The influence of the heme oxidation status on CBS activity
CBS, as purified in our laboratory, has a ferric heme status, and can be reduced using sodium dithionite. In our experiment, 400 μl of the enzyme solution (0.1 mg/ml) in 100 mM potassium phosphate buffer, pH 7.4, was placed in a quartz cuvette sealed with a rubber septum. Following deoxygenation [24], the sample was reduced by the addition of 5 μl of a deoxygenated saturated solution of sodium dithionite using a Hamilton gastight syringe. The success of heme iron reduction was monitored spectroscopically [25]. After the reduction was completed, the CBS activity was determined under anaerobic conditions at physiological pH 7.4.
Spectroscopic characterization
UV–visible spectra were measured on a Hewlett-Packard diode array model 8453 UV–visible spectrophotometer in 0.1M sodium phosphate buffer, pH 7.4, at 25 °C.
CBS activity assays
The CBS activity in the classical reaction was determined by a previously described radioisotope assay using [14C] L-serine as the labeled substrate [26]. The reaction was started by the addition of 2.5 μg of the wild type CBS, 5 μg of the 1–413 CBS or 25 μg of the 71–400 CBS, respectively, diluted in enzyme dilution buffer (0.5 mg/ml BSA, 50 μM PLP, 1 mM DTT in 1× PBS, pH 7.4).
For the determination of O-acetylserine sulfhydrase activity of CBS, we employed a colorimetric reaction specific for cysteine [27]. The 200 μl reaction mixture consisted of 25 mM O-acetylserine, 25 mM Na2S, 0.5 mg/ml BSA, 0.5 mM PLP, 10 mM DTT and 200 mM Bis–Tris propane buffer, pH 8.0. The reaction was started by the addition of 2.5 μg of the wild type CBS enzyme, 5 μg of the 1–413 CBS or 50 μg of the 71–400 CBS, respectively, diluted in enzyme dilution buffer (0.5 mg/ml BSA, 50 μM PLP, 1 mM DTT in 1× PBS, pH 7.4).
One unit of activity is defined as the amount of CBS that catalyzes the formation of 1 μmol of product in 1 h at 37 °C under standard assay conditions.
pH optimum determination
The pH profiles of the wild type and the two deletion mutants were determined using a two-buffer system. The pH of assays was varied from pH 6.5 to 11.25 in 0.25 pH increments (adjusted with 200 mM Bis–Tris propane buffer for pH 6.5–9.75, or 200 mM CAPS; for pH values 9.5–11.25).
Results and discussion
Expression and purification of wild type CBS, CBS 1–413 and CBS 71–400
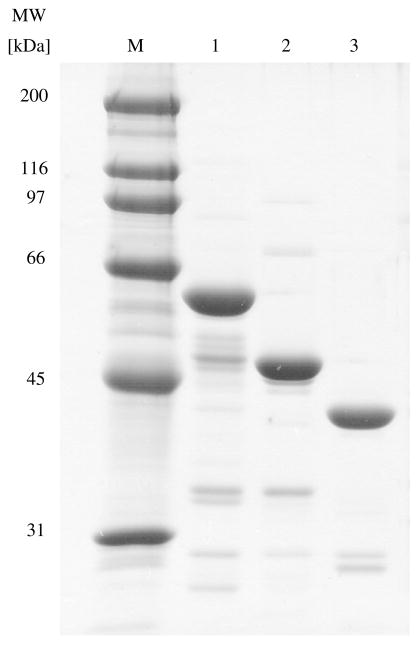
Wild type, 1–413 and 71–400 CBS were expressed as GST fusion proteins which were subsequently cleaved enabling us to rapidly purify large amounts of all three CBS species in an easy, two-step procedure. From a total of 6 l of culture, we were typically able to obtain between 20 and 40 mg of purified wild type CBS protein and ~60 mg of 1–413 CBS or 71–400 CBS at ~95% purity as judged by SDS–PAGE (Fig. 1). Compared to other methods for CBS purification, this system gave the added benefits of being able to obtain the full length enzyme with only an extra glycine residue at the N-terminus, having a long hinge region leaving enough space to allow for correct folding of both CBS and GST and rapid purification using a two step procedure which results in a very pure enzyme (Table 1).
Fig. 1.
