Abstract
Transcriptional regulation is a critical mechanism in the birth, specification, and differentiation of granule neurons in the adult hippocampus. One of the first negative-acting transcriptional regulators implicated in vertebrate development is repressor element 1-silencing transcription/neuron-restrictive silencer factor (REST/NRSF)—thought to regulate hundreds of neuron-specific genes—yet its function in the adult brain remains elusive. Here we report that REST/NRSF is required to maintain the adult neural stem cell (NSC) pool and orchestrate stage-specific differentiation. REST/NRSF recruits CoREST and mSin3A corepressors to stem cell chromatin for the regulation of pro-neuronal target genes to prevent precocious neuronal differentiation in cultured adult NSCs. Moreover, mice lacking REST/NRSF specifically in NSCs display a transient increase in adult neurogenesis that leads to a loss in the neurogenic capacity of NSCs and eventually diminished granule neurons. Our work identifies REST/NRSF as a master negative regulator of adult NSC differentiation and offers a potential molecular target for neuroregenerative approaches.
Introduction
Adult neurogenesis, a multistep process supporting neuronal turnover throughout life, represents a natural paradigm for maintaining tissue homeostasis from a resident population of quiescent and transit-amplifying neural stem cells (NSCs) (Kempermann and Gage, 1999; Gage, 2000; Lledo et al., 2006). In the mammalian hippocampal subgranular zone (SGZ) and subventricular zone (SVZ) that lines the lateral ventricles, both early and late events of adult neurogenesis are governed by multiple signaling pathways, such as BMP, Wnt, and Notch (Lie et al., 2005; Ables et al., 2010; Mira et al., 2010), and neurotransmitter signaling systems, such as GABA and glutamate (Deisseroth et al., 2004; Tozuka et al., 2005; Ge et al., 2006). How these signals are integrated to control the neuronal genome is essentially unknown. From quiescent NSCs to terminally differentiated neurons, both positively and negatively acting transcriptional regulators are critical to mediate stage-specific neuronal gene expression and the sequential stages of neurogenesis. Understanding and identifying cell-intrinsic regulators that integrate the complex repertoire of extracellular signals to control differentiation of quiescent and transit-amplifying NSCs is of fundamental importance toward harnessing the full potential of NSCs for patient therapies.
The GLI–Krüppel class C2H2 zinc finger protein repressor element 1 (RE1)-silencing transcription factor (REST) [also known as neuron-restrictive silencer factor (NRSF)] binds the highly conserved 21–23 bp DNA sequence called RE1 [also called neuron-restrictive silencer element (NRSE)] found in many neuronal genes (Chong et al., 1995; Schoenherr and Anderson, 1995). REST/NRSF recruits mSin3A/B (Huang et al., 1999) and REST corepressors (CoREST) (Ballas et al., 2001) to silence gene expression. It is highly expressed in embryonic stem (ES) cells and non-neural cells and declines as ES cells convert to NSCs and cortical progenitors differentiate into mature cortical neurons (Ballas et al., 2005). In contrast, adult hippocampal granule and pyramidal neurons express high levels of REST/NRSF (Palm et al., 1998; Calderone et al., 2003; Kuwabara et al., 2004; Sun et al., 2005), and adult postmitotic neurons upregulate REST/NRSF and select target genes in response to ischemic or seizure stimuli (Palm et al., 1998; Calderone et al., 2003; Jessberger et al., 2007). Because REST/NRSF null mice are embryonic lethal starting at E9.5, with delayed development and a malformed telencephalon, and do not show widespread precocious or ectopic expression of REST/NRSF target genes (Chen et al., 1998), it has been difficult to determine whether REST/NRSF plays distinct roles during embryonic and adult stages and whether there are tissue- or context-specific silencing mechanisms.
Here we provide compelling evidence that REST/NRSF is a master transcriptional repressor of the neurogenic program during adult hippocampal neurogenesis. REST/NRSF is required for the recruitment of CoREST and mSin3A to control stage-specific neuronal gene expression such as Ascl1 and NeuroD1 and maintain NSCs in an undifferentiated state. We generated REST/NRSF conditional knock-out (cKO) mice and revealed that inducible deletion of REST/NRSF in adult NSCs in vivo triggers an increase in neurogenesis whereby NSCs exit quiescence, pass through proliferation stages, and give rise to granule neurons. Ultimately, the loss of REST/NRSF leads to a functional depletion of the adult hippocampal NSC pool and decreased granule neurons. Our results indicate a fundamental role of REST/NRSF in maintaining adult NSCs in a quiescent state by restraining the neurogenic program.
Materials and Methods
Generation of REST/NRSF cKO mice.
REST/NRSF targeting vector was constructed using the pGKNEO–F2L2DTA vector (Hoch and Soriano, 2006), which contains two loxP and FRT sites flanking a neomycin resistance gene and a diphtheria toxin gene cassette at the 3′ end. The 5′ long arm, KO arm, and 3′ short arm of the targeting construct were generated with high-fidelity PCR amplification of 129SvEv genomic DNA and correlate to a 6.2 kb fragment containing the promoter region and the first three noncoding exons, a 1.8 kb fragment harboring the first coding exon 4, and a 2.3 kb fragment in intron 4, respectively (Palm et al., 1999). The targeting vector was linearized by BcgI and electroporated into 129SvEv ES cells. Two hundred ES cell clones were screened for homologous recombination, first by PCR and then confirmed by Southern blotting. 5′ loxP incorporation was confirmed using a 5′ probe after digestion with XbaI, and 3′ loxP incorporation was confirmed with a 3′ probe after digestion with HpaI. Five clones with a properly targeted allele were injected into 3.5-d-old C57BL/6 blastocysts. Four of the clones generated high percentage chimeras and achieved germ-line transmission when crossed to C57BL/6 females. Heterozygous REST/NRSFneo-loxP/+ mice were crossed with hACTB:FLPe transgenic mice (Rodríguez et al., 2000) to remove the neomycin resistance cassette. Global deletion of the first coding exon 4 was then achieved by breeding REST/NRSF+/loxP mice to CAG–Cre transgenic mice (Sakai and Miyazaki, 1997), the offspring of which recapitulated the embryonic lethal phenotype as observed with REST/NRSF conventional knock-out mice (Chen et al., 1998). Mouse genotypes were determined by PCR using tail DNA and primers specific to the REST locus. The primers used in this study (REST/NRSF primer 1, 5′- gagccgtttcctgtgatggcattc -3′; REST/NRSF primer 2, 5′-tgcaggtcgagggacctaataact-3′; REST/NRSF primer 3, 5′-acaggatctctaggagctcagactgg-3′; and REST/NRSF primer 4, 5′-ccagggttcagttctctacacccac-3′).
Animals.
Mice were housed on a 12 h light/dark cycle in an Association for Assessment and Accreditation of Laboratory Animal Care International-approved facility at University of Texas Southwestern. The Nestin–CreERT2/Rosa26 (R26R)–yellow fluorescent protein (YFP) mice, Nestin–eGFP reporter mice, and their genotyping have been described previously (Lagace et al., 2007; Gao et al., 2009). To generate REST/NRSF cKO mice, REST/NRSFloxP mice were crossed with Nestin–CreERT2/R26R–YFP mice. Mice (male and female) at 4–6 weeks of age were administered tamoxifen (TAM) intraperitoneally at 150 mg · kg−1 · d−1 for 6 d and killed at various time points after the last injection. TAM was dissolved in 10% EtOH/90% sunflower oil. For YFP-positive (YFP+) cell counts and phenotypic analysis, mice [n = 10–15 from 6–10 litters per wild-type (WT) and REST/NRSF cKO group] were killed 10, 20, 30, 70, 120, and 200 d after TAM. Mice were given BrdU 2 h (150 mg/kg, i.p.) before being killed or once a day for 14 d (50 mg/kg, i.p.). Efficiency of REST/NRSF takeout after TAM treatment was confirmed by the following: mice of 5 weeks of age were treated with TAM, and SVZ and hippocampal neurosphere cultures were derived 20 d after TAM treatment. Both DNA and RNA samples from a cKO line with >95% YFP+ cells and control primary NSC cultures were harvested and analyzed for genomic takeout. To semiquantify the takeout efficiency, we created an amplification standard by using templates mixed with DNA from non-TAM-treated REST/NRSFloxP NSCs and DNA from WT NSCs at ratios of 9:1, 1:2, 1:3, and 1:9. The REST/NRSF conditional takeout efficiency within the YFP+ population was determined using this standard as reference, and DNA from WT NSCs was used as a negative amplification control under identical PCR conditions.
Immunofluorescence staining.
Mice were anesthetized and perfused transcardially with cold 4% paraformaldehyde (PFA) in 0.1 m PBS. Brains were removed and postfixed in 4% PFA overnight and then cryoprotected in 30% sucrose in 0.1 m PBS. Brains were bisected, and half-brains were sectioned 30 μm thick on a freezing microtome in either the coronal or sagittal planes. Immunohistochemistry (IHC) was done either with free-floating sections or on mounted tissue as described previously (Gao et al., 2009). For free-floating IHC, sections were washed twice and then blocked with 3% normal donkey serum and 0.3% Triton X-100 in PBS for 30–60 min. All mounted sections underwent antigen retrieval using 0.01 m citric acid, pH 6.0, at ∼95°C for 15 min, followed by 10 min in PBS at room temperature. For Tyramide-Plus signal amplification, endogenous peroxidase activity was blocked by incubating sections with 0.3% H2O2 for 30 min, followed by 30–60 min blocking with 3% normal donkey serum and 0.3% Triton X-100. Primary antibodies used in this study included the following: mouse anti-Ascl1 (1:750; RDI Fitzgerald), rabbit anti-AC3 (1:500; Cell Signaling Technology), mouse anti-proliferating cell nuclear antigen (PCNA) (1:5000; Millipore), mouse anti-glutamine synthase (GS) (1:500; Millipore Bioscience Research Reagents), mouse anti-GSTπ (1:1300; BD Biosciences Transduction Laboratories), rat anti-platelet-derived growth factor receptor α (PDGFRα) (1:300; BD Pharmingen), rabbit anti-TubIII (Tuj1) (1:5000; Covance), rat anti-BrdU (1:500; Accurate Chemical & Scientific), goat anti-doublecortin (DCX) (1:5000; Santa Cruz Biotechnology), mouse, rabbit, or chicken anti-GFAP (1:4000; Advanced Immunochemical), chicken or rabbit anti-GFP (used for GFP or YFP detection; rabbit at 1:500; Invitrogen; chicken at 1:8000; Aves Lab), rabbit anti-Ki67 (1:500; Neomarkers), goat anti-NeuroD1 (1:1000; Santa Cruz Biotechnology), mouse anti-NeuN (1:1000; Santa Cruz Biotechnology), rabbit anti-Prox1 (1:500; Millipore Bioscience Research Reagents), and rabbit or goat anti-Sox2 (1:500; Millipore Bioscience Research Reagents or Santa Cruz Biotechnology). For double or triple labeling, primary antibodies were simultaneously incubated (e.g., AC3/YFP, BrdU/YFP, NeuroD1/YFP, Ki67/YFP, Ascl1/NeuroD1). Whenever amplification was performed, the staining for the first antigen, such as for YFP and DCX, was done individually on the first day, followed by a second (or third) antibody the next day (e.g., Sox2/GFAP, NeuroD1/Ki67, Prox1/NeuN, NeuroD1/NeuN). For AC3, BrdU, NeuN, Ki67, NeuroD1, Sox2, GFAP, Prox1, and GFP, a fluorescent-tagged secondary Ab was used (1:250 for 2 h or 1:600 overnight; Jackson ImmunoResearch). For YFP, DCX, and Ascl1, primary antibody incubation was followed with an appropriate biotin-tagged secondary Ab (1:250; Jackson ImmunoResearch), followed by ABC (Vector Laboratories) and Tyramide-Plus signal amplification (1:50; PerkinElmer Life and Analytical Science). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1:5000; Roche). For REST/NRSF antibody tissue staining, before proceeding with standard blocking and staining procedure, free-floating sections were freeze–thawed using dry ice twice each for 15 min and fixed for 10 min in cold methanol at −20°C. REST/NRSF antibody was used at 1:100 (catalog #Ab21635; Abcam).
