SUMMARY
Gene duplication is an important source of phenotypic change and adaptive evolution. We use a novel genomic approach to identify highly identical sequence missing from the reference genome, confirming the cortical development gene Slit-Robo Rho GTPase activating protein 2 (SRGAP2) duplicated three times in humans. We show that the promoter and first nine exons of SRGAP2 duplicated from 1q32.1 (SRGAP2A) to 1q21.1 (SRGAP2B) ~3.4 million years ago (mya). Two larger duplications later copied SRGAP2B to chromosome 1p12 (SRGAP2C) and to proximal 1q21.1 (SRGAP2D), ~2.4 and ~1 mya, respectively. Sequence and expression analysis shows SRGAP2C is the most likely duplicate to encode a functional protein and among the most fixed human-specific duplicate genes. Our data suggest a mechanism where incomplete duplication created a novel function —at birth, antagonizing parental SRGAP2 function 2–3 mya a time corresponding to the transition from Australopithecus to Homo and the beginning of neocortex expansion.
INTRODUCTION
Several genes have been implicated as being important in specifying unique aspects of evolution along the human lineage. These include genes involved with the development of language (FOXP2) (Enard et al., 2002), changes in the musculature of the jaw (MYH16) (Stedman et al., 2004), and limb and digit specializations (HACNS1) (Prabhakar et al., 2008). Despite these intriguing candidates, the bulk of the morphological and behavioral adaptations unique to the human lineage remain genetically unexplained. Not all genes, however, have been amenable to standard genetic analyses. This is particularly true for genes embedded within recently duplicated sequence (Bailey et al., 2002), which are frequently missing or misassembled from the reference genome (Eichler, 2001). Genes residing in these complex regions are important to consider for three reasons: (1) duplicated genes have been recognized as a primary source of evolutionary innovation (Lynch and Katju, 2004; Ohno, 1970); (2) the human and great-ape lineages have experienced a surge of genomic duplications over the last 10 million years (Marques-Bonet et al., 2009); and (3) human-specific duplications are significantly enriched in genes important in neurodevelopmental processes (Fortna et al., 2004; Sudmant et al., 2010).
Among these human-specific duplicated genes, Slit-Robo Rho GTPase activating protein 2 (SRGAP2) was recently shown to be important in cortical development (Guerrier et al., 2009; Guo and Bao, 2010). The gene encodes a highly conserved protein expressed early in development, where it acts as a regulator of neuronal migration and differentiation by inducing filopodia formation, branching of neurons, and neurite outgrowth. Analysis of the human reference genome revealed that SRGAP2 was misassembled and that most of its duplicate copies were not yet sequenced or characterized. We developed a novel approach using genomic material devoid of allelic variation [from a complete hydatidiform mole (Kajii and Ohama, 1977)] to completely sequence and characterize the missing loci corresponding to this human-specific gene family. These data allowed us to reconstruct the complex evolutionary history of this gene family since humans diverged from nonhuman primates [~6 million years ago, mya (Patterson et al., 2006)], understand the potential of these loci to generate functional transcripts, and assay the extent of human genetic variation. We put forward a model for gene evolution where incomplete segmental duplication creates derivative copies that antagonize the ancestral function.
RESULTS
Genome sequencing
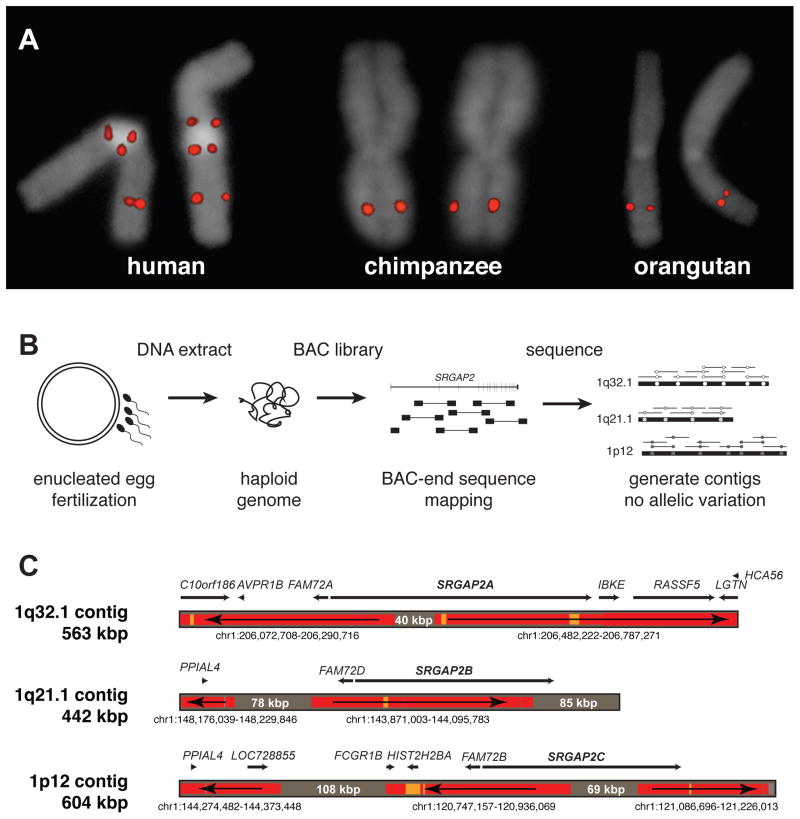
We confirmed that SRGAP2 was specifically duplicated in the human lineage by fluorescent in situ hybridization (FISH) using a probe corresponding to the human SRGAP2 (spanning exon 3, Table S1). We identified three map locations on chromosome 1 (1q32.1, 1q21.1, and 1p12) compared to a single chromosomal signal at chromosome 1q32.1 among other ape species (Figure 1A). An analysis of the segmental duplication content of 11 additional mammalian genomes (see Supplemental Experimental Procedures) showed no evidence of recent duplication in any lineage other than human and established chromosome 1q32.1 as the ancestral copy. FISH analysis of cell lines derived from humans of diverse ethnicity consistently showed a pattern of three distinct signals on each chromosome 1 corresponding to paralogs that were all incompletely sequenced in the human reference genome (GRCh37/hg19).
Figure 1. Genomic characterization and sequence resolution of SRGAP2 loci.