SDS–PAGE analysis of purified recombinant wild type, 1–413 and 71–400 human CBS proteins. Twenty micrograms of each protein was run on 9% SDS-containing polyacrylamide gel and stained with Coomassie brilliant blue. Lanes: M, protein marker (precision plus molecular weight standard, Bio-Rad); 1, wild type CBS (5 μg); 2, 1–413 CBS (5 μg); 3, 71– 551 CBS (5 μg), respectively.
Table 1.
Purification of human wild type, 1–413 and 71–400 CBS expressed in E. coli
| Step | Total activity (U)
|
Specific activity (U/mg protein)
|
Recovery (%)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | 1–413 | 71–400 | WT | 1–413 | 71–400 | WT | 1–413 | 71–400 | |
| 48,000g supernatanta | 6345 | 137288 | 11227 | 1.5 | 25.2 | 3.7 | 100 | 100 | 100 |
| GST affinity column | 3721 | 29989 | 8381 | 31.9 | 256.9 | 20.3 | 58.7 | 21.8 | 74.6 |
| Precision protease cleaved fusion protein | 6121 | 23472 | 15098 | 36.2 | 192 | 25.9 | 96.5 | 55 | 134.5 |
| Phenyl sepharose column | 3209 | 27229 | N/A | 138 | 849.9 | N/A | 50.6 | 39.7 | N/A |
| Sephadex G-100 size exclusion column | N/A | N/A | 1400 | N/A | N/A | 27 | N/A | N/A | 25 |
E. coli lacks CBS activity.
pH optimum of the wild type, 1–413 and 71–400 CBS in the classical reaction
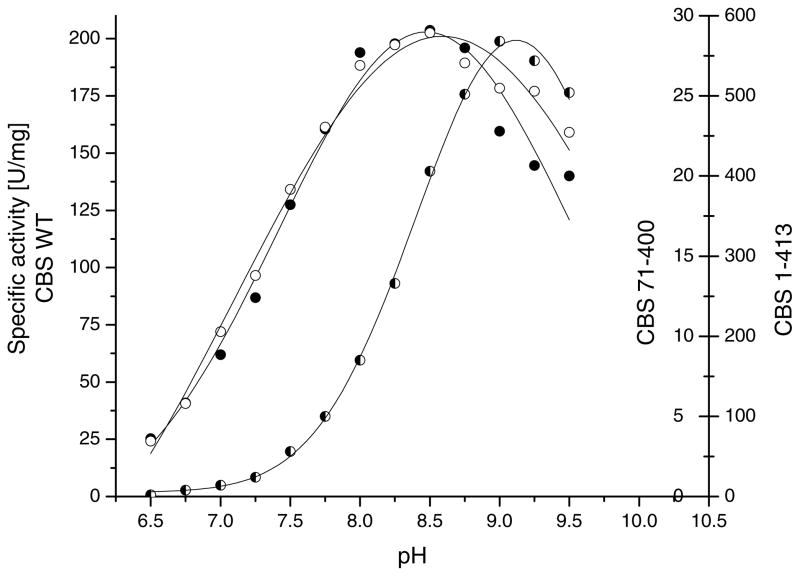
We have examined the effect of the CBS C- and N-terminal deletions on the pH optimum in the classical reaction. Our results show that the curves depicting the enzymatic activity of the recombinant wild type and 1–413 CBS as a function of pH were practically identical, showing the highest values at pH 8.5 (Fig. 2). The effect of pH on activity of the truncated human CBS has been studied previously. A broad optimum from pH 8.4 to 9.0 was observed for the proteolytically resistant active core, spanning residues 40- 413, purified from aged human liver lysate [28].
Fig. 2.
Effect of pH on activity of the wild type, 1–413 and 71–400 CBS in the serine and homocysteine condensation reaction. The pH was varied over a range of 6.5 to 9.5 in 0.25 pH increments using the 200 mM Bis–Tris propane buffer. The pH profiles shown are calculated curves from fitting the data.
The removal of both the C-terminal regulatory region and the N-terminal heme-binding region causes the pH curve to shift toward higher pH values, with a maximum at pH 9.0. It should be noted that the pH profile of the 71–400 CBS mutant is much sharper than those of the wild type and 1–413 mutant. The change in pH profile is probably due to a change in exposure of a catalytic base to the solvent.
Spectroscopic characterization of the wild type, 1–413 and 71–400 CBS
Comparison of spectroscopic properties of the purified wild type and the 1–413 deletion mutant confirmed the previous observation [21] that removal of the C-terminal regulatory region, spanning amino acids 414–551, does not impair heme binding (results not shown). However, the 71–400 deletion mutant, which is missing both the 70 amino acid N-terminal heme-binding domain and the entire C-terminal regulatory region cannot bind heme, which is illustrated by the lack of the Soret peak at 428 nm in its absorption profile [11].