REST/NRSF RNA in situ hybridization.
Brains from P15 and P60 mice were perfused, harvested, and fixed in 4% DEPC–paraformaldehyde at 4°C overnight and 30% sucrose–PBS (DEPC-treated) overnight and cryosectioned at 16 μm. Both sense and antisense RNA probes (Allen Brain Atlas) (Lein et al., 2007) were generated and labeled with digoxigenin (DIG) by in vitro T7 transcription with corresponding linearized construct templates, followed by RNA probe hybridization and incubation with anti-DIG antibody and visualization with nitro blue tetrazolium and 5-bromo-4-chloro-3-indoly phosphate (NBT/BCIP) in alkaline phosphatase (AP) buffer.
Microscopic analysis and quantification.
Microscopic analysis and quantification were completed as described previously (Gao et al., 2009). Slides were coded during IHC, and the code was not broken until analysis was complete. Briefly, quantification of cell number within the hippocampus was performed using the 20× objective of a Nikon TE2000-U inverted microscope by an observer blind to experimental groups. YFP+ cell quantification and morphology phenotyping were completed in every 12th 30 μm coronal section throughout the SGZ and outer portion of the granule cell layer (GCL) of the dentate gyrus (DG) (bregma, −0.82 to −4.24 mm). Cells were morphologically defined as immature neurons if they have a round soma and at least one long process that extends through GCL and as mature neurons if they have a round soma and processes extending up into molecular layer capped by a highly arborized dendritic tree (Lagace et al., 2007; Ables et al., 2010). Phenotypic analysis and colocalization of YFP+ cells with various markers in the DG was done in every 12th or 24th coronal section throughout the SGZ using a confocal microscope (Leica confocal; emission wavelengths of 488, 543, and 633 and 40× oil objective). Leica Confocal Software was used for scanning, optical sectioning in the Z plane, and 3D rendering.
Cell culture and in vitro quantification.
The hippocampal neural progenitor (HCN) cell line from adult rat hippocampus was cultured as described previously (Gao et al., 2009). Mouse NSCs were dissociated from dissected hippocampi and lateral ventricles from 3-week-old REST/NRSFloxP/loxP or WT littermate mice according to previous methods (Gao et al., 2009). NSCs (“neurospheres”) were cultured in DMEM/F-12 supplemented with N2 and B27 (Invitrogen) in the presence of FGF2 (20 ng/ml), EGF (20 ng/ml), and heparin (5 μg/ml) (proliferation conditions) on uncoated tissue culture plates. To delete REST/NRSF, neurospheres were trypsinized and infected (in either suspension or monolayer cultures) with GFP (control) or Cre–GFP adenovirus (1:10,000 of 1 × 1010 pfu/ml stock; University of Iowa) in N2/B27 medium containing FGF2/EGF/heparin. After 72 h, media was switched to conditions as indicated. For differentiation experiments, NSCs were switched to serum-free DMEM/F-12 media supplemented with N2 and treated with retinoic acid (RA) (1 nm) and forskolin (FSK) (5 nm) for indicated time points. In some experiments, cells were treated with defined media containing leukemia inhibitory factor (LIF) and BMP4 (50 ng/ml each) to promote astrocyte differentiation or retinoic acid (1 nm) and 1% FBS to promote mixed-lineage (neuron, astrocyte, and oligodendrocyte) differentiation. To inhibit p300/CBP histone acetyltransferase activity, 1 μm curcumin (Cur) was added to NSC cultures at the time of adenovirus infection (Kim et al., 2008). In some cultures, BrdU (10 mm; Sigma) was added to label dividing cells 1 h before fixation. For immunofluorescence staining, cells were plated onto laminin-coated four-well chamber slides and, after indicated treatments, fixed with 4% paraformaldehyde, and immunofluorescence staining was performed. Labeled cells were visualized using a Nikon TE2000-U inverted microscope and a CoolSnap digital camera (Photometrics). Quantification of cell phenotype was done by sampling 5–10 random fields in each well and counting a total of 250–500 cells at 20× magnification. Throughout the study, definitive neuronal or glial cells were scored on the basis of morphological criteria (elaboration of processes), as well as immunoreactivity with various markers (e.g., Tuj1 or GFAP staining). In some cases, DAPI or GFP was used to identify individual cells as counterstaining. The following primary antibodies were used: rabbit anti-Tuj1 (1:7500; Covance) or mouse anti-Tuj1 (1:500; Sigma), mouse anti-Map2ab (1:200; Sigma), guinea pig anti-GFAP (1:2500; Advanced Immunochemical), rabbit anti-Ki67 (1:400; Neomarkers), rat anti-BrdU (1:500; Covance), and chicken anti-GFP (1:2000; Aves Labs). Secondary antibodies were all from Jackson ImmunoResearch and used at 1:250 dilution. The detection of BrdU in cultured cells required treatment in 2N HCl at 37°C for 30 min.
Colony formation assay.
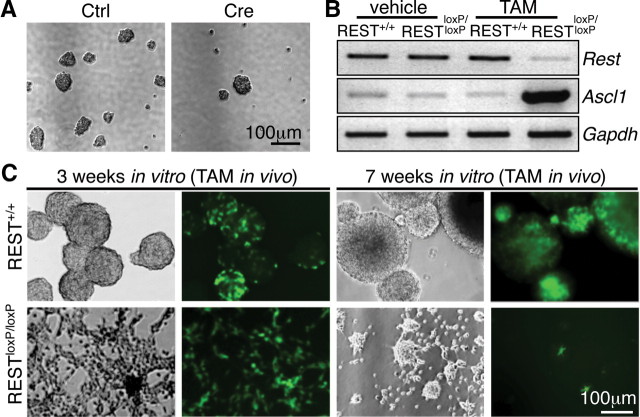
Primary WT and REST/NRSF cKO mouse neurospheres were infected with GFP control or Cre–GFP adenovirus and plated in six-well plates at single-cell low density (one cell per microliter) and cultured for 7–10 d without any movement of the plate (Coles-Takabe et al., 2008). Quantification of neurosphere colonies were done with bright-field microscopy at 10× using a Nikon TS100 inverted microscope. Afterward the sphere cultures were pooled to increase the density of the otherwise scattered spheres, and representative images of a randomly chosen field corresponding to WT or REST/NRSF cKO spheres were taken for Figure 8A. For neurosphere cultures derived from mice treated with TAM in vivo, different lines (n = 5 per WT and n = 6 per cKO), with the percentage of YFP+ cells ranging from 50% to >95%, were maintained in proliferating conditions and passaged once per week at equal density for all lines.
Figure 8.
REST is required for self-renewal of primary mouse NSCs in vitro. A, Ad–Cre-mediated REST ablation in primary mouse NSCs in vitro diminished their colony formation capacity. B, Confirmation of successful ablation of REST in mouse NSCs with TAM treatment in vivo accompanied by a dramatic upregulation of Ascl1 mRNA. C, TAM-induced REST deletion in NSCs in vivo abolished their ability to give rise to sustained neurosphere cultures in vitro. YFP+ NSCs from TAM-treated REST mice failed to form spheres and were depleted with passage over time. Shown in the figure are representative pictures from NSC cultures 3 weeks in vitro (left panels) and 7 weeks in vitro (right panels).
RT-PCR and Western blot analysis.
Total RNAs were isolated by Trizol reagent (Invitrogen), and RT-PCR was performed as described previously (Gao et al., 2009). Primer sequences are available on request. Whole protein lysates were prepared using RIPA buffer or precipitated in 10% trichloroacetic acid. After quantification, protein samples were resolved on 4–12% SDS-PAGE gels, and Western blotting was performed using standard procedures. Primary Abs (1:500) for protein blotting included the following: rabbit anti-REST/NRSF, anti-CoREST, anti-mSin3A, and anti-histone deacetylase 1 (HDAC1). Immunoreactive bands were visualized using AP-conjugated secondary antibodies, followed by BCIP/NBT detection (Kirkegaard & Perry Laboratories) or visualized by densitometry using HRP-conjugated secondary antibodies.
Chromatin immunoprecipitation and quantitative PCR analysis.
As described in Results, cells were cultured with the desired treatments and at the indicated time points were harvested and crosslinked. The chromatin immunoprecipitation (ChIP) assay was then performed following the instruction of the manufacturer (Millipore). Antibodies (10 μg) used for ChIP were as follows: REST/NRSF (Millipore), acetylated H4 (Millipore), methyl-CpG binding protein 2 (MeCP2) (Millipore), HDAC1 (Cell Signaling Technology), p300 (Santa Cruz Biotechnology), mSin3A (Santa Cruz Biotechnology), CoREST (Millipore), trimethylated-H3K27 (Cell Signaling Technology), dimethylated-H3K4 (Cell Signaling Technology), and dimethylated-H3K9 (Cell Signaling Technology). ChIP–PCR analyses were performed with independent chromatin preparations. For quantitative PCR (QPCR), all primer sets were subjected to a dissociation curve analysis and produced single peaks on a derivative plot of raw fluorescence, and analyses were done in triplicates or quadruplicates. The relative enrichment was determined by taking the absolute quantity ratios of specific IPs to normal rabbit IgG (IP/IgG) and taking that ratio and normalizing to the input groups. The ChIP primers used in this study were as follows: NeuroD1 ChIP forward, 5′- taactgattgcaccagcccttcct-3′; NeuroD1 ChIP reverse, 5′-actcggtggatggttcgtgtttga-3′; Ascl1 ChIP forward, 5′-aggattctcctgcctcagcttctt-3′; and Ascl1 ChIP reverse, 5′-aaaggtagggagacacgaacagga-3′).
Coimmunoprecipitation.
For coimmunoprecipitation (Co-IP) experiments, at the indicated time points described in Results, HCN cells were harvested in cell lysis buffer (Cell Signaling Technology: 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, and 1 μg/ml leupeptin) supplemented with protease inhibitor cocktail. Co-IP was then performed following standard protocols. Briefly, the cell lysate was rocked for 1 h at 4°C and centrifuged at 14,000 rpm for 15 min. The supernatants were transferred to new tubes and precleared with 50 μl of protein A Sepharose beads at 4°C for 30 min and then incubated with 10 μg of rabbit anti-REST/NRSF (Millipore) or IgG at 4°C overnight, followed by 1 h incubation with 50 μl beads per sample. All the incubation steps were done with gentle mixing. The protein A beads were collected with centrifugation of 1000 rpm at 4°C for 5 min and rinsed five times with 1× lysis buffer. After a 5 min boiling in 2× SDS loading buffer to elute the protein complexes, Western blot analysis was performed.
DNA cloning and virus production to evaluate REST/NRSF gain of function and loss of function.
Rat REST/NRSF short hairpin RNA (shRNA) constructs were cloned into a pLLU2G lentiviral vector following a published protocol (Dann and Garbers, 2008) and confirmed by DNA sequencing. The sequences are as follows: REST/NRSF shRNA1, 5′-tgtgtaacctgcagtaccatttcaagagaatggtactgcaggttacactttttt-3′; and REST/NRSF shRNA2, 5′-tgcggagtctgaagaacagtttcaagagaactgttcttcagactccgctttttt-3′. REST/NRSF–VP16, which consists of the one to eight zinc fingers (DNA binding domain) fused to the VP16 activation domain and lacks the N- and C-terminal repressor domains (Immaneni et al., 2000), were cloned into a pCAG–IRES–EGFP retroviral vector and confirmed by DNA sequencing. All constructs were confirmed by Western blot analyses for overexpression and/or knockdown. Lentivirus and retrovirus production were according to published protocols (Blömer et al., 1997; Zhao et al., 2006).
Luciferase reporter assay.