(A) FISH analysis shows three distinct copies of SRGAP2 on metaphase human chromosome 1, compared to a single copy in chimpanzee and orangutan (see Figure 2A for location of FISH probe; Figure S1 and Table S1 for details of additional FISH assays). (B) SRGAP2 genomic loci were sequenced and assembled using a BAC library (CHORI-17) created from human haploid genomic source material (complete hydatidiform mole). The absence of allelic variation allowed paralogous sequences to be resolved with high confidence based on near-perfect sequence identity overlap (>99.9%). (C) Regions highly identical to the reference genome (GRCh37/hg19) are colored in red (identity = 99.8–100%) and orange (99.6–99.8%), while regions completely absent from the current assembly are shaded gray (with region sizes indicated). Arrows show the orientation of the reference genome sequence with respect to the contigs (e.g., a left directional arrow indicates the reverse strand) indicating that even the ancestral (SRGAP2A) gene locus was missing sequence data, misassembled, and incorrectly orientated over 400 kbp of the current high-quality reference assembly. Genomic coordinates correspond to the representative human reference region with corresponding genes within these regions mapped along each contig.
We reasoned that the recent nature of the duplications resulted in high-identity duplications with little genetic variation; as a result, allelic and paralogous copies became difficult to disentangle during genome assembly (IHGSC, 2001). To resolve the different genomic copies, we constructed a large-insert bacterial artificial chromosome (BAC) library from DNA derived from a complete hydatidiform mole (CHORI-17). Because a complete hydatidiform mole originates from the fertilization of an enucleated human oocyte with a single spermatozoon (Fan et al., 2002; Kajii and Ohama, 1977), the corresponding DNA represents a haploid as opposed to a diploid equivalent of the human genome (Figure 1B). We leveraged the absence of allelic variation to unambiguously distinguish SRGAP2 copies despite their high sequence identity. We selected clones with homology to SRGAP2 and subjected them to high-quality capillary-based sequencing, requiring >99.9% sequence identity of the overlap between sequenced inserts for assembly into the same contig.
We generated three sequence contigs corresponding to SRGAP2 paralogs at 1q32.1 (562,704 bp; SRGAP2A), 1q21.1 (441,682 bp; SRGAP2B), and 1p12 (603,678 bp; SRGAP2C) (Figure 1C) generating over 1.6 Mbp of high-quality finished sequence. During the assembly process, we identified a single BAC clone (CH17-248H7) that harbored sequence for a SRGAP2 paralog (exons 7 through 9) but did not share >99.9% identity with any of the three contigs, suggesting a fourth SRGAP2 duplicate existed (SRGAP2D). Upon this discovery, we repeated our FISH analysis using a probe mapping across exon 1 of SRGAP2 and discovered four distinct signals on chromosome 1, with SRGAP2D mapping ~670 kbp proximally to SRGAP2B on chromosome 1q21.1 (Figure S1, Table S1). The absence of this signal from the initial FISH assay (Figure 1A) suggested that a genomic region containing exon 3 was deleted from SRGAP2D.
The new local assemblies resolved the sequence and structure of three copies adding 379,665 bp of new sequence completely absent from the human reference, including 40,233 bp within the ancestral SRGAP2A (Figure 1C). Additionally, we discovered 559,693 bp of sequence mapped incorrectly in orientation or chromosomal location within the human reference. Combined, we added or corrected over 0.4% of the human chromosome 1 euchromatic sequence (Gregory et al., 2006). All finished sequence data as well as the new human genome assemblies have been deposited into GenBank and will be integrated into subsequent human genome reference assemblies.
Comparisons between the three sequence contigs revealed large, interspersed segmental duplications of high sequence identity (99–99.5%) that were incomplete with respect to the ancestral locus (Table 1). We determined that the original duplication event (258,245 bp) encompassed the promoter, other cis regulatory elements, and the first nine exons of the 22-exon ancestral SRGAP2A (Figure 2A). Clusters of Alu repeat elements mapped precisely at the boundaries of this duplicated segment (Figure S2), confirming previous observations that Alu repeats are strongly associated with primate genomic duplications (Bailey et al., 2003; Zhou and Mishra, 2005). A larger, secondary duplication event (>515 kbp) was shared between the SRGAP2B (1q21.1) and SRGAP2C (1p12) loci and included the entirety of the original duplication, although the SRGAP2B locus was subjected to subsequent larger deletions (102.6 and 49.0 kbp) upstream of the gene (Figure S2). Using multicolor FISH assays, we determined that the ancestral SRGAP2A paralog at 1q32.1 is transcribed toward the telomere, whereas the duplicate paralogs SRGAP2B, SRGAP2C, and SRGAP2D are oriented such that gene transcription would proceed toward the centromere (Figure S1).
Table 1.
Percent sequence divergence of SRGAP2 paralogs
| SRGAP2A | SRAGP2B | SRGAP2C | SRGAP2D | |
|---|---|---|---|---|
| SRGAP2A | – | 0.015 | 0.016 | 0.069 |
| SRAGP2B | 0.525 | – | 0.014 | 0.038 |
| SRGAP2C | 0.584 | 0.451 | – | 0.065 |
| SRGAP2D | 0.452 | 0.136 | 0.400 | – |
Kimura two-parameter model of genetic distance computed as base substitutions per site (left diagonal) and standard error (right diagonal). Pairwise distances are computed across 244,200 sites representing the complete shared genomic region between SRGAP2 paralogs. Values for SRGAP2D represent pairwise distances computed across 9,541 sites. As a reference, the genetic distance between SRGAP2A and its chimpanzee ortholog locus is 0.852 +/−0.019 while that of chimpanzee to human paralogs SRGAP2B and SRGAP2C (0.901+/−0.019 and 0.960 +/− 0.020) are consistent with the accelerated mutation rate for these chromosomal regions.
Figure 2. Evolutionary characterization of SRGAP2 duplications.
(A) A depiction of the gene structure of SRGAP2 with respect to the three assembled contigs. Homologous segments are shown using Miropeats (Parsons, 1995) where green lines indicate nearly identical segments (s = 1,000) shared between SRGAP2A and the duplicate SRGAP2 paralogs; blue lines delineate the larger (>515 kbp) extent of homology between SRGAP2B and SRGAP2C. The 244.2 kbp genomic region shared among all three contigs is highlighted (red box) with clusters of Alu repeats at the breakpoints (arrows). Also see Figure S2 for detailed representation of Alu elements and segmental duplications across duplicated regions. (B) An unrooted neighbor-joining tree was constructed based on a 244.2 kbp multiple sequence alignment of the three loci. Both 1p12 and 1q21.1 branches show accelerated rates of substitution (p = 0.00001 and p = 0.0249; Tajima’s relative rate test). The actual (no parentheses) and adjusted (parentheses) number of substitutions for locus-specific acceleration is indicated above each branch along with the bootstrap support at each node. We estimate the timing assuming chimpanzee and human diverged 6 mya. Also see Table S2 for molecular evolution of the shared SRGAP2 coding regions. (C) FISH experiments on metaphase human chromosome 1, as well as the orthologous chimpanzee and orangutan chromosomes, were performed to discern the order of duplication events. Locations of probes with respect to the contigs are shown in part (A). A probe (yellow) targeting sequence adjacent to the original SRGAP2 duplicate region hybridizes to 1q21.1 in chimpanzee and orangutan, suggesting the original SRGAP2 duplicate paralog maps to the region homologous with nonhuman primate 1q21.1. A probe (green) targeting unique sequence on the p-arm of chromosome 1 proximal to SRGAP2C hybridizes to the chromosome 1p-arm in orangutan, refuting the possibility that SRGAP2C moved to the p-arm via a simple pericentromeric inversion (Szamalek et al., 2006) and distinguishing the p-arm from the genomic region at 1q21.1 where the original SRGAP2 duplicate paralog maps. A probe (blue) was used to distinguish the chromosome 1q-arm.