Since there is no comprehensive information about the values of absorption coefficients for wild type and 71–400 CBS proteins and the value for the 71–400 mutant has not yet been published, we determined these constants for all three protein variants (Table 2). In order to compensate for variability in purity of the enzymes and their heme and PLP saturations, we performed multiple measurements of three or more independent protein preparations.
Table 2.
Extinction coefficients of wild type, 1–413 and 71–400 CBS
| CBS variant | ε280 (mM−1 cm−1 Theoretical value | ε280 (mM−1 cm−1 | ε280 (l g−1 cm−1 | ε412 (mM−1 cm−1 | ε412 (l g−1 cm−1 | ε428 (mM−1 cm−1 | ε428 (l g−1 cm−1 |
|---|---|---|---|---|---|---|---|
| WT | 57.08 | 83.75 ± 3.66 | 1.38 ± 0.06 | — | — | 89.93 ± 8.73 | 1.49 ± 0.14 |
| 1–413 | 54.40 | 82.59 ± 6.71 | 1.82 ± 0.15 | — | — | 94.13 ± 12.04 | 2.08 ± 0.27 |
| 71–400 | 25.71 | 31.96 ± 3.49 | 0.84 ± 0.05 | 7.27 ± 1.91 | 0.20 ± 0.05 | — | — |
Because heme largely masks the spectroscopic properties of the PLP coenzyme in the wild type and 1–413 proteins, only the ε280 and ε428 values were determined. Conversely, due to the lack of the heme cofactor in the 71–400 CBS mutant, only ε280 and ε412 were determined.
Full-length human CBS contains eight tryptophans, eight tyrosines and 15 phenylalanines per each subunit. Due to this chromophore content, the theoretical UV maximum of the denatured and oxidized enzyme could be calculated, using the Peptide Property Calculator software (http://www.basic.northwestern.edu/biotools/ProteinCalc. html). Theoretical and measured values of extinction coefficients for the wild type and both truncation CBS mutants are listed in Table 2. The measured UV maximum at 280 nm for the wild type enzyme was 83.75 mM−1 cm−1, which is much higher than the calculated theoretical value of 57.08 mM−1 cm−1. This phenomenon has been observed previously [21] and attributed to the significant contribution of the heme and PLP cofactors to the UV absorbance [21]. Truncation of the wild type CBS by 138 C-terminal amino acids (CBS 1–413) removes five phenylalanine residues and one tyrosine, but has no effect on the heme and PLP content when compared to its wild type counterpart. The decrease in the chromophore content should cause the extinction coefficient at 280 nm to drop by 2.68 mM−1 cm−1. We observed a decrease of the ε280 by 1.16 mM−1 cm−1 accompanied by a slight increase in the absorption maximum at 428 nm, both of which suggest that the 1–413 CBS binds the heme cofactor slightly better than the wild type enzyme. The shortest deletion mutant (CBS 71–400) is missing five phenylalanine, five tryptophan and one tyrosine residue, when compared to the wild type enzyme. In addition, due to the absence of the heme-binding domain, it is lacking the heme cofactor. The PLP saturation of this enzyme is very low, usually not more than 30% [11] suggesting that one of the functions of the heme in the human enzyme may be to increase its affinity for the PLP cofactor. The decreased PLP content together with the lack of the heme cofactor leads to a very small contribution in UV absorbance and results in a relatively small difference between the theoretical (25.71 mM−1 cm−1) and the measured (31.96 mM−1 cm−1) extinction coefficients.
Heme cofactor studies
Heme content
The degree of heme saturation was determined in numerous wild type, 1–413 and 71–400 CBS preparations using the previously described pyridine hemochromogen method [23]. The protein concentration of these preparations, needed for the calculation of the heme saturation, was determined both by the Lowry method [29] and spectroscopically using the extinction coefficients ε280 corresponding to each protein variant (Table 2). We have observed that the wild type CBS in general has a slightly lower heme saturation (91.5 ± 3.9%) than the 1–413 CBS mutant (94.4 ± 8.8%). These results, based on numerous heme and protein determinations of multiple enzyme preparations confirm our earlier observations that both wild type and 1–413 CBS bind one molecule of heme per one enzyme subunit [7,22]. This finding is also supported by the determination of the iron content of CBS. Using the method of Graphite Furnace Atomic Absorption Spectrometry we found 0.75 moles of Fe per one mole of CBS subunit, a value consistent with the above determination of one heme per subunit.
