Abstract
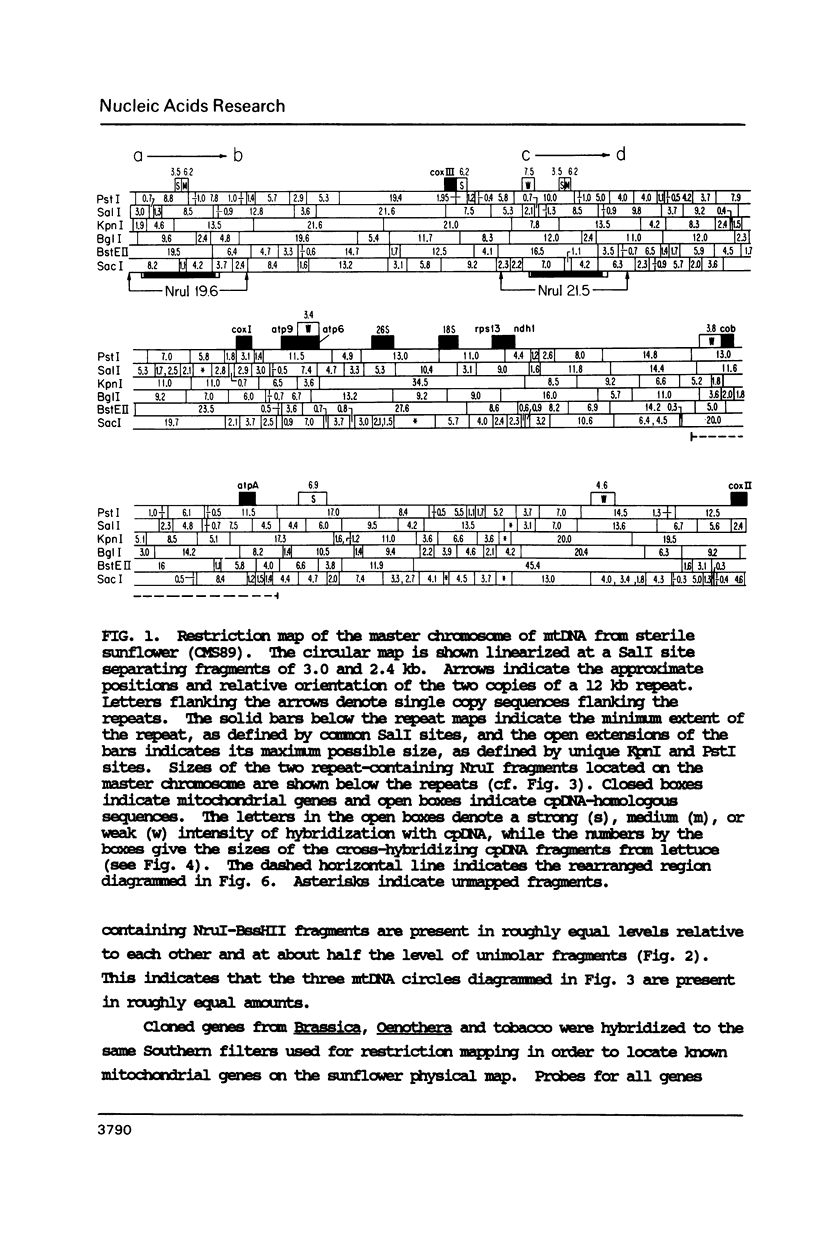