The wild-type NR1 and GluR2 reporters have been reported previously with RE1/NRSE mutants constructed by PCR mutagenesis (Schneider et al., 2008). All reporter gene assays were done in 96-well format, and each data point represents the average of 12 replicates. Reporter genes included NR1--luciferase and GluR2--luciferase, together with indicated constructs, and each well contained ∼25,000 cells in a volume of 100 μl. Five × 106 NSCs were transfected with 5 μg of DNA by electroporation (Amaxa) and plated in proliferation media. Twenty-four hours after transfection, cells were switched to conditions as indicated, and luciferase assays were performed 24 h later.
NeuroD1 DsiRNA knockdown experiments.
To determine whether NeuroD1 knockdown interferes with the precocious differentiation resulting from REST/NRSF ablation, a TriFECTa kit of predesigned NeuroD1 and control DsiRNAs (27-mer Dicer substrate siRNAs) was ordered from IDT. For transfection, NSCs were plated onto coated coverslips in a 24-well plate, and DsiRNAs were transfected at a final concentration of 2 nm with FuGENE HD transfection reagent (Roche) 8–12 h after adenovirus infection to ablate REST/NRSF. Three days after transfection, cells were fixed for immunofluorescence staining. NeuroD1 DsiRNA sequences are as follows: NeuroD1 DsiRNA control, 5′-rCrGrU rUrArA rUrCrG rCrGrU rArUrA rArUrA rCrGrC rGrUA T-3′ and 5′-rArUrA rCrGrC rGrUrA rUrUrA rUrArC rGrCrG rArUrU rArArC rGrArC-3; NeuroD1 DsiRNA1, 5′-rCrCrA rArArU rCrArU rArCrA rGrCrG rArGrA rGrCrG rGrGC T-3′ and 5′-rArGrC rCrCrG rCrUrC rUrCrG rCrUrG rUrArU rGrArU rUrUrG rGrUrC-3′; and NeuroD1 DsiRNA2, 5′-rGrCrA rArUrA rArUrU rArGrG rArUrC rUrArU rGrCrA rArUT T-3′ and 5′-rArArA rUrUrG rCrArU rArGrA rUrCrC rUrArA rUrUrA rUrUrG rCrUrU-3′.
Statistics.
All experiments were analyzed for statistical significance using an unpaired, two-tailed Student's t test, with all error bars expressed as ±SEM. Values of p ≤ 0.05 were considered significant.
Results
The progression from quiescent NSCs to mature granule neurons is associated with biphasic expression of REST/NRSF
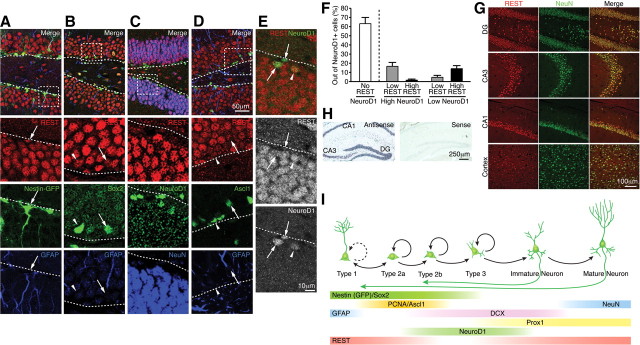
During the course of adult neurogenesis, NSCs transition through a number of stages that retain the ability to proliferate, including slowly dividing (“quiescent”) NSCs (type 1 or radial cells), transit-amplifying progenitors (type 2 or non-radial cells), and neuroblasts (type 3) (Hayes and Nowakowski, 2002; Kempermann et al., 2004; Encinas and Enikolopov, 2008). To precisely define the neural precursor population in the adult SGZ that expresses REST/NRSF, we immunostained REST/NRSF and a selection of cell-type-specific markers in WT and Nestin–GFP transgenic reporter mice (Yamaguchi et al., 2000). REST/NRSF is expressed in type 1 NSCs (Nestin–GFP+/GFAP+) and type 2a transit-amplifying progenitors (Nestin–GFP+/GFAP−/DCX−) (Fig. 1A). In support of this, REST/NRSF colocalized with Sox2+/GFAP+ type 1 NSCs and Sox2+/GFAP− type 2a transit-amplifying progenitors (Fig. 1B). As type 2a cells transition to type 2b cells (Nestin–GFP+/GFAP−/DCX+), the essential basic helix–loop–helix transcription factor NeuroD1 is upregulated (Gao et al., 2009). With the exception of a subset of type 2b and 3 cells, REST/NRSF staining was not detected in the majority of NeuroD1+ cells (Fig. 1C,E,F). To examine the overlap of REST/NRSF and NeuroD1 expression, we grouped cells into high or low NeuroD1 staining and assessed the level of REST/NRSF staining. Strikingly, there was an inverse correlation between REST/NRSF and NeuroD1 levels, consistent with the downregulation of REST/NRSF in NeuroD1+ late-stage progenitors and neuroblasts (Fig. 1C,E,F). During the transition of type 1 cells to type 2 cells, a subset of type 1 cells exit quiescence and undergo proliferation (Lugert et al., 2010) and express the earlier progenitor marker Ascl1 (Mash1) (Kim et al., 2007; Ables et al., 2010). Using Ascl1–GFP transgenic reporter mice (Kim et al., 2007; Leung et al., 2007), we observed that more than half of the Ascl1–GFP+ cells expressed REST/NRSF (Fig. 1D). Among the coexpressing cells, the level of REST/NRSF was inversely correlated with that of Ascl1–GFP+ cells, with ∼61% of them expressing low levels of REST/NRSF (Fig. 1D), consistent with downregulation of REST/NRSF during the transition of type 1 and 2 cells to granule neurons. Nevertheless, of the rare Ascl1–GFP+/GFAP+ type 1 cells, >95% were positive for REST/NRSF (Fig. 1D). Finally, as type 2b and 3 cells differentiate into immature (DCX+) and mature (NeuN+) granule neurons, REST/NRSF expression is restored (Fig. 1C,G and data not shown).
Figure 1.
REST has biphasic expression in adult hippocampal NSCs and mature granule neurons. A, B, Immunostaining of REST, Nestin–GFP, and Sox2 (markers of quiescent NSCs and transit-amplifying progenitors) and GFAP (a marker of quiescent NSCs). Insets show colocalization of REST with these markers: REST+/Nestin--GFP+/GFAP+ and REST+/Sox2+/GFAP+ type 1 NSCs (A, B, arrows); REST+/Sox2+/GFAP− type 2a transit-amplifying progenitor cell (B, arrowhead). C, Immunostaining of REST, NeuroD1, and NeuN (a marker of mature neurons). D, Immunostaining of REST, Ascl1–GFP, and GFAP. REST is expressed in a subset of Ascl1–GFP+ cells. Inset shows REST+/Ascl1–GFP+/GFAP+ cells (arrow) and REST+/Ascl1–GFP+/GFAP− cells (arrowhead). E, REST is highly expressed in mature granule neurons and some low NeuroD1+ cells (arrowhead), whereas it is only expressed in a minor subset of high NeuroD1+ cells (arrows). Dotted lines indicate border between SGZ and hilus. F, Assessment of REST and NeuroD1 colocalization demonstrated an inverse relationship between the expression of REST and NeuroD1, implying a repressive role for REST in NeuroD1 expression during adult neurogenesis. Error bars represent ± SEM. G, H, Immunostaining (G) and RNA in situ hybridization (H) of 6-week-old adult mouse brain showing that REST is broadly expressed in postmitotic neurons (NeuN+) in DG, CA3, CA1, and cortex. I, Summary of REST expression with different markers during adult hippocampal neurogenesis.
In contrast to previous reports indicating that REST/NRSF is downregulated in primary cultures of E16 cortical neurons (Ballas et al., 2005), we observed high REST/NRSF expression in both adult hippocampal and cortical neurons (Fig. 1G), which was further confirmed with RNA in situ hybridization analysis (Fig. 1H) and similar to previous work (Palm et al., 1998; Calderone et al., 2003; Kuwabara et al., 2004; Sun et al., 2005; Jessberger et al., 2007). These results suggest that REST/NRSF is expressed in distinct populations of neurons in embryonic and adult stages. Consistent with REST/NRSF being a transcriptional repressor in non-neuronal cells (Chong et al., 1995; Schoenherr and Anderson, 1995), we detected REST/NRSF expression in astrocytes (GFAP+/GS+) and oligodendrocytes (GSTπ+/PDGFRα+) within the adult hippocampus (data not shown). A summary of the biphasic REST/NRSF expression dynamics with respect to stage-specific neuronal genes such as Ascl1, NeuroD1, and other markers expressed during adult hippocampal neurogenesis is shown (Fig. 1I).
REST/NRSF and its corepressors CoREST and mSin3A repress pro-neuronal target genes in HCN cells
To begin to investigate the role of REST/NRSF in adult NSCs, we used a neural stem/progenitor cell line (HCN) derived from adult rat hippocampus that can be maintained for many passages in the presence of FGF2 and are reminiscent of transit-amplifying progenitors (Gage et al., 1995). Unlike NSCs isolated from adult mouse brain, which mainly differentiate into astrocytes with RA (Ray and Gage, 2006), HCN cells undergo robust neuronal differentiation with RA and FSK treatment (Hsieh et al., 2004). Although REST/NRSF expression persisted during the first 2 d of RA- and FSK-induced neuronal differentiation (Fig. 2A,D), the expression of RE1/NRSE site-containing neuronal target genes of REST/NRSF, including NeuroD1, NR1, Tuj1, SCG10, Syn1, and BDNF, was upregulated during neuronal induction (Fig. 2B). Conversely, the REST/NRSF target gene Ascl1 (Ballas et al., 2005) levels already declined by 12 h after neuronal induction (Fig. 2B,C). Additional examination revealed that Ascl1 was upregulated at 1 h after neuronal induction and then downregulated by 12 h compared with the expression of NeuroD1, which peaked at 3 and 24 h and then downregulated by 2 d (Fig. 2C), consistent with the stage-dependent pattern of Ascl1 and NeuroD1 during adult hippocampal neurogenesis in vivo (Gao et al., 2009) (Fig. 1H). These results suggest that REST/NRSF target genes are de-repressed in a stage-specific manner, whereas global levels of REST/NRSF remained constant, at least during the first 2 d of neuronal differentiation.
Figure 2.
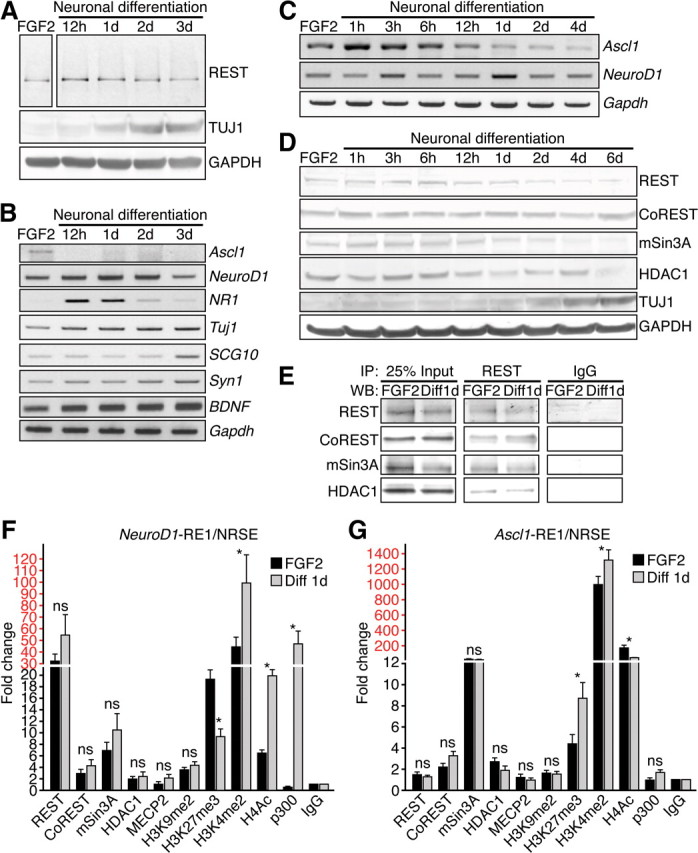
The REST complex represses neuronal genes in HCN cells. A–C, Western blot analysis (A) or RT-PCR analysis (B, C) of REST and/or its target genes after RA-induced (1 nm) and FSK-induced (5 nm) neuronal differentiation. Gapdh was used as a normalization control. D, Western blot analysis of REST and its corepressors CoREST, mSin3A, and HDAC1 during neuronal differentiation. TUJ1 was used as a positive control for neuronal differentiation, and GAPDH was used as a loading control. E, Coimmunoprecipitation experiments showing the interaction of REST with its corepressors CoREST, mSin3A, and HDAC1. WB, Western blot. F, G, The occupancy of REST, cofactors, and histone modifications at the NeuroD1– and Ascl1–RE1/NRSE sites in HCN cells under proliferation conditions (FGF2) and under differentiation conditions (RA–FSK for 24 h). Relative enrichment of factor binding was calculated as fold changes relative to IgG control (n = 3) under the same conditions after normalizing to the respective input control. Different scales are demarked by different colors (black and red). Error bars represent ± SEM. *p ≤ 0.05 by two-tailed Student's t test. NS, Not significant.