This stoichiometry is different from the results of Banerjee and co-workers, who detected only two hemes per wild type tetramer, suggesting that heme binds in a pocket formed by each of the two neighboring subunits [12,13]. The crystal structure of the truncated CBS (1–413) has been since solved [30,31] and equimolar binding of heme to CBS subunits confirmed.
The influence of the heme oxidation status on the wild type CBS activity
Although it has been known for over 10 years that CBS is a heme protein [7], the function of the heme group remains unknown. Catalytic, regulatory and structural roles have been considered for the heme in CBS. A previous study [18] brought up the possibility of heme functioning as a sensor responding to changes in the cellular redox environment. This group examined the effect of heme oxidation state on CBS activity and observed that under reducing conditions, the enzyme exhibits a 1.7-fold lower activity than under oxidizing conditions, and that this inhibition was reversible upon re-oxidation. They concluded that heme in CBS is redox-active and reversibly regulates the activity of the human enzyme [12,13]. However attractive this theory seems to be one has to consider the validity of these results in the in vivo scenario. In vitro, the reducing conditions are generated by 5 mM titanium citrate and the reoxidation of the ferrous to ferric heme is achieved by addition of 5 mM potassium ferricyanide. Since it is unlikely that such a strong redox potential would normally occur in the cell, further experiments need to be performed to confirm the plausibility of this theory in the in vivo environment.
We too have examined the effect of heme oxidative status on the activity of wild type CBS. The activity of the enzyme with the heme iron in both the ferric and ferrous state was measured at the physiological pH 7.4. In contrast to the results of Banerjee and colleagues, we have observed only modest changes in enzymatic activity with the changes in the redox status of the heme iron. Under oxidizing conditions, the activity of the wild type CBS was ~1.1 ± 0.05- fold higher than under reducing conditions (results not shown). Further experiments by our collaborators [32] have demonstrated that FeII CBS initially has the same enzyme activity as FeIII CBS, but FeII CBS loses activity due to a heme ligand exchange.
Activation of wild type CBS by AdoMet and AdoMet analogs
AdoMet activates the mammalian CBS enzyme by as much as 5-fold with an apparent Kact value of 15–18 μM [20,33]. The level of activation is comparable to that induced by partial thermal denaturation [20] or limited proteolysis [21].
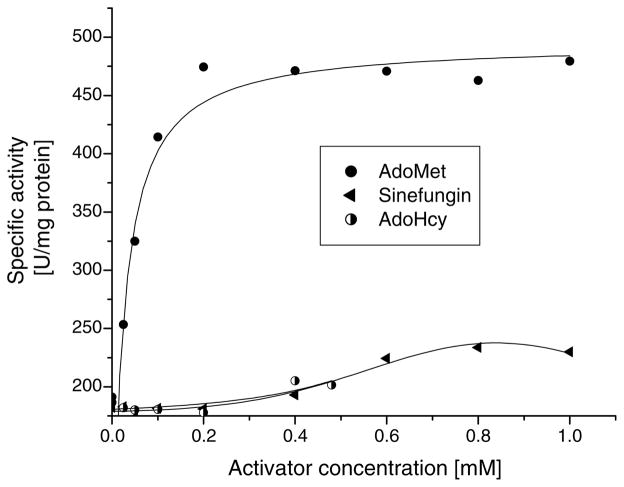
Another related PLP enzyme, threonine synthase (TS) from Arabidopsis thaliana is also strongly stimulated by AdoMet [34]; however, the mode of activation seems to be different from that of CBS. In TS, binding of the effector results in an 8-fold increase of the catalytic rate and a 25- fold decrease in the Km for the substrate O-phosphohomoserine [34]. In contrast, CBS exhibits no significant change in the Km for either substrate in the presence of AdoMet (Table 3); the activation is achieved via 3- to 5-fold increase of the catalytic rate. To date, the location of the AdoMet binding site, the identification of the key element(s) for molecular recognition and the mechanism of activation of both TS and CBS enzymes remain unknown. It has been reported over 30 years ago that the activity of CBS in crude extracts was activated not only by AdoMet but also by S-adenosylhomocysteine (AdoHcy) and adenosylornithine (Sinefungin) [35,36]. We have retested these compounds using a homogeneous human wild type CBS in an attempt to determine whether it is the amino acid moiety or the adenosine group that enters the effector-binding site. Our results show that the strongest activator is AdoMet (Kact 23.1 μM), achieving full activation at a concentration of ~0.2 mM (Fig. 3). This result is very similar to the ones we previously published [20,33]. At the same concentration, neither Sinefungin, nor AdoHcy had any effect on CBS activity (Fig. 3). Sinefungin and AdoHcy stimulated CBS activity only 1.28-fold and 1.1-fold at 1 mM and 0.48 mM concentrations, respectively.