Evolutionary history of SRGAP2
To reconstruct the evolution of the duplication events, we generated a multiple-sequence alignment for a 244.2 kbp region shared among the three contigs using orthologous sequence from chimpanzee (build GGSC 2.1.3/panTro3) and orangutan (build WUGSC 2.0.2/ponAbe2) as outgroups (Figure 2B). Phylogenetic analysis provides strong support (>99%) for two distinct duplication events occurring at different time points during human evolution. Notably, we find that the duplicated sequences have evolved much more rapidly (Tajima’s relative rate test; p = 0.00001–0.0249) than the ancestral 1q32.1 locus (p = 0.5345). Mutation rates are known to vary significantly depending on chromosomal location and context (CSAC, 2005). Based on analysis of unique orthologous sequence adjacent to the SRGAP2C duplicate region, we determined that the distal 1p12 region shows a 20–46% higher substitution rate when compared to 1q32.1. If we adjust for this difference, calibrating to the estimated 1q32.1 substitution rate, we predict the initial duplication occurred ~3.4 mya with the secondary event occurring ~2.4 mya. We note that estimates of molecular divergence between the paralogs are robust (e.g., 0.451 +/− 0.014% substitutions per site between the SRGAP2B and SRGAP2C loci) owing to the large number of substitutions discovered in the high-quality sequence used in these comparisons (Table 1). Some uncertainty in our estimates comes from our correction factor for differing substitution rates, but most arises from ambiguity in the evolutionary timing of the divergence of chimpanzee and human (estimated at ~6 mya) (Patterson et al., 2006). If we take into account previously reported human and chimpanzee divergence times ranging from ~5–7 mya, based on fossil records (Brunet et al., 2005; Brunet et al., 2002; Vignaud et al., 2002) as well as recent genetic analyses (Patterson et al., 2006), we estimate the initial duplication occurred 2.8–3.9 mya followed by the secondary duplication at 2.0–2.8 mya. We also performed phylogenetic analysis of the 9,541 bp region shared among the SRGAP2A-C paralogs and the incompletely sequenced SRGAP2D and determined that this copy was derived from the SRGAP2B locus ~1 mya (0.4–1.3 mya assuming a 6 mya divergence time for human and chimpanzee). Using comparative FISH analysis and probes mapping outside of the original duplication (Figure 2C), we determined the likely order of events: the ancestral SRGAP2A region duplicated first to 1q21.1 (SRGAP2B), and later the 1q21.1 copy duplicated to chromosome 1p12 (SRGAP2C) and within 1q21.1 (SRGAP2D).
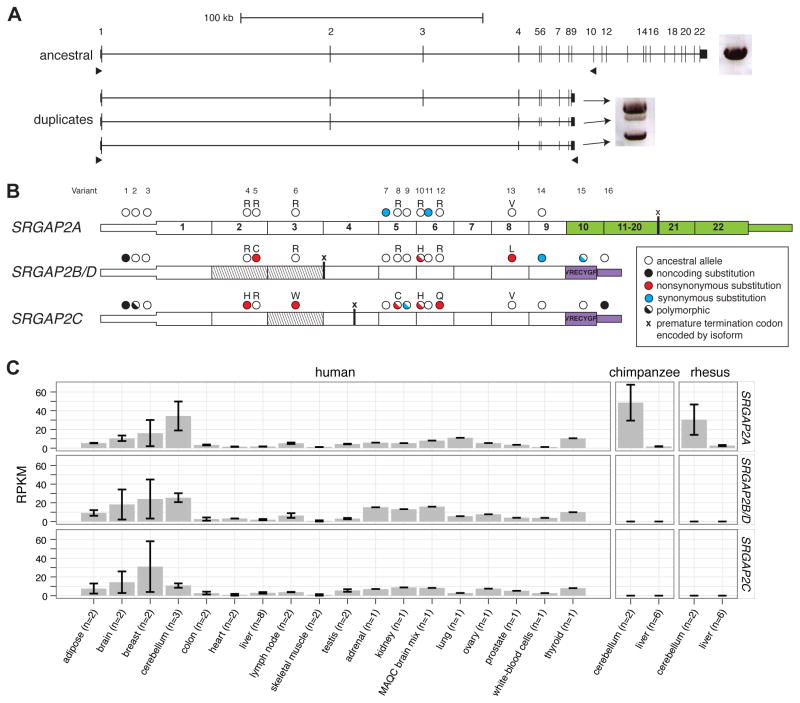
Based on the gene structure of the ancestral SRGAP2A, sequence analysis predicts that SRGAP2B and SRGAP2C would produce transcripts maintaining an open-reading frame (ORF). These two duplicate copies, however, are predicted to produce a truncated form of SRGAP2 carrying nearly the entire F-BAR domain, lacking the final 49 amino acids (Figure 2A) (Guerrier et al., 2009). The ancestral SRGAP2 protein sequence is highly constrained based on our analysis of 10 mammalian lineages. We find only a single amino-acid change between human and mouse and no changes among nonhuman primates within the first nine exons of the SRGAP2 orthologs. This is in stark contrast to the duplicate copies, which diverged from ancestral SRGAP2A less than 4 mya, but have accumulated as many as seven amino-acid replacements (five for SRGAP2C and two for SRGAP2B) compared to one synonymous change.
We used a likelihood ratio test (Yang, 2007) to evaluate differences in selective pressures acting on SRGAP2 and found that the best model of selection allows an increased nonsynonymous (dN) to synonymous substitution (dS) ratio of the SRGAP2 duplicate paralogs while maintaining purifying selection in the remaining lineages (compared with the fixed dN/dS model, p = 1.32 × 10−11, Table S2). This difference is consistent with an increased substitution rate of the 1q21.1 and 1p12 chromosomal regions and a relaxation of selective pressure on the duplicate copies. Overall, this mechanism provides a means for rapid evolutionary change of an otherwise constrained developmental gene (Lynch and Conery, 2000).