The mechanism of transcriptional repression by REST/NRSF is mediated through its association with corepressors CoREST and mSin3A/B and HDACs, such as HDAC1 (Huang et al., 1999; Ballas et al., 2001, 2005). The protein levels of CoREST and HDAC1 remained unchanged during neuronal differentiation, whereas REST/NRSF and mSin3A declined by 4 d (Fig. 2D). Coimmunoprecipitation experiments confirmed REST/NRSF interaction with CoREST, mSin3A, and HDAC1 in HCN cells under proliferating and neuronal differentiation conditions (Fig. 2E), consistent with the ability of REST/NRSF to recruit specific corepressors in other neural and non-neuronal cell types (Huang et al., 1999, Roopra et al., 2000; Ballas et al., 2001, 2005; Lunyak et al., 2002).
Next, we performed ChIP assays followed by QPCR. Under proliferation conditions, REST, CoREST, mSin3A, and HDAC1 were all bound at the RE1/NRSE site of the NeuroD1 gene (NeuroD1–RE1/NRSE) (Johnson et al., 2007) in HCN cells, and their binding did not significantly change during 24 h of neuronal induction (Fig. 2F). Consistent with upregulation of NeuroD1, histone H4 acetylation (H4Ac), and histone H3 dimethylation on lysine 4 (H3K4me2), modifications associated with gene activation, were significantly upregulated during neuronal induction (Fig. 2F). In contrast, although there was no change in histone H3 dimethylation on lysine 9 (H3K9me2), we did observe decreased histone H3 trimethylation on lysine 27 (H3K27me3), histone modifications correlated with gene repression (Fig. 2F). The binding of histone acetyltransferase p300 was also highly enriched at the NeuroD1–RE1/NRSE after neuronal induction, consistent with the important role of p300 during gene activation (Fig. 2F).
To determine whether REST/NRSF controlled stage-specific neuronal gene expression, we surveyed a second REST/NRSF target gene Ascl1 (Ascl1–RE1/NRSE) (Ballas et al., 2005). In contrast to the NeuroD1–RE1/NRSE site and consistent with the downregulation of Ascl1 at 12 h after neuronal induction, we observed an enrichment of H3K27me3, decreased enrichment of H4Ac, and lack of enrichment of p300 binding at the Ascl1–RE1/NRSE 24 h after neuronal induction (Fig. 2G). Surprisingly, H3K4me2 was also enriched at the Ascl1–RE1/NRSE (Fig. 2G), perhaps reflecting a bivalent chromatin state containing both active and silent histone modifications (Bernstein et al., 2006) or representing a transitional state before the silencing of Ascl1. Together with the biphasic expression of REST/NRSF in vivo, these results suggest that REST/NRSF and its corepressors CoREST and mSin3A regulate stage-specific target gene expression, such as Ascl1 expression before NeuroD1, by controlling the epigenetic chromatin landscape in HCN cells undergoing neuronal differentiation.
Knockdown of REST/NRSF enhances neuronal differentiation in vitro
To functionally dissect the role of REST/NRSF in HCN cells, we performed a loss-of-function analysis using two independent REST/NRSF-specific shRNAs and showed effective knockdown of endogenous REST/NRSF protein in HCN cells (Fig. 3A). REST/NRSF knockdown induced expression of pro-neuronal genes NeuroD1, Tuj1, and DCX (Fig. 3B) and enhanced the REST/NRSF target gene promoter NR1–luciferase reporter activity in both proliferation and differentiation conditions (Fig. 3D), suggesting that REST/NRSF knockdown is sufficient to de-repress target genes even in the absence of neuronal induction.
Figure 3.
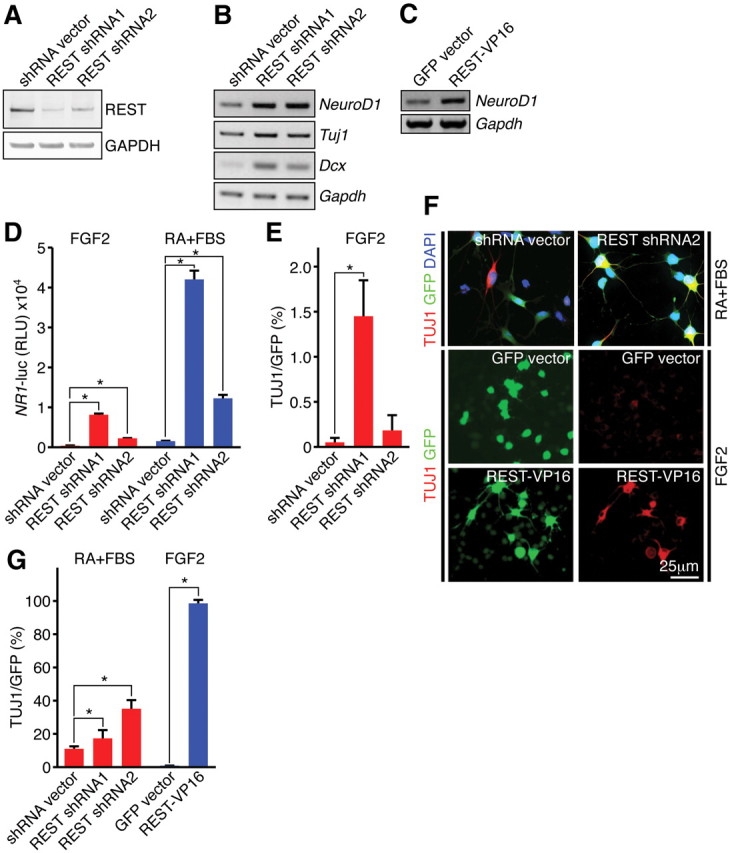
Knockdown of REST results in de-repression of pro-neuronal genes and accelerates the neuronal phenotype. A, Confirmation of REST shRNA knockdown in HCN cells. Two different REST shRNAs resulted in efficient knockdown of endogenous REST protein. GAPDH was used as a normalization control. B, De-repression of neuronal target genes during REST shRNA knockdown in differentiation conditions (RA–FSK for 24 h). C, Upregulation of NeuroD1 during expression of REST–VP16 under proliferation conditions (FGF2). D, Luciferase reporter assays showing REST shRNA-mediated de-repression of promoter reporter activity of NR1 (a REST target gene) in both proliferating (FGF2) and differentiation (RA–FBS for 24 h) conditions. E, Quantification of REST shRNA-mediated neuronal differentiation (percentage TUJ1) in proliferation conditions (FGF2). F, shRNA knockdown of REST enhanced neuronal differentiation when HCN cells were primed with RA and FBS for 24 h, and introduction of REST–VP16 resulted in precocious differentiation under proliferating conditions (FGF2). G, Quantification of REST shRNA- or REST–VP16-mediated neuronal differentiation (percentage TUJ1) in differentiation conditions (RA–FBS) and proliferation conditions (FGF2), respectively. Error bars represent ± SEM. *p ≤ 0.05 by two-tailed Student's t test.
To determine the phenotypic consequences of REST/NRSF knockdown in HCN cells, we analyzed individual cells infected by our lentiviral constructs, which were engineered to simultaneously express REST/NRSF shRNAs and GFP. Under proliferation conditions, we observed extremely modest effects on the differentiation of Tuj1+ cells, suggesting that REST/NRSF knockdown is not sufficient to induce robust neuronal differentiation (Fig. 3E). Notably, when HCN cells were primed to differentiate with RA and FBS for 24 h, we observed a marked increase in morphologic- and marker-positive Tuj1+ neurons in REST/NRSF shRNA–GFP-expressing cells compared with cells expressing shRNA–GFP vector alone (Fig. 3F,G). These findings indicate that REST/NRSF represses neuronal gene expression important for maintaining the undifferentiated state. Nevertheless, downregulation of REST/NRSF under proliferation conditions was not sufficient to promote robust neuronal differentiation, suggesting that the release of other repressors (e.g., MeCP2) (Ballas et al., 2005) or recruitment of other activators (e.g., Ascl1) is needed. In support of this, expression of a REST/NRSF mutant variant (REST/NRSF–VP16), which lacks the N- and C-terminal repressor domains and contains the strong activation domain of the viral activator VP16 (Immaneni et al., 2000), induced NeuroD1 expression and resulted in nearly 100% Tuj1+ cells even under conditions that promote proliferation (Fig. 3C,F,G).
Adult NSC-specific deletion of REST/NRSF in vivo results in premature exit of quiescence and transient acceleration of adult neurogenesis
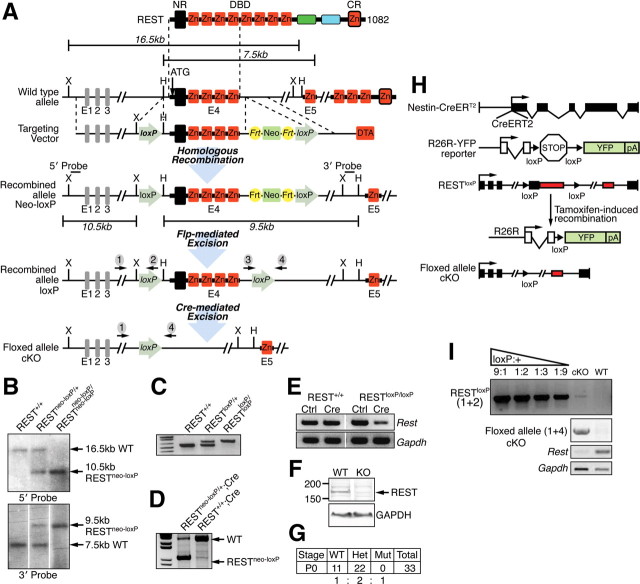
To investigate the significance of REST/NRSF during adult neurogenesis, we generated a REST/NRSF mutant allele that contained loxP sites in the introns flanking the first coding exon (REST/NRSFloxP), because mice lacking REST/NRSF die during early embryonic development starting at E9.5 (Chen et al., 1998) (Fig. 4A). The same exon was deleted in the original null allele that failed to produce REST/NRSF and related RE1/NRSE binding activity in ES cells from homozygous mutants (Chen et al., 1998). Successful gene targeting was confirmed by Southern blot, RT-PCR, and Western blot analysis (Fig. 4B–F). Furthermore, when REST/NRSFloxP mice were crossed with a germ-line Cre deleter strain (CAG–Cre), WT and heterozygous REST/NRSFloxP mice were born at predicted Mendelian ratios, but there were no homozygous REST/NRSFloxP mice born, similar to the reported REST/NRSF-null mutant (Chen et al., 1998) (Fig. 4G).
Figure 4.