Table 3.
Steady state kinetics of wild type and 1–413 CBS in the classical reaction
| CBS WT tetramer
|
CBS 1–413 dimer | ||||
|---|---|---|---|---|---|
| L-Homocysteine – AdoMet | L-Homocysteine + AdoMet | L-Homocysteine − AdoMet | |||
| Vmax (μmol mg−1 h−1) | 277 ± 40 | Vmax (μmol mg−1 h−1) | 756 ± 75 | Vmax (μmol mg−1 h−1) | 864 ± 57 |
| Km (mM) | 1.04 ± 0.24 | Km (mM) | 1.00 ± 0.06 | Km (mM) | 0.33 ± 0.07 |
| kcat (s−1 | 4.66 ± 0.67 | kcat (s −1) | 12.70 ± 1.25 | kcat (s −1) | 10.88 ± 0.72 |
| kcat/Km (mM−1 s−1) | 4.61 ± 0.94 | kcat/Km (mM −1 s−1) | 12.74 ± 1.22 | kcat/Km (mM −1 s−1) | 26.97 ± 5.87 |
| L-Serine – AdoMet | |||||
| Vmax (μmol mg−1 h−1) | 218 ± 40 | Vmax (μmol mg−1 h−1) | 823 ± 58 | Vmax (μmol mg−1 h−1) | 1096 ± 70 |
| Km (mM) | 1.41 ± 0.35 | Km (mM) | 2.13 ± 0.98 | Km (mM) | 2.2 ± 0.46 |
| kcat (s−1) | 3.67 ± 0.67 | kcat (s−1) | 14.01 ± 0.80 | kcat (s−1) | 13.81 ± 0.88 |
Fig. 3.
Effect of AdoMet and its structural analogues, Sinefungin and AdoHcy, on the activity of wild type CBS in the classical reaction.
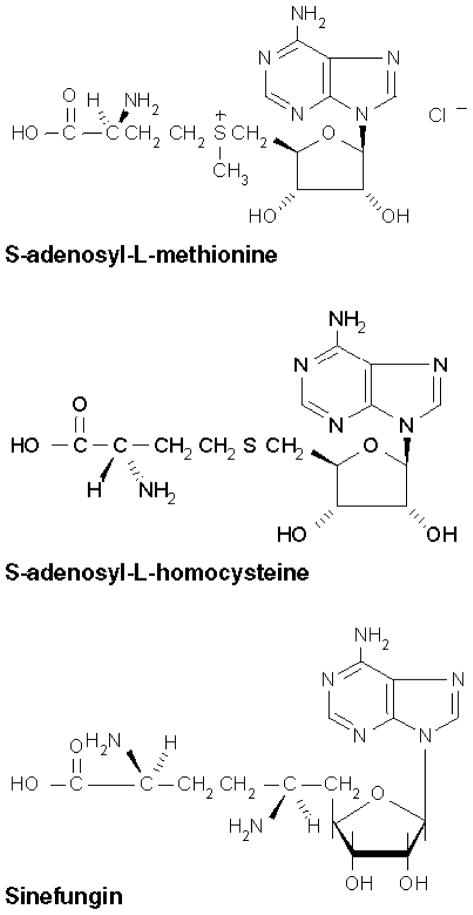
AdoMet, AdoHcy and Sinefungin all consist of adenosine linked at the 5′ position to the side chain of an α-amino acid (Fig. 4) (Sδ of methionine and homocysteine, and Cδ of ornithine, respectively). Sinefungin is isoelectronic with AdoMet. In AdoMet, the positive charge is located at the Sδ atom, in Sinefungin at the Ne atom. In the corresponding position, AdoHcy is neither branched, nor charged. These charge properties are reflected in the activating properties of these three compounds. AdoMet is the strongest activator; the relatively similar compound Sinefungin is weaker and AdoHcy is the weakest effector.
Fig. 4.
Chemical structures of AdoMet and its two analogues.