SRGAP2 mRNA expression and paralog gene structure
We assayed for expression of SRGAP2 paralogs by designing specific reverse-transcriptase PCR (RT-PCR) assays that distinguish the duplicate paralogs from the ancestral copy based on the presence of a duplicate-specific 3′ UTR present in a previously sequenced cDNA mapping to the SRGAP2C locus (GenBank accession: BC112927). A total of 96 transcripts were sequenced from RNA derived from the SH-SY5Y neuronal cell line, pooled fetal brain, a single fetal brain, and a single adult brain (Figure 3A, Table S3). Comparing genomic and cDNA sequences, we assigned the transcripts to their respective copies and identified the exon/intron structure, alternative splice forms, as well as fixed and polymorphic paralog-specific variants (PSVs) (Figure 3B). We found that all SRGAP2 paralogs are transcribed, though at different relative proportions. We identified transcripts containing exons 1 through 9 mapping specifically to SRGAP2C (N = 47) and SRGAP2B or SRGAP2D (N = 4). Using capillary sequencing of these transcripts, focusing our analysis on two fixed PSVs, we show that relative expression of the SRGAP2B/D transcript is markedly low (14–25% and 30–72% of SRGAPC transcript abundance in fetal and adult brain, respectively) (Figure S3). The most abundant duplicate transcript is expressed from SRGAP2C and predicts an ORF that would encode a truncated SRGAP2 protein (458 amino acids), including a partial F-BAR domain (Guerrier et al., 2009) and seven unique residues at the carboxyl terminus.
Figure 3. Paralog-specific SRGAP2 gene expression.
(A) Long-range RT-PCR products from pooled fetal brain RNA are shown next to the gene models. A single band was amplified from the ancestral paralog, while three bands were amplified from duplicate paralogs using primers designed to target alternative isoforms. 96 cDNA transcripts were cloned and sequenced. (B) Fixed paralog-specific variants were used to assign transcripts to respective genomic loci allowing both polymorphic and fixed putative amino acid changes to be deduced. Exonic sequence specific to the ancestral copy (SRGAP2A; green) and the duplicate loci (SRGAP2B/C/D; purple) are shown. The locations of stop codons encoded by isoforms missing exons are represented with an “x”. Exons missing from transcripts are indicated (diagonal lines) and likely correspond to the genomic deletion within SRGAP2D in the case of the exon 2–3 deleted isoform. (C) Paralog-specific expression profiling was performed using RNA-Seq data mapped to unique sequence identifiers. The specificity of next-generation sequence data and the determination of fixed single base-pair difference between the copies was necessary to tease apart the expression profiles of these virtually identical copies. Chimpanzee and macaque RNA-Seq data affirm the specificity of this assay. Also see Figure S3 and Table S3 for additional expression results.
We also observed numerous transcripts and putative splice isoforms that are unlikely to encode functional proteins. The most abundant of these map to SRGAP2B/D (N = 31) missing exons 2 and 3 and resulting in a transcript that would encode a premature truncated protein (23 amino acids). These transcripts are consistent with our genomic sequence analysis indicating that SRGAP2D has acquired a 115 kbp deletion including exons 2 and 3 (described later). Moreover, our analysis suggests that this transcript may be subjected to nonsense-mediated decay.
Using diagnostic PSVs to distinguish copies, we interrogated the expression of specific SRGAP2 paralogs in various human and nonhuman primate tissues using RT-PCR (Figure S3) and RNA-Seq data (Figure 3C). The tissue profile reveals that the paralogs show similar broad patterns of expression including expression in the developing human fetal brain concurrently with SRGAP2A. We observe higher expression in multiple regions of the human cortex and cerebellum when compared to other tissues including lung, kidney, and testis. As expected, we did not detect expression of the duplicate copies in any of the nonhuman primate-derived tissues.
SRGAP2 copy-number variation
Since SRGAP2 has been shown to play an important role in brain development, we initially focused on the ancestral SRGAP2A gene by examining a large cohort of pediatric cases with developmental delay [1,602 individuals tested using a quantitative PCR (qPCR) assay specifically targeting SRGAP2A and 15,767 individuals reported by Cooper et al. (2011)] for potential copy-number variation. We identified six large (>1 Mbp) copy-number variants (CNVs), including three deletions of the ancestral 1q32.1 region (Table 2), with no similar large CNVs observed among 10,123 controls [1,794 individuals (NIMH, N = 962; ClinSeq, N = 832 (Biesecker et al., 2009)) tested using the qPCR assay and 8,329 individuals from the Cooper et al. (2011) study]. Since the CNVs are large and encompass multiple candidate genes, this observation does not prove pathogenicity of dosage imbalance of SRGAP2A. We note, however, that in one patient the proximal breakpoint maps within the first intron of SRGAP2A potentially disrupting the gene (Figure S4, Table S4). The patient is a ten-year-old child with a history of seizures, attention deficit disorder, and learning disabilities. An MRI of this patient also indicates several brain malformations, including hypoplasia of the posterior body of the corpus callosum. Recently, a de novo balanced translocation t(1;9)(q32;q13) breaking within intron 6 of SRGAP2A was reported in a five-year-old girl diagnosed with West syndrome and exhibiting epileptic seizures, intellectual disability, cortical atrophy, and a thin corpus callosum (Saitsu et al., 2011). While much more work needs to be done, the neurological phenotypes observed in these two cases are consistent with neuronal migration deficits implicated in forms of developmental delay and epileptic encephalopathies (Saitsu et al., 2011).
Table 2.
SRGAP2A and SRGAP2C copy-number variation genotyping of cases and controls
| Genotype method | Size resolution | Cohort1 | Total genotyped | Deletions | Duplications |
|---|---|---|---|---|---|
| SRGAP2A (Cases, N=17,369; Controls, N=10,784) | |||||
| Custom array CGH platforms | >50 kbp | Intellectual disability (Signature Genomics)* (Cooper et al., 2011) | 15,767 | 3 | 3 |
| SNP arrays | >50 kbp | Controls (Cooper et al., 2011) | 8,329 | None | None |
| qPCR2 | >0.5 kbp | Intellectual disability | 1,602 | None | None |
| Controls (NIMH and ClinSeq) | 1,794 | None | None | ||
| Illumina sequencing | >1 kbp | Controls (1000 Genomes Project) | 661 | None | None |
| SRGAP2C (Cases, N=4,475; Controls, N=2,662) | |||||
| qPCR3 | >0.5 kbp | Intellectual disability | 1,602 | None | 1 |
| Controls NIMH and ClinSeq | 1,794 | None | 1 | ||
| Custom Agilent array CGH4 | >300 kbp | Idiopathic autism (SSC) | 2,294 | None | 2 |
| Familial autism (AGRE) | 579 | None | None | ||
| Controls (NIMH and ClinSeq5) | 580 | None | None | ||
| Illumina sequencing | >1 kbp | Controls (1000 Genomes Project | 661 | None | None |
All detected deletions and duplications of SRGAP2A and SRGAP2C were >1 Mbp and include additional genes. Data from the Cooper et al. (2011) study could not be used to assess CNVs for SRGAP2C as there was insufficient probe coverage on the microarrays used in those studies. See also Figure S4 and Table S4 for details of CNV breakpoints, phenotypes, and inheritance status.