Generation of REST cKO mice. A, Strategy to generate a REST cKO allele. Protein, corresponding exonic structure, targeting vector, and targeted allele are depicted. The FRT site flanked neomycin resistance cassette was removed by crossing to transgenic animals expressing hACTB:FLPe in the germ line. H, HpaI; X, XbaI; NR, N-terminal repressor domain; CR, C-terminal repressor domain; DBD, DNA binding domain. B, Southern blot analysis of WT, RESTneo-loxP/+, and RESTneo-loxP/neo-loxP. Tail DNA was digested with XbaI (5′ probe) and HpaI (3′ probe), and the corresponding WT (16.5/7.5 kb) and targeted (10.5/9.5 kb) bands are indicated for the 5′/3′ probes. C, Genotyping of RESTloxP mice by genomic PCR. The primer set (3 + 4) flanks the 3′ loxP site, resulting in one ∼550 bp product for the WT allele and one ∼650 bp band for the targeted allele. D, Successful recombination with global deletion by CAG–Cre. The primer set (1 + 4) flanks both the 5′ and 3′ loxP sites, resulting in one ∼2.8 kb product for the WT allele and ∼1.1 kb band for the targeted allele. E, F, RT-PCR and Western blot analyses to confirm successful deletion of the REST cKO allele by Ad–Cre-mediated recombination in vitro in RESTloxP/loxP mouse embryonic fibroblast cells (MEFs) and NSCs (data not shown). G, REST+/+ or REST+/loxP; CAG–Cre mice segregate according to Mendelian ratios, whereas RESTloxP/loxP; CAG–Cre mutant is embryonic lethal, recapitulating the previously reported global KO phenotype (Chen et al., 1998). H, Strategy to generate an NSC-specific deletion of REST using an inducible form of Nestin–CreERT2. I, Assessment of TAM-induced recombination frequency in vivo by establishing adult NSC cultures from TAM-treated mice. An NSC line of >95% YFP+ cells was used to assess REST recombination frequency within the YFP+ population during TAM treatment using REST genomic PCR (top 2 panels) and RT-PCR (bottom 2 panels). DNA from non-TAM-treated REST/NRSFloxP/loxP NSCs and DNA from REST/NRSF+/+ NSCs were mixed at ratios (loxP:+) of 9:1, 1:2, 1:3, and 1:9, and PCR was performed with primer set (1 + 2) to produce an amplification standard. DNA from TAM-treated REST/NRSFloxP/loxP NSCs (cKO lane) showed barely detectable amplification (>90% recombination efficiency), and DNA from REST/NRSF+/+ NSCs (WT lane) showed absence of product with primer set (1 + 2) under identical PCR conditions. We confirmed positive amplification of the floxed allele cKO with primer set (1 + 4) in DNA from TAM-treated REST/NRSFloxP/loxP NSCs but not from REST/NRSF+/+ NSCs.
To determine the role of REST/NRSF in the adult SGZ, we generated NSC-specific REST/NRSF cKO mice by crossing REST/NRSFloxP mice with a mouse line that contains a TAM-inducible Cre transgene under the control of the Nestin promoter (Nestin–CreERT2) (Lagace et al., 2007) and R26R–YFP reporter mice to track Cre-mediated recombination events (Fig. 4H). PCR analysis from genomic DNA and cDNA from >95% YFP+ NSCs isolated from TAM-treated REST/NRSF cKO mice revealed loss of REST/NRSF expression, confirming efficient Cre-mediated deletion (>90% recombination efficiency) (Fig. 4I).
Because REST/NRSF is expressed in both type 1 and type 2 cells and differentiation of adult NSCs to mature granule neurons occurs within a reported time frame of 28 d (Kempermann et al., 2004), REST/NRSF cKO mice were killed at 10, 20, and 30 d after TAM (Fig. 5A). First, no significant difference in the total number of YFP+ cells was detected between REST/NRSF cKO and WT littermates between 10 and 30 d after TAM (Fig. 5B). We next assessed markers of neuronal differentiation in REST/NRSF cKO mice because REST/NRSF deletion in vitro leads to precocious neuronal differentiation. Although we failed to observe a significant difference in NeuroD1+ cells between 10 and 30 d after TAM (data not shown), remarkably, we observed a significant but transient increase in the proportion of Ascl1+/YFP+ cells in total YFP+ cells in the REST/NRSF cKO at 20 d but not at 10 or 30 d after TAM (Fig. 5C,D).
Figure 5.
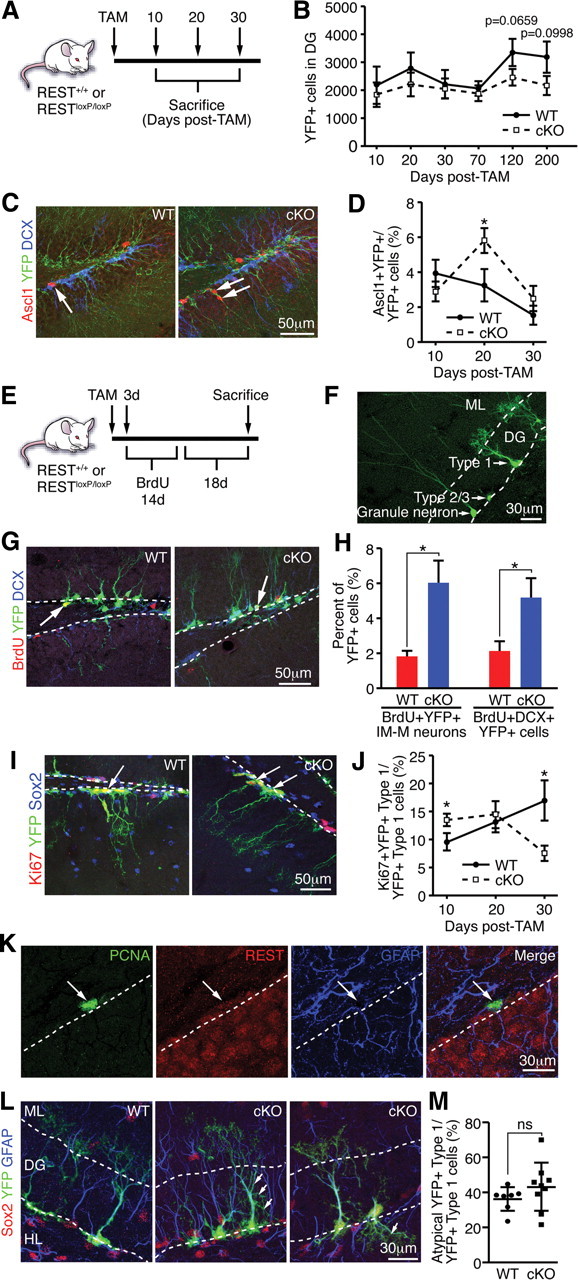
Deletion of REST in vivo leads to precocious neuronal differentiation and impaired maintenance of type 1 cells. A, Schematic of TAM treatment and harvesting of brain tissue. B, No significant change in the number of total YFP+ cells was observed at 10 d (p = 0.3618), 20 d (p = 0.4252), 30 d (p = 0.7903), and 70 d (p = 0.6281) after TAM, but there was a strong decreasing trend in REST cKO compared with WT at 120 d (p = 0.0659) and 200 d (p = 0.0998) after TAM. C, Representative images of an Ascl1+/YFP+ cell in WT (arrow, left) and Ascl1+/YFP+ cells in REST cKO (arrows, right). DCX (blue) is a marker for immature granule neurons. D, REST ablation transiently increases the percentage of Ascl1+/YFP+ cells at 20 d, but not at 10 or 30 d, after TAM. E, Schematic of TAM and BrdU treatment. F, Representative morphology of YFP+ type 1, 2/3 cells, and granule neurons in the DG. ML, Molecular layer. G, Representative images of a BrdU+/YFP+ cell in WT (arrow, left) and a BrdU+/DCX+/YFP+ cell in REST cKO (arrow, right). H, REST ablation accelerates the progression NSCs to immature and mature (IM-M) neurons as shown by morphological phenotyping of BrdU+/YFP+ cells and increased percentage of BrdU+/DCX+/YFP+ cells of the total YFP+ cells in REST cKO mice compared with WT. I, Representative images of a Ki67+/YFP+ type 1 cell in WT (arrow, left) and Ki67+/YFP+ cells in REST cKO (arrows, right). Sox2 (blue) stains type 1 and 2 cells. J, REST ablation in adult NSCs transiently increases type 1 cell proliferation at 10 d after TAM, determined by quantifying Ki67+/YFP+ type 1 cells, which are quickly depleted, ultimately resulting in a decrease in type 1 cell proliferation 30 d after TAM. K, Immunostaining of REST and cell-type-specific markers showing that REST is expressed in PCNA+/GFAP+ proliferating type 1 cells (arrow). Dotted lines indicate border of SGZ and hilus. L, REST cKO type 1 cells display atypical morphological characteristics (see Results). Representative images of YFP+ type 1 cells in WT and REST cKO mice are shown. Arrows indicate examples of atypical type 1 cell morphology in REST cKO. We confirmed that GFAP (blue) and Sox2 (red) are expressed in YFP+ type 1 cells in both WT and REST cKO. Dotted lines indicate the boundaries of the GCL. ML, Molecular layer; HL, hilus. M, Quantification of the percentage of atypical YFP+ type 1 cells in WT (n = 7) compared with REST cKO (n = 9). Error bars represent ±SEM. *p ≤ 0.05 by two-tailed Student's t test. If not indicated by * or ns, then not significant.
The increased percentage of Ascl1+/YFP+ cells could reflect the de-repression of REST/NRSF target genes, leading to increased hippocampal neurogenesis. To further examine this, TAM administration was combined with the administration of BrdU for 14 d, followed by 18 d without BrdU, which allowed us to label a discrete set of progenitors and assess the fate of proliferating YFP+ type 1 and 2 cells (Fig. 5E). We determined the presence of immature and mature neurons by the morphological assessment of YFP+ cells as described previously (Gao et al., 2009; Ables et al., 2010) (Fig. 5F). Compared with WT, the proportion of BrdU+/YFP+ immature and mature neurons was increased in REST/NRSF cKO mice (Fig. 5G,H). We also stained for the immature neuron marker DCX and confirmed that hippocampal neurogenesis is accelerated in REST/NRSF cKO mice (Fig. 5G,H). Thus, conditional deletion of REST/NRSF transiently augments hippocampal neurogenesis in vivo, consistent with its effects in vitro.
The transient increase in Ascl1+/YFP+ cells observed in the REST/NRSF cKO could also be attributed to proportionally more quiescent NSCs becoming Ascl1+ because Ascl1 expression is found in a subset of type 1 cells (Kim et al., 2007; Ables et al., 2010). However, we did not observe an increase in the proportion of Ascl1+/YFP+ of YFP+ type 1 cells at early time points after TAM (data not shown). Alternatively, type 1 cells may be exiting quiescence and transitioning to proliferating Ascl1+ type 2 cells (Kim et al., 2007; Suh et al., 2007). Surprisingly, we found the percentage of proliferating type 1 cells (Ki67+/YFP+ type 1 cells of YFP+ type 1 cells) was significantly increased in REST/NRSF cKO mice at 10 d after TAM (Fig. 5I,J), whereas the proportion of proliferating type 2 and 3 cells did not significantly change (data not shown), implicating REST/NRSF in the control of NSC quiescence. Consistent with the increase in proliferating type 1 cells, we observed proportionally more GFAP+/YFP+ type 1 cells in REST cKO (37.1 ± 2.8%) than in WT (27.0 ± 1.8%) mice at 20 d after TAM. As demonstrated by its colocalization with PCNA (a proliferation marker) in GFAP+ type 1 cells, REST/NRSF is indeed expressed in proliferating type 1 cells as well as non-proliferating type 1 cells (Fig. 5K). However, by 20 d after TAM, there was no appreciable change in the proportion of proliferating type 1 cells, and, by 30 d after TAM, the proliferation of YFP+ type 1 cells was significantly decreased in REST/NRSF cKO mice (Fig. 5J), suggesting premature exit of quiescence that leads to a loss of adult NSCs.