Since these three compounds differ only in their amino acid moieties, it is probable that the key elements for molecular recognition of AdoMet and its analogs are the amino acid moieties rather than the adenosine groups. Interestingly, ATP itself showed no CBS activation (result not shown). We conclude that the two AdoMet analogs do not fit sterically into the binding site as well as AdoMet does.
Kinetic characterization of the wild type and 1–413 CBS proteins in the classical reaction
Kinetic properties of both the full-length and 1–413 CBS forms were investigated with respect to both serine and homocysteine substrates. Table 3 summarizes the steady state kinetic data.
Purified recombinant wild type CBS exhibits nearly identical Km for serine and homocysteine (1.41 mM and 1.04 mM, respectively). AdoMet, being the V-type allosteric effector, has practically no effect on Km for either substrate. In contrast, AdoMet binding to the wild type CBS results in ~3- to 4-fold increase of the Vmax (Table 3). The removal of the C-terminal regulatory domain 1–413 causes a decrease of Km for homocysteine of ~3-fold without having the same effect on the mutant’s affinity for the second substrate, serine. The kcat values, representing the turnover number per catalytic site, are comparable for both the AdoMet-induced wild type and the 1–413 CBS. This observation suggests that with respect to the catalytic efficiency (kcat/Km), the absence of the C-terminal regulatory domain has the same effect as the AdoMet binding, although the activation mechanisms might be slightly different. The kinetic parameters for the 71–400 CBS yielded inconsistent measurements and are not shown.
The Km values observed for wild type, AdoMet-stimulated wild type and the 1–413 deletion mutant are very similar to the values published by our group in 1998 [21]. However, the kinetic values published later for wild type CBS [20] are very different from both our previous [21] and the current study. We attribute the differences in kinetic values to the variations in the N-terminal regions of these recombinant proteins. Both our previous [21] and current studies used proteins that were almost identical to the wild type (in each case, there was only one amino acid substitution when compared to the native sequence; either in the first or second position from the N-terminus). On the contrary, the recombinant protein used in the study published by our group in 2001 [20] contained an extra 23- amino acid “spacer” that was introduced to the N-terminus of the CBS protein to ensure that both the GST tag and the CBS protein have sufficient space for independent folding. The introduction of this spacer resulted in a dramatic (~10-fold) decrease of Km for the L-Hcy substrate and an unusually high rate of AdoMet activation (~5.5- fold). We have also demonstrated previously [11] that the 71–400 CBS mutant yielded an active enzyme only when it was folded in the presence of the N-terminal GST tag. Thus, results obtained with fusion proteins or with N-terminal extensions may affect both the kinetic parameters as well as folding of the enzymes.
O-Acetyl-L-serine (OAS) is a poor substrate for both the wild type and the 71–400 CBS
Since the acetyl group significantly increases the size of the substrate, we expected that the wild type protein would not be able to accommodate OAS in the serine-binding site. Interestingly, the wild type enzyme was capable of OAS sulfhydration, although the catalytic efficiency was very low (kcat/Km = 0.135 s−1 mM−1). We predicted the 71– 400 CBS truncation mutant would be able to interact with OAS more efficiently in the OAS sulfhydrase reaction than its wild type counterpart. This mutant, with both the N-terminal heme-binding and C-terminal regulatory regions truncated, represents the catalytic core of the enzyme and spans the conserved region referred to as the “CBS/CS domain”. This region is shared by all CBS and CS enzymes from various organisms [11,21]. When expressed as a fusion protein with GST, this deletion mutant possesses ~15% of the wild type activity in the classical reaction. Surprisingly, the 71–400 CBS turned out to be impaired in both the cystathionine forming reaction, and in the OAS sulfhydrase reaction, with the catalytic rate below the detection limit of the method (results not shown). This suggests that the deletion of both N- and C- terminal regions significantly impairs the folding of the catalytic core and has a deleterious effect on the catalytic potency of the enzyme in both classical and alternative reactions.
