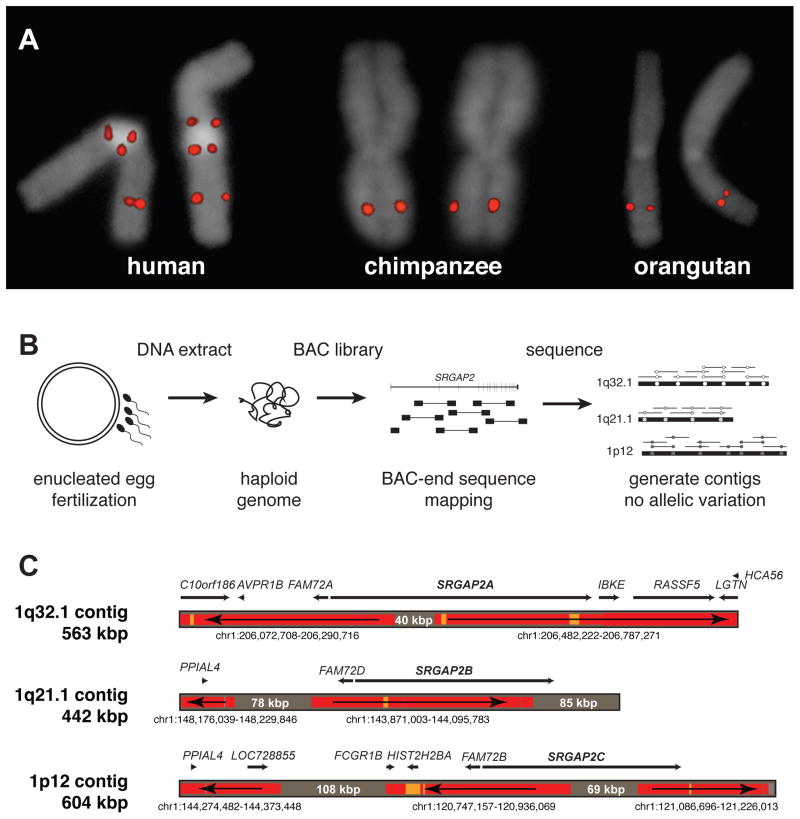
Figure 1. Genomic characterization and sequence resolution of SRGAP2 loci.
(A) FISH analysis shows three distinct copies of SRGAP2 on metaphase human chromosome 1, compared to a single copy in chimpanzee and orangutan (see Figure 2A for location of FISH probe; Figure S1 and Table S1 for details of additional FISH assays). (B) SRGAP2 genomic loci were sequenced and assembled using a BAC library (CHORI-17) created from human haploid genomic source material (complete hydatidiform mole). The absence of allelic variation allowed paralogous sequences to be resolved with high confidence based on near-perfect sequence identity overlap (>99.9%). (C) Regions highly identical to the reference genome (GRCh37/hg19) are colored in red (identity = 99.8–100%) and orange (99.6–99.8%), while regions completely absent from the current assembly are shaded gray (with region sizes indicated). Arrows show the orientation of the reference genome sequence with respect to the contigs (e.g., a left directional arrow indicates the reverse strand) indicating that even the ancestral (SRGAP2A) gene locus was missing sequence data, misassembled, and incorrectly orientated over 400 kbp of the current high-quality reference assembly. Genomic coordinates correspond to the representative human reference region with corresponding genes within these regions mapped along each contig.