Abbreviations: SSC, Simons Simplex Collection (Fischbach and Lord, 2010); AGRE, Autism Genetic Resource Exchange (Geschwind et al., 2001); NIMH, National Institute of Mental Health (https://www.nimhgenetics.org/available_data/controls/); ClinSeq, Clinical Sequencing Pilot Project (Biesecker et al., 2009)
The assay targeted intron 11 of SRGAP2A.
Two assays were used targeting introns 6 and 7 of SRGAP2C, respectively.
Using probes targeting the chromosome 1p11.2 region proximal to SRGAP2C, we identified duplications and determined that a subset of them extended into SRGAP2C by using qPCR assays. Notably, all duplications of SRGAP2C identified from the qPCR assay alone extended into the 1p11.2 proximal region and would have been detected using this same method.
ClinSeq controls (N=373) were screened both with the Agilent array and SRGAP2C qPCR assay.
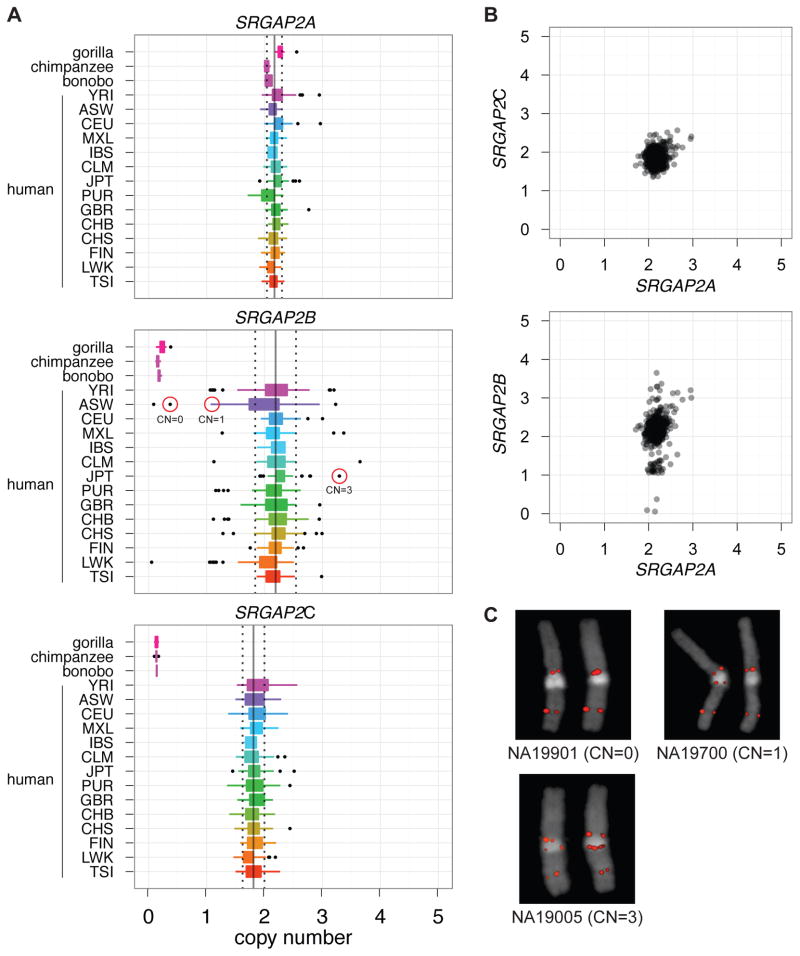
We next focused on assessing copy-number variation of each SRGAP2 paralog in the human population. This is particularly challenging because most recently duplicated genes are typically highly copy-number polymorphic (Sharp et al., 2005; Sudmant et al., 2010) and experimental assays for accurately predicting copy number are problematic. For this purpose, we took advantage of diagnostic singly unique nucleotide (SUN) identifiers (N = 3,535) determined using our high-quality sequence of the three loci (see above). We mapped genome-sequencing data from 661 human individuals corresponding to 14 populations (1000 Genomes Project) and estimated the diploid copy number for each paralog by measuring read-depth to these SUNs (Figure 4A) (Sudmant et al., 2010).
Figure 4. SRGAP2 copy-number diversity in human populations.
(A) Diploid copy-number estimates of SRGAP2 paralogs for 661 sequenced human genomes from 14 distinct populations (1000 Genomes Project) and from nonhuman primates. (B) SRGAP2A and SRGAP2C paralogs clearly are fixed at a copy number of 2, while SRGAP2B is polymorphic showing four distinct copy-number states. Note, we also detect polymorphism for SRGAP2D and have identified individuals homozygously deleted for this paralog. (C) FISH validation of three HapMap individuals genotyped for SRGAP2B [circled in red in part (A)]. All samples falling at the lower and upper tails of copy-number distributions for all three paralogs were experimentally genotyped using a paralog-specific qPCR assay; in all cases, SRGAP2A and SRGAP2C were validated as diploid copy number 2. Also refer to Figure S5.
We find that both the ancestral SRGAP2A and the derived SRGAP2C copy are fixed at diploid copy number 2 across all humans assayed. In contrast, the SRGAP2B and SRGAP2D copies varied from 0–4 copies among the individuals tested (Figure 4B–C). Importantly, we identified three individuals that are homozygously deleted for SRGAP2B. Note, we also identified normal individuals that were homozygously deleted for SRGAP2D the granddaughter copy with an acquired internal deletion of exons 2–3 (see Figure S5 for identification of deletion). We prepared cDNA from lymphoblastoid cells corresponding to one of these SRGAP2B-deletion homozygotes and observed no full-length SRGAP2B transcript by RT-PCR in contrast to samples carrying the paralog (Figure S3). Since the frequency of homozygotes is consistent with Hardy-Weinberg Equilibrium expectation and these individuals are representatives of the sample populations, the discovery of SRGAP2B-homozygous deletions in a —normal population argues against a critical functional role of this copy in brain development. We additionally applied our method to 34 nonhuman primates and the Denisova and Neanderthal genomes (Green et al., 2010; Reich et al., 2010) and found that, consistent with our sequence-based estimations of the timing of the duplication events, SRGAP2B, SRGAP2C, and SRGAP2D copies are absent from all assayed nonhuman great apes, yet are present in both the Neanderthal and Denisova genomes. We conclude that no new SRGAP2 duplications have occurred since Homo sapiens and Homo neanderthalensis diverged about one million years ago.