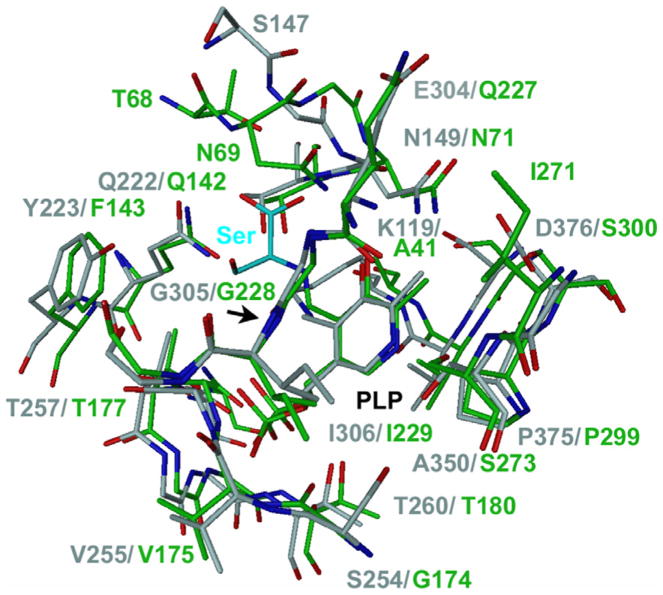
No structure of CBS with a bound ligand is available today. However, the crystal structure of CS of Salmonella typhimurium with the substrate analogue methionine covalently linked to PLP as an external aldimine [37] can be used to draw preliminary conclusions. The residues and the structure active sites of CBS and CS are very much conserved. In the immediate vicinity of the bound methionine the only different residue is F143, which is Y223 in human CBS. CS undergoes a large structural change during ligand binding and data suggest that this is also true for CBS [38]. Fig. 5 shows a superposition of the active site structures of CBS [22] and CS. For illustration, the original substrate analogue methionine has been replaced by a serine in this picture. In this crude model, the hydroxyl group of serine might possibly form a hydrogen bond to either the main chain oxygen atom of G305 or to the hydroxyl group of Y223 or both. The sulfhydryl group of cysteine cannot form hydrogen bonds. Such additional interactions of serine with the active site might explain the higher affinity of this substrate for CBS compared to cysteine.
Fig. 5.
Superposition of the active site structures of CBS and the CS. CBS is colored gray and CS green. The active site of CBS is in the open, ligand-free conformation and CS in the closed conformation with a ligand bound. The image was adapted from Fig 3a in [22]. Here, the original substrate analogue methionine in CS has been replaced by a serine in a model-building program. The figure illustrates a putative serine/cysteine-binding site for CBS. Serine and cysteine are similar in size and adopt similar conformations. The methionine in the structure of CS is covalently linked to the PLP as external aldimine. Also the substrate serine or cysteine forms an external aldimine during catalysis. It is therefore likely that they bind at a same location like methionine, considering the high conservation of the active site residues and structure between CBS and CS. For discussion of the active site please see the text.
Acknowledgments
We thank Dr. Philip E. Brandish for measuring CBS activity containing the heme moiety in a ferrous or ferric state. We also acknowledge the help of Dr. Jiri Pavel (Solvias Inc., Basel, Switzerland) with the iron determinations. This work was supported by Grants from the American Heart Association (AHA 2-5-80663) and Jerome Lejeune foundation to J.P.K.
Footnotes
Abbreviations used: CBS, Cystathionine β-synthase; GST, glutathione S-transferase; PLP, pyridoxal 5′-phosphate; AdoMet, S-adenosyl-L-methionine; CS, cysteine synthase; OASS, O-acetylserine sulfhydrase; β-gal, β-galactosidase; OAS, O-acetyl serine; AdoHcy, S-adenosylhomocysteine.