Common features associated with type 1 cells exiting quiescence are changes in dendritic morphology and migration in the GCL (Kempermann et al., 2004). Although ∼60% of YFP+ type 1 cells in WT mice exhibit stereotypic morphology—a single thick process (central stalk) through the DG with a tuft of dendritic branches in the molecular layer—approximately 40% display a range of atypical morphologies, such as short lateral branching or small, hair-like projections from the central stalk, loss of central stalk, loss or variable tufts, or migration away from the SGZ. Interestingly, of the remaining YFP+ type 1 cells in the REST/NRSF cKO, we observed an increased percentage of YFP+ type 1 cells with atypical morphology, although this did not reach significance (Fig. 5L,M). Together, the transiently increased proliferation and atypical morphology reminiscent of YFP+ type 1 cells in the REST/NRSF cKO strongly support the model that loss of REST/NRSF leads to failure of type 1 NSCs to maintain their integrity impacting the adult NSC pool.
REST/NRSF deletion ultimately leads to decreased production of adult-generated granule neurons
If loss of REST/NRSF in type 1 cells leads to the exit of quiescence and failure to maintain a functional adult NSC pool, the production of granule neurons may be compromised over time. In adult mouse SGZ and SVZ, the contribution of NSCs to granule neurons reaches a plateau between 2 and 6 months depending on the strain and the promoter strategies used (Lagace et al., 2007; Imayoshi et al., 2008; Ables et al., 2010). We therefore extended our phenotypic analysis to 200 d after TAM (Fig. 6A). Similar to 10–30 d after TAM, we also did not observe a significant difference in the total number of YFP+ cells between 30 and 200 d after TAM, although there was a trend toward decrease in REST/NRSF cKO compared with WT at 120 d (p = 0.0659) and 200 d (p = 0.0998) after TAM (Fig. 5B). Importantly, we observed a decrease in immature and mature granule neurons (based on morphology) at 120 and 200 d but no significant change at 10, 20, 30, and 70 d after TAM (Fig. 6B). Consistent with our morphological analysis, quantification with markers Prox1 and NeuN, together labeling mature granule neurons, confirmed that the percentage as well as the absolute number of mature granule neurons in REST/NRSF cKO mice were significantly decreased at 120 and 200 d after TAM (Fig. 6C–E). Moreover, we did not observe any change in the total number of cleaved caspase 3 (AC3+) cells at 30 or 70 d after TAM in the SGZ, suggesting that enhanced cell death was not likely the major cause of the decline in granule neuron production (Fig. 6F,G). Collectively, our results demonstrate that removal of REST/NRSF leads to the eventual loss of the neurogenic capacity of adult NSCs and decreased neurogenesis over time.
Figure 6.
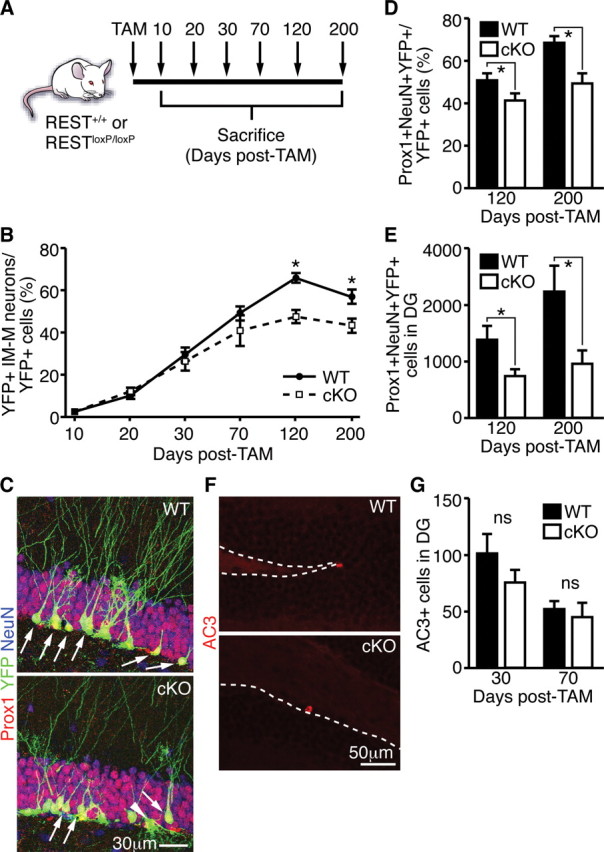
Decreased granule neuron production in REST cKO mice over time. A, Schematic of TAM treatment and harvesting of brain tissue. B, Ablation of REST in adult NSCs resulted in decreased production of immature and mature granule neurons (IM-M) at 120 and 200 d after TAM. There was no significant change between REST cKO and WT at 10, 20, 30, and 70 d after TAM. C, Representative images of Prox1+/NeuN+/YFP+ cells in the SGZ of WT (arrows, top) or REST cKO (arrows, bottom). Note an YFP+ type 1 cell with atypical morphology in REST cKO (arrowhead). D, E, In contrast to the earlier transient increase in immature and mature neurons (Fig. 5H) and consistent with a functional depletion of the stem cell pool, REST ablation in adult NSCs resulted in decreased production of Prox1+/NeuN+/YFP+ mature granule neurons at 120 and 200 d after TAM. F, Representative images of AC3+ cells in WT and REST cKO mice. Dotted line indicates the border between SGZ and hilus. G, No change in AC3+ cells was observed between WT and REST cKO mice. Error bars represent ±SEM. *p ≤ 0.05 by two-tailed Student's t test. If not indicated by * or ns, then not significant.
REST/NRSF is required for the recruitment of its corepressor complex to NSC chromatin to control neuronal target genes and maintain the undifferentiated state
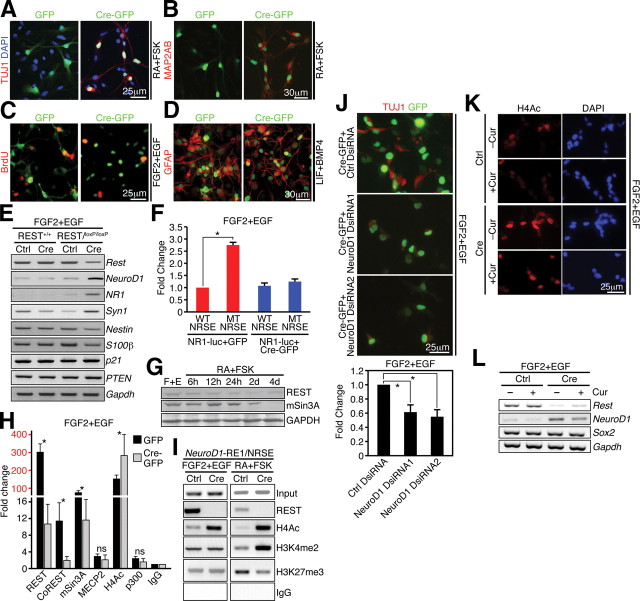
To expand on our findings in HCN cells and gain additional mechanistic insight regarding REST/NRSF, we established primary hippocampal and SVZ neurosphere cultures from 3-week-old WT and REST/NRSFloxP mice and infected cells with adenovirus expressing GFP (control) or Cre–GFP. When cells were primed to differentiate with RA and FSK for 24 h, Tuj1+ cells increased from 5 ± 1.2% in Ad–GFP-infected cKO cells to 88 ± 10% Tuj1+ cells in Ad–Cre–GFP-infected cKO NSCs (Fig. 7A). We also confirmed robust neuronal differentiation after REST/NRSF deletion with a second neuronal marker, Map2ab (Fig. 7B). Strikingly, even in proliferation conditions, the percentage of Tuj1+ cells increased from 2.7 ± 2.0 to 28.8 ± 12.2% in REST/NRSF cKO NSCs. Moreover, under conditions that promote proliferation, loss of REST/NRSF decreased NSC proliferation, from 31.3 ± 6.5% in WT to 13.8 ± 3.2% in REST/NRSF cKO, as determined by diminished BrdU incorporation, likely an indirect effect coupled to neuronal differentiation (Fig. 7C). Finally, REST/NRSF deletion altered GFAP+ astrocyte morphology, but not the number of GFAP+ cells per se, when cKO NSCs were treated with LIF and BMP4 for 4 d (Fig. 7D). Loss of REST/NRSF also induced ectopic Tuj1+ expression in many GFAP+ astrocytes (data not shown), consistent with the important role of REST/NRSF to preserve astrocyte identity (Kohyama et al., 2010).
Figure 7.
REST is required in adult NSCs to control stage-specific neuronal gene expression in primary NSCs. A, B, Ad–Cre-mediated ablation of REST in SVZ and hippocampal NSCs promotes precocious neuronal differentiation when primed with RA–FSK as indicated by morphology and neuronal marker staining (TUJ1+ and MAP2AB+). C, Neural progenitor cell proliferation was decreased in REST cKO after Ad–Cre-mediated ablation in proliferating conditions (FGF2+EGF) compared with either Ad–GFP-infected cKO cells or Ad–Cre-infected WT cells. D, Deletion of REST in astrocyte differentiation media (LIF–BMP-4 for 4 d) results in morphological changes but does not significantly compromise the number of GFAP+ astrocytes. E, During REST deletion, many of its target neuronal genes (NeuroD1, NR1, and Syn1) are upregulated, whereas some NSC (Nestin), glial (S100β), or cell cycle genes (p21 and PTEN) are downregulated or remain unchanged. Gapdh was used as an internal control. F, Luciferase reporter assays showing RE1/NRSE-dependent regulation of NR1 expression, a known REST target. G, REST and mSin3A levels remained unchanged immediately after neuronal induction with RA–FSK but declined over time when compared with cells grown in proliferation conditions (FGF2–EGF). H, ChIP–QPCR analysis showing that REST ablation leads to changes in corepressor binding at the NeuroD1–RE1/NRSE. The occupancy of respective factors was calculated as fold changes relative to IgG control (n = 3) in the same conditions (Ad–GFP or Ad–Cre–GFP infected) after normalizing to the respective input control. Different scales are demarked by different colors (black and red). I, ChIP–PCR of REST and various histone modifications in WT and cKO NSCs. J, NeuroD1 knockdown partially attenuates the precocious differentiation phenotype resulting from REST ablation. For quantification, the level of differentiation in the control (Ctrl) DsiRNA group was set as 1, and the experimental DsiRNA groups (NeuroD1 DsiRNA1 and NeuroD1 DsiRNA2) were then normalized accordingly. K, L, Treatment with curcumin (p300/CBP-specific HAT inhibitor) blocks histone acetylation as indicated by H4Ac immunostaining (K) and upregulation of NeuroD1 after REST ablation (L). Error bars represent ±SEM. *p ≤ 0.05 by two-tailed Student's t test.
We confirmed that Rest/Nrsf mRNA levels was reduced in Ad–Cre-infected cKO NSCs (Fig. 7E). Concurrently, the transcription of REST/NRSF target genes NeuroD1, NR1, and Syn1 was de-repressed, consistent with our results in HCN cells (Fig. 7E). In contrast, the NSC gene Nestin and the astrocyte gene S100β were both slightly decreased after REST/NRSF deletion, whereas the cell cycle regulatory genes p21 and phosphatase and tensin homolog (PTEN) were unaffected (Fig. 7E), supporting that REST/NRSF functions to directly control the neurogenic lineage program. Moreover, we transfected WT or REST/NRSFloxP NSCs with a WT or mutated (MT) RE1/NRSE NR1–luciferase reporter, together with either GFP− or Cre–GFP plasmids, and observed >2.5-fold de-repression of MT RE1/NRSE normalized to WT RE1/NRSE in GFP-expressing cells, which was not observed after REST/NRSF deletion (Fig. 7F). These results are consistent with REST/NRSF-dependent repression of target genes in NSCs requiring an intact RE1/NRSE site.