While it is common to observe a functional progenitor duplicated gene fixed in copy, the discovery that SRGAP2C is fixed at a diploid copy-number state is striking. When compared to the 23 genes duplicated specifically in the human lineage, we previously found that SRGAP2 is among the six least copy-number polymorphic gene families under a naïve analysis that does not distinguish paralogs (Sudmant et al., 2010). When we extend this analysis to human-specific duplicates where complete sequence is available and limiting our analysis solely to those genes (N = 23), we find that SRGAP2C is the least copy-number variable gene duplicate. Using qPCR assays that specifically assess copy-number variation of SRGAP2C, we investigated this experimentally and found one individual harboring a ~1 Mbp duplication containing numerous genes in an additional set of 1,794 controls (Table 2, Figure S4). Applying this same assay to patients with intellectual disability and/or autism spectrum disorder (N = 4,475), we identified three additional individuals carrying large duplications of this locus. Strikingly, in our cumulative analysis of 7,137 individuals (cases and controls), we detected no deletions of SRGAP2C. In total, our combined analyses indicate that both SRGAP2A and SRGAP2C copies are nearly fixed at a copy number of 2 in all human populations assayed, with rare deletions and duplications observed only in cases with intellectual disability for SRGAP2A (p = 0.055, Fisher’s exact test), and rare duplications observed ata frequency of ~0.06% for SRGAP2C.
DISCUSSION
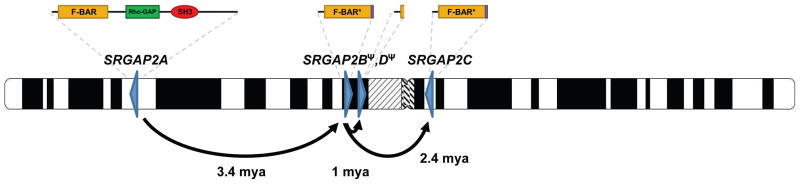
SRGAP2 has been highly conserved over mammalian evolution and human is the only lineage wherein gene duplications have occurred. Our analysis indicates that the duplications spread across 80 Mbp of chromosome 1 at a time corresponding to the transition from Australopithecus to Homo (Figure 5). This included an initial large interspersed duplication (258 kbp) from chromosome 1q32.1 to 1q21.1, creating SRGAP2B ~3.4 mya. The initial duplication was followed by larger (>500 kbp), secondary duplications of the 1q21.1 locus, creating SRGAP2C and SRGAP2D (~2.4 and 1 mya, respectively). Consistent with these timing estimates, archaic Homo species, including Neanderthal and Denisova, carry these SRGAP2 paralogs (Figure S5). It is intriguing that the general timing of the potentially functional copies, SRGAP2B and SRGAP2C, corresponds to the emergence of the genus Homo from Australopithecus (2–3 mya). This period of human evolution has been associated with the expansion of the neocortex, use of stone tools, as well as dramatic changes in behavior and culture (Jobling et al., 2004).
Figure 5. Model for SRGAP2 evolution.
Schematic depicts location and orientation (blue triangles) of SRGAP2 paralogs on human chromosome 1 with putative protein products indicated above each based on cDNA sequencing. Arrows trace the evolutionary history of SRGAP2 duplication events. Copy-number polymorphism and expression analyses suggest both paralogs at 1q21.1 (SRGAP2B and SRGAP2D) are pseudogenes, whereas the 1q32.1 (SRGAP2A) and 1p12 (SRGAP2C) paralogs are likely to encode functional proteins.
Our analysis provides insight into one mechanism by which gene duplicates evolve. We find that the initial genomic duplication of SRGAP2 was incomplete, encompassing the promoter and first nine exons of a 22-exon gene. Since SRGAP2 has been shown to homodimerize via its F-BAR domain (Guerrier et al., 2009), we propose that incomplete segmental duplication of the gene ~3.4 mya created an antagonistic functional state. In fact, functional evidence suggests that these partial SRGAP2 copies produce protein with a nearly complete F-BAR domain, but missing other functional domains, and heterodimerize with the full-length SRGAP2, creating a de facto dominant negative interaction equivalent to a knockdown of the ancestral copy (Charrier et. al., companion). The large size of the segmental duplication included the putative cis regulatory machinery of this gene and ensured that the duplicate genes would be developmentally co-expressed with the parental copy. Experimental analyses indicate (Guerrier et al., 2009; Charrier et al., companion) that if the segmental duplication had been slightly larger (i.e., included exon 10) such antagonism would not be possible.
The incomplete nature of the segmental duplication was, therefore, ideal to establish this new function by virtue of its structure, which arose at the time of its “birth”. This model of gene duplication involving an “instantaneous” dominant negative function at birth stands in stark contrast to the favored model involving duplication of a complete gene followed by the gradual accumulation of adaptive mutational events leading toward subfunctionalization or neofunctionalization (Lynch and Katju, 2004). We suggest that SRGAP2C ultimately assumed the antagonistic function of the SRGAP2B duplicate, which shows evidence of pseudogenization in contemporary humans. While all four SRGAP2 paralogs show evidence of transcription, it is unlikely that the two copies at 1q21.1 are now functional for several reasons. SRGAP2B has a markedly reduced expression in human brain compared to SRGAP2C. Likewise, the transcripts produced by SRGAP2D lack two internal exons leading to a premature termination codon, hence this copy is unlikely to produce a functional protein. Both SRGAP2B and SRGAP2D are highly copy-number polymorphic, with normal individuals identified that completely lack these paralogs. This argues that if there is a phenotypic consequence to their complete deletion, it is likely to be relatively minor.
In stark contrast, both the SRGAP2A (progenitor) and SRGAP2C (granddaughter) paralogs are nearly fixed at a diploid state based on our analysis of 28,153 and 7,137 human DNA samples, respectively. If we assume that the original SRGAP2B function was acquired by SRGAP2C, there is a possibility that both paralogs were functional at some point during human evolution. It is interesting that the comparison of the >515 kbp of duplicated sequence shared between SRGAP2B and SRGAP2C indicates that SRGAP2B has been subjected to large upstream deletions (103 kbp and 49 kbp in size) while SRGAP2C has not. Thus, the genomic instability of the SRGAP2B locus and its reduced expression in the contemporary human brain imply that the chromosome 1q21.1 locus may have been a suboptimal environment for gene transcription. The duplication event that yielded SRGAP2C, ~2.4 mya, may have provided a means of escape, transporting this truncated gene to a much more stable genomic environment for robust, long-term expression. One cannot, of course, definitively exclude the possibility that SRGAP2B/D transcripts may still confer some function (Charrier et al., companion), perhaps via transcript regulation, but the finding of apparently normal individuals completely missing these duplicate copies would suggest they are not critical for normal development.