References
- 1.Mudd SH, Levy HL, Kraus JP. Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler K, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- 2.Swaroop M, Bradley K, Ohura T, Tahara T, Roper MD, Rosenberg LE, Kraus JP. J Biol Chem. 1992;267:11455–11461. [PubMed] [Google Scholar]
- 3.Mino K, Ishikawa K. J Bacteriol. 2003;185:2277–2284. doi: 10.1128/JB.185.7.2277-2284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nozaki T, Shigeta Y, Saito-Nakano Y, Imada M, Kruger WD. J Biol Chem. 2001;276:6516–6523. doi: 10.1074/jbc.M009774200. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein AE, Goryachenkova EV. Biochimie. 1976;58:5–17. doi: 10.1016/s0300-9084(76)80351-3. [DOI] [PubMed] [Google Scholar]
- 6.Miles EW, Kraus JP. J Biol Chem. 2004;279:29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- 7.Kery V, Bukovska G, Kraus JP. J Biol Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- 8.Maclean KN, Janosik M, Oliveriusova J, Kery V, Kraus JP. J Inorg Biochem. 2000;81:161–171. doi: 10.1016/s0162-0134(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 9.Jhee KH, McPhie P, Miles EW. J Biol Chem. 2000;275:11541–11544. doi: 10.1074/jbc.c000056200. [DOI] [PubMed] [Google Scholar]
- 10.Bruno S, Schiaretti F, Burkhard P, Kraus JP, Janosik M, Mozzarelli A. J Biol Chem. 2001;276:16–19. doi: 10.1074/jbc.C000588200. [DOI] [PubMed] [Google Scholar]
- 11.Oliveriusova J, Kery V, Maclean KN, Kraus JP. J Biol Chem. 2002;277:48386–48394. doi: 10.1074/jbc.M207087200. [DOI] [PubMed] [Google Scholar]
- 12.Taoka S, West M, Banerjee R. Biochemistry. 1999;38:2738–2744. doi: 10.1021/bi9826052. [DOI] [PubMed] [Google Scholar]
- 13.Taoka S, Widjaja L, Banerjee R. Biochemistry. 1999;38:13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 14.Finkelstein JD, Kyle WE, Martin JJ, Pick AM. Biochem Biophys Res Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- 15.Kraus JP, Rosenberg LE. Arch Biochem Biophys. 1983;222:44–52. doi: 10.1016/0003-9861(83)90500-3. [DOI] [PubMed] [Google Scholar]
- 16.Bukovska G, Kery V, Kraus JP. Protein Expr Purif. 1994;5:442–448. doi: 10.1006/prep.1994.1063. [DOI] [PubMed] [Google Scholar]
- 17.Shan X, Kruger WD. Nat Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 18.Taoka S, Ojha S, Shan X, Kruger WD, Banerjee R. J Biol Chem. 1998;273:25179–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 19.Janosik M, Meier M, Kery V, Oliveriusova J, Burkhard P, Kraus JP. Acta Crystallogr D Biol Crystallogr. 2001;57:289–291. doi: 10.1107/s0907444900017893. [DOI] [PubMed] [Google Scholar]
- 20.Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 21.Kery V, Poneleit L, Kraus JP. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 22.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. EMBO J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison M, Horie S. Anal Biochem. 1965;12:77–82. doi: 10.1016/0003-2697(65)90144-2. [DOI] [PubMed] [Google Scholar]
- 24.Brandish PE, Buechler W, Marletta MA. Biochemistry. 1998;37:16898–16907. doi: 10.1021/bi9814989. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa T, Sadano H, Omura T. J Biochem (Tokyo) 1984;96:265–268. doi: 10.1093/oxfordjournals.jbchem.a134822. [DOI] [PubMed] [Google Scholar]
- 26.Kraus JP. Methods Enzymol. 1987;143:388–394. doi: 10.1016/0076-6879(87)43068-1. [DOI] [PubMed] [Google Scholar]
- 27.Gaitonde MK. Biochem J. 1967;104:627–633. doi: 10.1042/bj1040627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraus JP, Packman S, Fowler B, Rosenberg LE. J Biol Chem. 1978;253:6523–6528. [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Meier M, Oliveriusova J, Kraus JP, Burkhard P. Biochim Biophys Acta. 2003;1647:206–213. doi: 10.1016/s1570-9639(03)00048-7. [DOI] [PubMed] [Google Scholar]
- 31.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 32.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 33.Kozich V, Kraus JP. Hum Mutat. 1992;1:113–123. doi: 10.1002/humu.1380010206. [DOI] [PubMed] [Google Scholar]
- 34.Curien G, Job D, Douce R, Dumas R. Biochemistry. 1998;37:13212–13221. doi: 10.1021/bi980068f. [DOI] [PubMed] [Google Scholar]
- 35.Finkelstein JD, Harris B. Arch Biochem Biophys. 1975;171:282–286. doi: 10.1016/0003-9861(75)90034-x. [DOI] [PubMed] [Google Scholar]
- 36.Koracevic D, Djordjevic V. Experientia. 1977;33:1010–1011. doi: 10.1007/BF01945937. [DOI] [PubMed] [Google Scholar]
- 37.Burkhard P, Tai CH, Ristroph CM, Cook PF, Jansonius JN. J Mol Biol. 1999;291:941–953. doi: 10.1006/jmbi.1999.3002. [DOI] [PubMed] [Google Scholar]
- 38.Dong A, Kery V, Matsuura J, Manning MC, Kraus JP, Carpenter JF. Arch Biochem Biophys. 1997;344:125–132. doi: 10.1006/abbi.1997.0202. [DOI] [PubMed] [Google Scholar]