To examine the nature of REST-dependent changes in its corepressor complex and histone modifications, we performed ChIP assays in primary mouse NSC cultures. First, REST/NRSF and mSin3A levels remained unchanged immediately after neuronal induction with RA–FSK but declined over time when compared with cells grown in proliferation conditions (Fig. 7G). Consistent with the mechanism of REST/NRSF-mediated repression of neuronal gene expression, we confirmed that REST/NRSF is associated with the NeuroD1–RE1/NRSE in GFP-infected cKO NSCs and is significantly reduced during REST/NRSF deletion in Cre-infected cKO NSCs under proliferation conditions (Fig. 7H,I). Importantly, the binding of its corepressors CoREST and mSin3A, but not MeCP2, is dramatically reduced after REST/NRSF deletion (Fig. 7H). Concurrently, there was increased H4Ac binding, which correlated with the upregulation of NeuroD1 in Cre-infected cKO NSCs (Fig. 7E,H,I). Nevertheless, no significant change in p300 binding was observed at the NeuroD1–RE1/NRSE (Fig. 7H), possibility reflecting the transient nature of p300 binding and its acetylation activity or alternatively suggesting that REST/NRSF removal alone is not sufficient to stimulate the recruitment of p300 to neuronal gene promoters (Chen et al., 1999). When REST/NRSF cKO NSCs were primed to differentiate into neurons with RA–FSK, we confirmed increased H4Ac binding at the NeuroD1–RE1/NRSE site (Fig. 7I). We observed increased H3K4me2 and decreased H3K27me3 in REST/NRSF cKO NSCs only after neuronal induction but not in cells under proliferation conditions (Fig. 7I), consistent with the role of REST/NRSF-dependent acetylation/deacetylation in the recruitment of histone methylases/demethylases (Ooi and Wood, 2007).
To provide additional evidence that REST/NRSF is essential for the repression of neuronal target genes such as NeuroD1, we introduced DsiRNAs to knockdown NeuroD1 in Cre-expressing REST/NRSFloxP NSCs. Notably, we observed an attenuation of spontaneous neuronal differentiation with both NeuroD1 DsiRNAs in cKO NSCs (Fig. 7J), suggesting that the precocious neuronal differentiation phenotype observed in REST cKO NSCs could be rescued, at least in part, by downregulation of NeuroD1. To further examine whether histone acetylation changes are involved in REST-dependent NeuroD1 repression, we treated Ad–GFP- and Cre-infected REST cKO NSCs with the p300/CBP-specific HAT inhibitor curcumin (Balasubramanyam et al., 2004). First, we confirmed that REST cKO NSCs treated with Cur displayed decreased levels of H4Ac compared with vehicle-treated cells (Fig. 7K). Interestingly, REST/NRSF deletion effectively induced global increase in H4Ac by immunostaining as well as upregulated NeuroD1 expression in Cre-infected REST/NRSF cKO NSCs compared with Ad–GFP-infected cells (Fig. 7K,L). These changes were blocked by Cur treatment, at least in part, consistent with histone acetylation changes being necessary for expression of NeuroD1.
Together, our findings in both HCN cells and primary mouse NSCs demonstrate the following: (1) removal of REST/NRSF leads to precocious neuronal differentiation, suggesting that REST/NRSF is essential for maintaining the undifferentiated state; (2) REST/NRSF is required to recruit corepressors CoREST and mSin3A to NSC chromatin to maintain a bivalent chromatin state and regulate the activation of stage-specific target neuronal genes; and (3) REST/NRSF function requires an intact RE1/NRSE site(s), although other silencing mechanisms may exist depending on the local regulatory context.
REST/NRSF is required for self-renewal of primary mouse NSCs in vitro
To provide additional support that REST/NRSF plays a key role in maintaining the NSC pool in vivo, we next examined whether REST/NRSF plays a role in the self-renewal of neurosphere cultures. Primary WT and REST cKO NSCs were grown in the absence of tissue substrate (neurospheres) and the conventional neurosphere assay was performed, whereas the colony size from single cells indicates the behavior of the more limited neural progenitor cells (Coles-Takabe et al., 2008). After 1 week in culture, REST/NRSF cKO cells deleted in vitro using Cre-expressing adenovirus formed significantly fewer colonies (37 ± 5.6 per 2000 cells seeded) compared with cells infected with control adenovirus (132 ± 9.2 per 2000 cells seeded) (Fig. 8A). In contrast to WT cells, the majority of REST/NRSF cKO cells exhibited limited proliferation and either persisted as smaller colonies or died off. The dramatic reduction in REST/NRSF cKO cell colony formation capacity strongly indicates that the NSC population is undergoing decreased self-renewal after loss of REST/NRSF.
As a complementary approach, we deleted REST/NRSF in vivo by TAM administration and assessed whether recombined YFP+ NSCs from 2-month-old WT and REST/NRSFloxP SVZ and hippocampus could give rise to sustained NSC cultures in vitro. We confirmed deletion of the REST/NRSF allele in neurosphere cultures by genomic DNA and RT-PCR and upregulation of Ascl1 by RT-PCR (Fig. 8B and data not shown). Rather than proliferating to form neurospheres as observed with YFP+ NSCs from TAM-treated WT mice, YFP+ NSCs from TAM-treated REST/NRSF cKO mice failed to form spheres and were distributed throughout the culture dish as a monolayer of cells (Fig. 8C). With continued passage, there was no appreciable change in the percentage of YFP+ WT NSCs. In contrast, for the line of REST/NRSF cKO cells that initially contained >95% YFP+ cells, essentially no (<1%) YFP+ cells were observed at the end of 7 weeks in vitro, whereas the residual non-recombined (YFP−) cells replaced the YFP+ cells and formed neurospheres (Fig. 8C). Therefore, REST/NRSF is required for self-renewal in vitro because it is critical to preserve the NSC pool in vivo.
Discussion
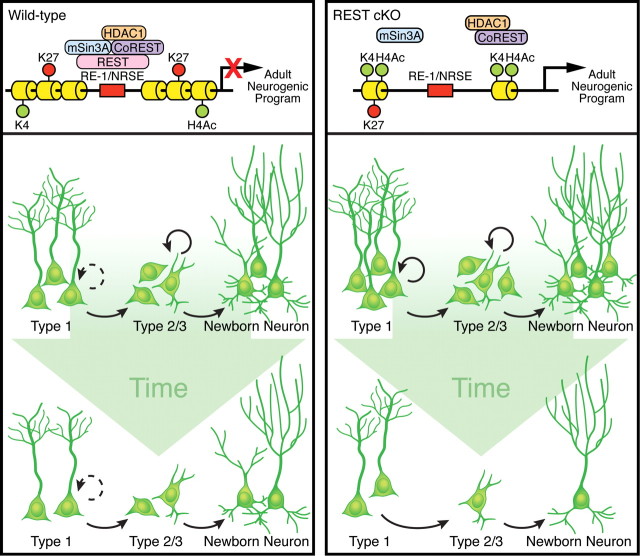
The initiation of the adult neurogenic program in resident NSCs is considered to be an active process. Equally important is the prevention of precocious differentiation in quiescent stem cells and the coordination of the neuronal gene program with the exit of quiescence and the transition to proliferative stages. Our data establish REST/NRSF as a master negative regulator of adult neurogenesis, as schematized in Figure 9.
Figure 9.
Model of REST function during adult neurogenesis. Left, In wild-type mice, REST and its corepressor complexes containing mSin3A, CoREST, and HDAC1 restrict the adult neurogenic program (e.g., Ascl1, NeuroD1, NR1, and other neuronal genes). REST functions to restrain the neurogenic program in quiescent and proliferating NSCs and regulate stage-specific neuronal gene expression, thus ensuring continued neurogenesis throughout life. Right, Loss of REST mediates changes in histone acetylation (H4Ac) and bivalent chromatin (H3K4me2 and H3K9me3) that lead to the activation/dysregulation of neuronal gene expression in NSCs. Consequently, adult NSCs prematurely exit quiescence and precociously differentiate into granule neurons. Over time, the neurogenic capacity of adult NSCs becomes exhausted, ultimately leading to decreased neurogenesis.
REST/NRSF plays an essential role to control NSC quiescence and adult neurogenesis
Loss of REST/NRSF promotes precocious neuronal differentiation and premature exit from NSC quiescence, leading to the diminished production of granule neurons over time. We propose that REST/NRSF serves as a guardian of the neuronal genome controlling the timing of neuronal gene activation downstream of diverse signaling pathways that regulates the balance of neuronal differentiation and maintenance of a functional adult NSC pool. In support of this, the biphasic expression of REST/NRSF in NSCs and mature neurons parallels the sequential stages of adult hippocampal neurogenesis. The exit from quiescence and initiation of neuronal differentiation is likely coupled to the downregulation of REST/NRSF because loss of REST/NRSF resulted in de-repression of a cohort of REST/NRSF target genes and precocious neuronal differentiation. Multiple signaling molecules, including Notch, Wnt, and Shh, have been reported to crosstalk with REST/NRSF in other systems, suggesting that REST/NRSF may integrate diverse signaling pathways to control the neuronal genome (Nishihara et al., 2003; Lie et al., 2005; Tsuda et al., 2006; Ables et al., 2010; Gates et al., 2010). Notably, BMP signaling plays a crucial role in the control of quiescence in the adult hippocampal niche (Mira et al., 2010). The BMP antagonist Noggin-treated and BMP signaling-defective mice display enhanced neurogenesis and a premature exit of quiescence, similar to our REST/NRSF cKO phenotype (Mira et al., 2010). Meanwhile, BMP signaling induces REST/NRSF expression to maintain astrocyte identity (Kohyama et al., 2010). Therefore, it is plausible that the initial downregulation of REST/NRSF may be controlled by BMP signaling that modulates NSC fate decisions.
The observation that REST/NRSF expression is restored in immature and mature neurons suggests that REST/NRSF may come back to repress target genes such as Ascl1 and NeuroD1 in a stage-specific manner, thus ensuring the proper progression of adult neurogenesis and maturation of granule neurons. In mature neurons, REST/NRSF may regulate activity-dependent neuronal gene expression, as reported for other transcriptional regulators (Mao et al., 1999; Chahrour et al., 2008). One provocative hypothesis is that the restoration of REST/NRSF expression is attributable to the same signaling pathways that drive the initial downregulation. In fact, Wnt and BMP signaling pathways manifest biphasic activity during adult hippocampal neurogenesis (Doe, 2008; Mira et al., 2010). Alternatively, REST/NRSF may play a direct role in the survival of granule neurons. Although we did not observe any change in the total number of AC3+ cells at 30 or 70 d after TAM in the SGZ, which are time points preceding the decrease in granule neurons, we cannot exclude that cell death is not involved in the reduction of YFP+ neurons at 120 and 200 d because of the transient nature of cell death and the low number of AC3+ cells in our study. Given that maturation of granule neurons takes place between 2 and 6 weeks (Tashiro et al., 2006; Zhao et al., 2006), if REST/NRSF has a major role in neuronal survival during neurogenesis, a persistent change in the net production of granule neurons would have been observed throughout our time course of 30–200 d after TAM. However, a decrease in the production of adult-generated neurons was not observed until 3–4 months after TAM, arguing against a major effect on cell survival. Importantly, there was a transient increase in neuronal differentiation at early time points preceding the ultimate reduction in granule neuron production, again arguing against a major effect on cell survival but in agreement with a NSC depletion phenotype.
Interestingly, type 1 cells persisted in the REST/NRSF cKO at later time points. During aging, the production of transit-amplifying cells from quiescent NSCs declines, leading to decreased rates of neurogenesis (Hattiangady and Shetty, 2008). A non-exclusive model is that quiescent NSCs undergo terminal differentiation after a finite number of cell divisions restricting their proliferative and neurogenic capacity (Pixley and de Vellis, 1984). Thus, the remaining YFP+ type 1 cells in REST/NRSF mutants at later time points may reflect functionally inert type 1 cells in terms of their neurogenic capacity, reminiscent of NSCs in the aged brain. Indeed, these functionally inert type 1 cells in the aged brain could be reactivated after stimulation, such as seizure activity (Lugert et al., 2010). It would be interesting to determine whether REST/NRSF deletion can block the activity-mediated reactivation of these cells.