We have identified larger deletions of the ancestral locus, SRGAP2A, only among children with developmental delay. While the deletion intervals are large and other genes contributing to the disease phenotype cannot be excluded at this time, the absence of structural variation in the normal population and the discovery of a de novo translocation (Saitsu et al., 2011), as well as a second patient with a duplication breakpoint mapping within SRGAP2, provide some evidence of its role in brain development. In this light, the fixation of the duplicated SRGAP2C is especially noteworthy. SRGAP2C was found to be the least copy-number polymorphic of all human-specific duplicate genes despite the fact that it is embedded in a complex region prone to non-allelic homologous recombination. Our data, thus, point to two functional SRGAP2 copies at 1p12 and 1q32.1, consistent with experimental characterization (Charrier et al., companion). Based on these data, we propose more systematic screening of these genes for mutations in children with developmental delay and brain malformations, including West Syndrome, agenesis of the corpus callosum, and epileptic encephalopathies. This will be particularly challenging since most commercial SNP microarrays have failed to include probes from these duplicated regions, and reads from next-generation sequencing platforms are typically too short to assign to a specific paralog (Eichler et al., 2010). Nevertheless, final proof of the functional significance of these genes will rest on the discovery of disruptive mutations associated with human phenotypes.
Finally, we emphasize that much of the genomic sequence corresponding to the ancestral and duplicate gene copies was missing or misassembled in the current human reference genome. In this study, we sequenced, corrected, and annotated ~0.4% of the euchromatin of chromosome 1 more than six years after the “finished” human genome was declared (IHGSC, 2004). This was possible because the clone-based resource we developed using a complete hydatidiform mole essentially provides a haploid version of the human genome. Since this resource is devoid of allelic variation, we can rapidly distinguish even highly identical duplicate genes, thus providing a clear path forward for the characterization of other complex duplicated regions. It is worthwhile noting that we ensured the hydatidiform mole primary cell line (Ch1hTERT) we used did not contain any large CNVs that could confound our analysis (Fan et al., 2002). It is especially intriguing that SRGAP2 is only one of several human-specific duplicate genes missing or incompletely assembled in the human genome (Sudmant et al., 2010). A number of remaining genes (e.g., GPRIN2, GTF2IRD2, and HYDIN) in this category have been implicated in neurodevelopment, neurite outgrowth, and behavior (Brunetti-Pierri et al., 2008; Chen et al., 1999; Dai et al., 2009). Additionally, human-specific protein-coding genes derived de novo from non-coding DNA merit further exploration (Wu et al., 2011). We propose that these uncharacterized human-specific genes constitute important pieces in the puzzle underlying the genetic basis of human brain evolution.
EXPERIMENTAL PROCEDURES
Fluorescent in situ hybridization (FISH)
Metaphase spreads were prepared from lymphoblastoid human cell lines (NA12878, NA19317, NA20334, NA19901, NA19700, and NA19005; Coriell Cell Repository, Camden, NJ); a chimpanzee cell line (Douglas, provided by Dr. Mariano Rocchi); and an orangutan cell line (PR01109 a.k.a. Susie; Coriell Cell Repository, Camden, NJ). FISH experiments were performed using fosmid clones (Supplemental Experimental Procedures) as described previously (Antonacci et al., 2010).
Cloning using a complete hydatidiform mole library
A large-insert BAC library (CHORI-17) was generated from a well-characterized complete hydatidiform mole primary cell culture (CHM1hTERT) using a modified protocol (Osoegawa et al., 1998) (http://bacpac.chori.org/library.php?id=231). To ensure the quality of CHM1hTERT, a karyotype analysis and extensive SNP genotyping with 1,494 SNP markers (Fan et al., 2002) and array comparative genomic hybridization (CGH) using the NimbleGen 2.1 M whole-genome array were performed. We generated paired-end sequences (N = 169,022) using Sanger dideoxy methods and mapped sequence reads to the human reference genome. This provided a haplotype-resolved tiling path of clones for selection and sequencing (Kidd et al., 2008).
Sequencing and assembly
We selected BAC clones with at least one sequenced end mapping to a SRGAP2 region in the human reference genome and completely sequenced and assembled the insert (see Supplemental Experimental Procedures for detailed clone order, sequence assembly, and annotation). Inserts overlapping with >99.9% sequence identity were assembled into distinct contigs corresponding to SRGAP2 loci at 1q32.1, 1q21.1, and 1p12.
Phylogenetic analysis
We created a 244.2 kbp multiple sequence alignment from three completely sequenced SRGAP2 genomic loci [ClustalW (Thompson et al., 2002)] and constructed an unrooted phylogenetic tree [MEGA (Tamura et al., 2011)] using the neighbor-joining method (Saitou and Nei, 1987) with the complete-deletion option. Genetic distances were computed using the Kimura two-parameter method (Kimura, 1980) with standard error estimates [an interior branch test of phylogeny (Dopazo, 1994; Rzhetsky and Nei, 1994); N = 500 bootstrap replicates]. For the incompletely sequenced SRGAP2D paralog and the 1p12 chromosomal distal region, we created phylogenetic trees using a 9.5 kbp and 50 kbp multiple species alignment, respectively (see Supplemental Experimental Procedures for details). The orthologous SRGAP2 exons were extracted from different mammalian reference genomes without segmental duplications and were used to test various models of selection using a maximum-likelihood framework [codemL; PAML statistical software package (Yang, 2007)].
SRGAP2 transcript analysis
Total RNA was isolated using Trizol reagent (Invitrogen) and the RNeasy Mini Kit (Qiagen) from SH-SY5Y neuronal cell line. Total RNA was analyzed from human fetal brain (collected from spontaneously aborted fetuses, 50–60 pooled samples, 20–33 weeks of development; ClonTech S2437) as well as a single fetal (R1244035, BioChain) and adult brain sample (M1234035, BioChain) (see Supplemental Experimental Procedures for details regarding RT-PCR, cDNA cloning, and sequencing). We also analyzed RNA-Seq data from 17 different human tissues (Illumina’s Human BodyMap2.0), seven human cell lines (Wang et al., 2008), and both chimpanzee and macaque cerebellum and liver tissues (Blekhman et al., 2010). Briefly, RNA-Seq datasets were mapped to the human reference genome (NCBI36/hg18) and our described SRGAP2 contigs. Expression levels for specific paralogs were calculated in units of RPKM (Liu et al., 2011) using transcribed PSVs, which allowed RNA-Seq data to be unambiguously assigned to a specific paralog.
Paralog-specific copy-number genotyping
CNVs in cases with intellectual disability and controls for SRGAP2A were identified from previously published array CGH data and SNP microarray data, respectively (Cooper et al., 2011). Copy-number estimates of specific SRGAP2 paralogs using SUNs were determined using previously described methods (Sudmant et al., 2010). Custom qPCR assays were performed in triplicate using variants specific to each SRGAP2 paralogous locus; see Supplemental Experimental Procedures for a description of variant detection and primer sequences). Validations of deletions and duplications, as well as identification of CNVs in the autism cohorts and some controls, were performed by array CGH using custom microarrays (Agilent) and a HapMap individual (NA18507) as a reference.
Acknowledgments
We thank B. Coe for assistance in CNV analysis and the 1000 Genomes Project for access to sequence data from the SRGAP2 locus. For DNA samples used in paralog-specific CNV screening, we would like to thank C. Romano, M. Fichera, J. Gecz, B. Devries, the Simons Foundation, Autism Speaks, the National Institute of Mental Health, and the ClinSeq Project. We acknowledge C. Baker, L. Vives, and J. Huddleston for technical assistance, T. Brown for manuscript editing, and the laboratory of S. Fields for use of their Roche LC480. We also thank J. Akey, T. Bonet-Marques, A. Andres, S. Girirajan, and K. Meltz Steinberg for helpful discussion, as well as the laboratory of F. Polleux for comments and kindly sharing human RNA samples for expression studies. The BAC clones from the complete hydatidiform mole were derived from a cell line created by U. Surti. M.Y.D. is supported by U.S. National Institutes of Health (NIH) Ruth L. Kirchstein National Research Service Award (NRSA) Fellowship (1F32HD071698-01). X.N. is supported by an NIH NRSA Genome Training Grant to the University of Washington (2T32HG000035-16). P.H.S. is a Howard Hughes Medical Institute International Student Research Fellow. This work was supported by NIH Grants HG002385 and GM58815. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
CONFLICTS OF INTEREST
J.A.R. and L.S. are employees of Signature Genomic Laboratories, a subsidiary of PerkinElmer, Inc. E.E.E. is on the scientific advisory boards for Pacific Biosciences, Inc. and SynapDx Corp.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, Girirajan S, Alkan C, Campbell CD, Vives L, Malig M, et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010;20:180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet M, Guy F, Pilbeam D, Lieberman DE, Likius A, Mackaye HT, Ponce de Leon MS, Zollikofer CP, Vignaud P. New material of the earliest hominid from the Upper Miocene of Chad. Nature. 2005;434:752–755. doi: 10.1038/nature03392. [DOI] [PubMed] [Google Scholar]
- Brunet M, Guy F, Pilbeam D, Mackaye HT, Likius A, Ahounta D, Beauvilain A, Blondel C, Bocherens H, Boisserie JR, et al. A new hominid from the Upper Miocene of Chad, Central Africa. Nature. 2002;418:145–151. doi: 10.1038/nature00879. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LT, Gilman AG, Kozasa T. A candidate target for G protein action in brain. J Biol Chem. 1999;274:26931–26938. doi: 10.1074/jbc.274.38.26931. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Dai L, Bellugi U, Chen XN, Pulst-Korenberg AM, Jarvinen-Pasley A, Tirosh-Wagner T, Eis PS, Graham J, Mills D, Searcy Y, et al. Is it Williams syndrome? GTF2IRD1 implicated in visual-spatial construction and GTF2I in sociability revealed by high resolution arrays. Am J Med Genet A. 2009;149A:302–314. doi: 10.1002/ajmg.a.32652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo J. Estimating errors and confidence intervals for branch lengths in phylogenetic trees by a bootstrap approach. J Mol Evol. 1994;38:300–304. doi: 10.1007/BF00176092. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Segmental duplications: what’s missing, misassigned, and misassembled--and should we care? Genome Res. 2001;11:653–656. doi: 10.1101/gr.188901. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Fan JB, Surti U, Taillon-Miller P, Hsie L, Kennedy GC, Hoffner L, Ryder T, Mutch DG, Kwok PY. Paternal origins of complete hydatidiform moles proven by whole genome single-nucleotide polymorphism haplotyping. Genomics. 2002;79:58–62. doi: 10.1006/geno.2001.6676. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, Dunham A, Scott CE, Howe KL, Woodfine K, Spencer CC, et al. The DNA sequence and biological annotation of human chromosome 1. Nature. 2006;441:315–321. doi: 10.1038/nature04727. [DOI] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Bao S. srGAP2 arginine methylation regulates cell migration and cell spreading through promoting dimerization. J Biol Chem. 2010;285:35133–35141. doi: 10.1074/jbc.M110.153429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jobling M, Hurles M, Tyler-Smith C. Human Evolutionary Genomics. New York: Garland Science; 2004. [Google Scholar]
- Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature. 1977;268:633–634. doi: 10.1038/268633a0. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin-Heidelberg-New York: Springer-Verlag; 1970. [Google Scholar]
- Osoegawa K, Woon PY, Zhao B, Frengen E, Tateno M, Catanese JJ, de Jong PJ. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, Fitzpatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzhetsky A, Nei M. METREE: a program package for inferring and testing minimum-evolution trees. Comput Appl Biosci. 1994;10:409–412. doi: 10.1093/bioinformatics/10.4.409. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Osaka H, Sugiyama S, Kurosawa K, Mizuguchi T, Nishiyama K, Nishimura A, Tsurusaki Y, Doi H, Miyake N, et al. Early infantile epileptic encephalopathy associated with the disrupted gene encoding Slit-Robo Rho GTPase activating protein 2 (SRGAP2) Am J Med Genet A. 2011 doi: 10.1002/ajmg.a.34363. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman HH, Kozyak BW, Nelson A, Thesier DM, Su LT, Low DW, Bridges CR, Shrager JB, Minugh-Purvis N, Mitchell MA. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. doi: 10.1038/nature02358. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamalek JM, Goidts V, Cooper DN, Hameister H, Kehrer-Sawatzki H. Characterization of the human lineage-specific pericentric inversion that distinguishes human chromosome 1 from the homologous chromosomes of the great apes. Hum Genet. 2006;120:126–138. doi: 10.1007/s00439-006-0209-y. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2(Unit 23) doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Vignaud P, Duringer P, Mackaye HT, Likius A, Blondel C, Boisserie JR, De Bonis L, Eisenmann V, Etienne ME, Geraads D, et al. Geology and palaeontology of the Upper Miocene Toros-Menalla hominid locality, Chad. Nature. 2002;418:152–155. doi: 10.1038/nature00880. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP. De novo origin of human protein-coding genes. PLoS Genet. 2011;7:e1002379. doi: 10.1371/journal.pgen.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mishra B. Quantifying the mechanisms for segmental duplications in mammalian genomes by statistical analysis and modeling. Proc Natl Acad Sci USA. 2005;102:4051–4056. doi: 10.1073/pnas.0407957102. [DOI] [PMC free article] [PubMed] [Google Scholar]