REST/NRSF recruits CoREST and mSin3A to adult NSC chromatin to control stage-specific neuronal gene expression
REST/NRSF is required for the recruitment of CoREST and mSin3A to repress target genes in adult NSCs. Under proliferation conditions, the binding of REST/NRSF and its corepressors at neuronal target genes is associated with bivalent histone modifications characteristic of a poised chromatin state (Bernstein et al., 2006). During neuronal induction, there is a dramatic increase in p300 and H4Ac binding at NeuroD1 but a decrease of H4Ac binding at Ascl1, consistent with stage-specific neuronal gene regulation. Concurrently, bivalent histone modifications undergo dynamic but distinct changes at individual REST/NRSF target genes exemplified by Ascl1 and NeuroD1. Importantly, the loss of REST/NRSF resulted in precocious neuronal differentiation and failure to maintain the undifferentiated state, thus underscoring its essential role in maintaining this chromatin landscape and controlling stage-specific gene expression. Moreover, loss of REST/NRSF increased the histone acetylation at NeuroD1 RE1/NRSE sites even under proliferation conditions. When induced to differentiate, the acetylation state was further enhanced, accompanied by a change in the bivalent state at RE1/NRSE sites (i.e., increase in H3K4me2 and decrease in H3K27me3).
Intriguingly, REST/NRSF complexes were not displaced from target genes during neuronal differentiation, unlike a previous study in progenitors and neurons derived from embryonic cortex (Ballas et al., 2005). Several explanations exist, such as the dynamic and transient nature of these associated histone modifications, differences in the experimental systems, or the lack of quiescence in the cultured adult NSC system. Alternatively, there could be a context-dependent role of REST/NRSF based on the binding of partners with diverse functions such as chromatin- and/or histone-modifying enzymes (e.g., G9a, LSD1, and BRAF–iBRAF–MLL) (Roopra et al., 2004; Lee et al., 2005) or even non-coding RNAs (e.g., small modulatory RNA) that convert REST/NRSF from a repressor to an activator (Kuwabara et al., 2004). The observation that loss of REST/NRSF results in upregulation, not downregulation, of target genes is consistent with REST/NRSF acting as a repressor in quiescent and proliferating NSCs to restrain neuronal gene expression. Nevertheless, we cannot exclude that differential regulation could occur at select REST/NRSF target genes. One favorable hypothesis is that REST/NRSF does not act exclusively as a repressor or activator but as a permissive factor controlling access through stage-specific neuronal gene expression. Genome-wide binding studies have revealed thousands of REST/NRSF target genes, but their physiological relevance remains to be determined (Bruce et al., 2004; Johnson et al., 2007; Abrajano et al., 2009). Thus, REST/NRSF ChIP-sequencing studies combined with transcriptome analysis between WT and REST/NRSF-deficient NSCs may provide a more comprehensive view of the REST/NRSF-dependent regulatory mechanism underlying adult neurogenesis.
Dysregulation of REST/NRSF has broad disease implications
REST/NRSF shows hallmarks of dysregulation in a host of pathological conditions, such as Huntington's disease (Zuccato et al., 2003) and cancer (Lawinger et al., 2000), and after traumatic brain insults, such as ischemia (Calderone et al., 2003) and seizure activity (Jessberger et al., 2007). Surprisingly, very little is known about whether dysregulated REST/NRSF has a causative or correlative role in any of these pathological states. Our studies of REST/NRSF during neurogenesis may provide useful clues regarding its roles in brain tumors. We hypothesize that dysregulation of REST/NRSF and select target genes that control normal neuronal growth and differentiation from NSCs mediates the aberrant changes associated with tumor progression, in particular cancers of neural origin such as medulloblastomas, gliomas, and neuroblastomas that could arise from cancer stem cells. Thus, elucidating the molecular events underlying REST/NRSF function in normal NSCs has the potential to shed light on some of these brain tumors in which cancer stem cells are suspected to lie at the root.
On the reprogramming front, direct fibroblast-to-neuron conversion was achieved with just three neural cell-fate-determining transcription factors (e.g., Ascl1, Brn2, and Myt1l) (Vierbuchen et al., 2010). Along these lines, REST/NRSF knockdown could promote mesenchymal to neuronal lineage conversion (Yang et al., 2008), and REST/NRSF–VP16 has been shown to mediate direct myoblast-to-neuron conversion (Gopalakrishnan et al., 2010). Thus, by restraining the neurogenic program, REST/NRSF may underlie the stochastic and inefficient nature of somatic cell reprogramming, representing a barrier to direct reprogramming to neurons. In summary, our work highlights REST/NRSF as a potential therapeutic target in pathological states related to dysregulated neuronal gene activation and premature exit from quiescence, leading to depletion of the NSC pool, such as after brain injury or during aging. Moreover, our work identifies REST/NRSF as an attractive molecular target for applications of reprogramming technology in research and therapy.
Footnotes
This work was supported by a Epilepsy Foundation Predoctoral Fellowship (K.U.), Welch Foundation Grant I-1660 (J.H.), Cancer Prevention and Research Institute of Texas Grant RP100674 (J.H.), and National Institutes of Health Grant AG032383 (J.H.). We thank Ling Zhang, Yan Jiang, Yindi Jiang, and Ridhima Chadha for technical assistance and Jose Cabrera for graphic support. We also thank Amelia Eisch for the Nestin–CreERT2 and Nestin–GFP mice, Jane Johnson for the Ascl1–GFP mice, Chun-li Zhang for assistance with in situ hybridization, and members of the Eisch, Johnson, and Zhang laboratories for helpful discussions on this manuscript. We are grateful to David Anderson for the 12C11 mouse monoclonal REST antibody and Sadhan Majumder for the REST–VP16 construct. We thank Fred Gage for exchanging information before publication and Eric Olson, Tomoko Kuwabara, and Ondine Cleaver for critical comments on this manuscript.
References
- Ables JL, Decarolis NA, Johnson MA, Rivera PD, Gao Z, Cooper DC, Radtke F, Hsieh J, Eisch AJ. Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J Neurosci. 2010;30:10484–10492. doi: 10.1523/JNEUROSCI.4721-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009;4:e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blömer U, Naldini L, Kafri T, Trono D, Verma IM, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Coles-Takabe BL, Brain I, Purpura KA, Karpowicz P, Zandstra PW, Morshead CM, van der Kooy D. Don't look: growing clonal versus nonclonal neural stem cell colonies. Stem Cells. 2008;26:2938–2944. doi: 10.1634/stemcells.2008-0558. [DOI] [PubMed] [Google Scholar]
- Dann CT, Garbers DL. Production of knockdown rats by lentiviral transduction of embryos with short hairpin RNA transgenes. Methods Mol Biol. 2008;450:193–209. doi: 10.1007/978-1-60327-214-8_14. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Singla S, Toda H, Monje M, Palmer TD, Malenka RC. Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron. 2004;42:535–552. doi: 10.1016/s0896-6273(04)00266-1. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Encinas JM, Enikolopov G. Identifying and quantitating neural stem and progenitor cells in the adult brain. Methods Cell Biol. 2008;85:243–272. doi: 10.1016/S0091-679X(08)85011-X. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc Natl Acad Sci U S A. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates KP, Mentzer L, Karlstrom RO, Sirotkin HI. The transcriptional repressor REST/NRSF modulates hedgehog signaling. Dev Biol. 2010;340:293–305. doi: 10.1016/j.ydbio.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Bie B, Sinnappah-Kang ND, Adams H, Fuller GN, Pan ZZ, Majumder S. Myoblast-derived neuronal cells form glutamatergic neurons in the mouse cerebellum. Stem Cells. 2010;28:1839–1847. doi: 10.1002/stem.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattiangady B, Shetty AK. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol Aging. 2008;29:129–147. doi: 10.1016/j.neurobiolaging.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes NL, Nowakowski RS. Dynamics of cell proliferation in the adult dentate gyrus of two inbred strains of mice. Brain Res Dev Brain Res. 2002;134:77–85. doi: 10.1016/s0165-3806(01)00324-8. [DOI] [PubMed] [Google Scholar]
- Hoch RV, Soriano P. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development. 2006;133:663–673. doi: 10.1242/dev.02242. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Aimone JB, Kaspar BK, Kuwabara T, Nakashima K, Gage FH. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Ohtsuka T, Takao K, Miyakawa T, Yamaguchi M, Mori K, Ikeda T, Itohara S, Kageyama R. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11:1153–1161. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- Immaneni A, Lawinger P, Zhao Z, Lu W, Rastelli L, Morris JH, Majumder S. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000;28:3403–3410. doi: 10.1093/nar/28.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S, Nakashima K, Clemenson GD, Jr, Mejia E, Mathews E, Ure K, Ogawa S, Sinton CM, Gage FH, Hsieh J. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J Neurosci. 2007;27:5967–5975. doi: 10.1523/JNEUROSCI.0110-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Gage FH. Experience-dependent regulation of adult hippocampal neurogenesis: effects of long-term stimulation and stimulus withdrawal. Hippocampus. 1999;9:321–332. doi: 10.1002/(SICI)1098-1063(1999)9:3<321::AID-HIPO11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27:447–452. doi: 10.1016/j.tins.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Leung CT, Reed RR, Johnson JE. In vivo analysis of Ascl1 defined progenitors reveals distinct developmental dynamics during adult neurogenesis and gliogenesis. J Neurosci. 2007;27:12764–12774. doi: 10.1523/JNEUROSCI.3178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Son TG, Park HR, Park M, Kim MS, Kim HS, Chung HY, Mattson MP, Lee J. Curcumin stimulates proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus. J Biol Chem. 2008;283:14497–14505. doi: 10.1074/jbc.M708373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, Nakashima K. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J Cell Biol. 2010;189:159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, DiLeone RJ, Greer CA, Mandyam CD, Eisch AJ. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J Neurosci. 2007;27:12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, Rastelli L, Marin Dias Carneiro A, Levin V, Fuller GN, Echelard Y, Majumder S. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Lugert S, Basak O, Knuckles P, Haussler U, Fabel K, Götz M, Haas CA, Kempermann G, Taylor V, Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Mira H, Andreu Z, Suh H, Lie DC, Jessberger S, Consiglio A, San Emeterio J, Hortigüela R, Marqués-Torrejón MA, Nakashima K, Colak D, Götz M, Fariñas I, Gage FH. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell. 2010;7:78–89. doi: 10.1016/j.stem.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Nishihara S, Tsuda L, Ogura T. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem Biophys Res Commun. 2003;311:55–63. doi: 10.1016/j.bbrc.2003.09.158. [DOI] [PubMed] [Google Scholar]
- Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Metsis M, Timmusk T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res Mol Brain Res. 1999;72:30–39. doi: 10.1016/s0169-328x(99)00196-5. [DOI] [PubMed] [Google Scholar]
- Pixley SK, de Vellis J. Transition between immature radial glia and mature astrocytes studied with a monoclonal antibody to vimentin. Brain Res. 1984;317:201–209. doi: 10.1016/0165-3806(84)90097-x. [DOI] [PubMed] [Google Scholar]
- Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560–573. doi: 10.1016/j.mcn.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Rodríguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, Buckley NJ. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol Cell Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- Schneider JW, Gao Z, Li S, Farooqi M, Tang TS, Bezprozvanny I, Frantz DE, Hsieh J. Small-molecule activation of neuronal cell fate. Nat Chem Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D'Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YM, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, Bee T, Wilde S, Buckley NJ. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol Biol Cell. 2005;16:5630–5638. doi: 10.1091/mbc.E05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Sandler VM, Toni N, Zhao C, Gage FH. NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature. 2006;442:929–933. doi: 10.1038/nature05028. [DOI] [PubMed] [Google Scholar]
- Tozuka Y, Fukuda S, Namba T, Seki T, Hisatsune T. GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron. 2005;47:803–815. doi: 10.1016/j.neuron.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Kaido M, Lim YM, Kato K, Aigaki T, Hayashi S. An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J. 2006;25:3191–3202. doi: 10.1038/sj.emboj.7601179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Saito H, Suzuki M, Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li Y, Lv Y, Zhang S, Chen L, Bai C, Nan X, Yue W, Pei X. NRSF silencing induces neuronal differentiation of human mesenchymal stem cells. Exp Cell Res. 2008;314:2257–2265. doi: 10.1016/j.yexcr.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Zhao C, Teng EM, Summers RG, Jr, Ming GL, Gage FH. Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J Neurosci. 2006;26:3–11. doi: 10.1523/JNEUROSCI.3648-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]