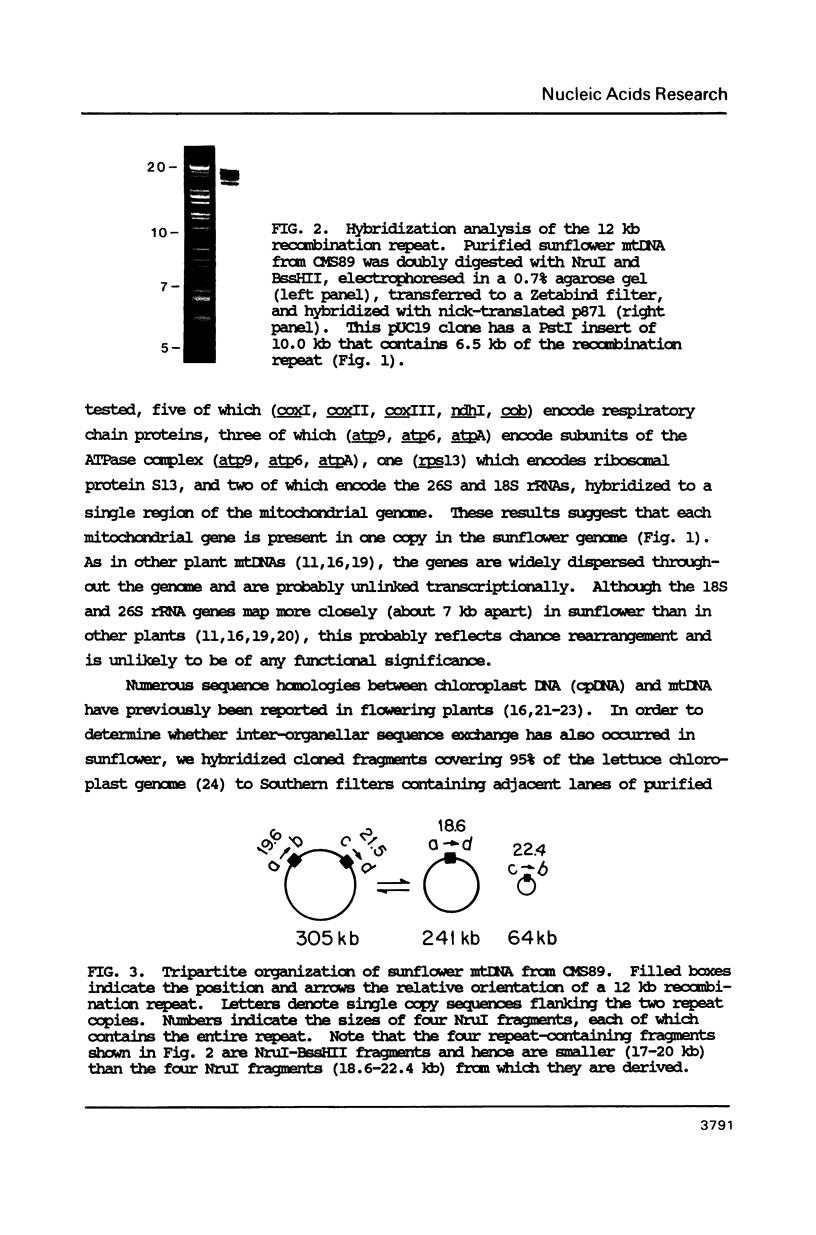
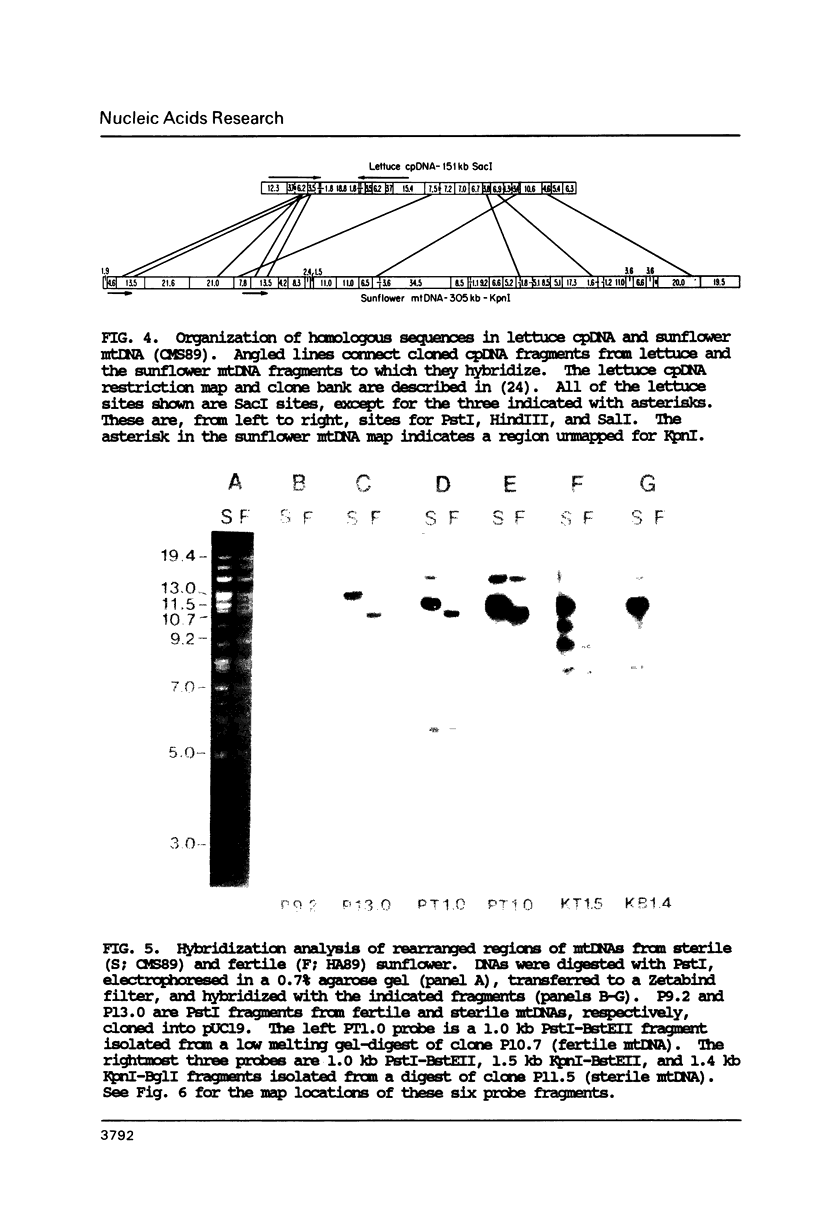
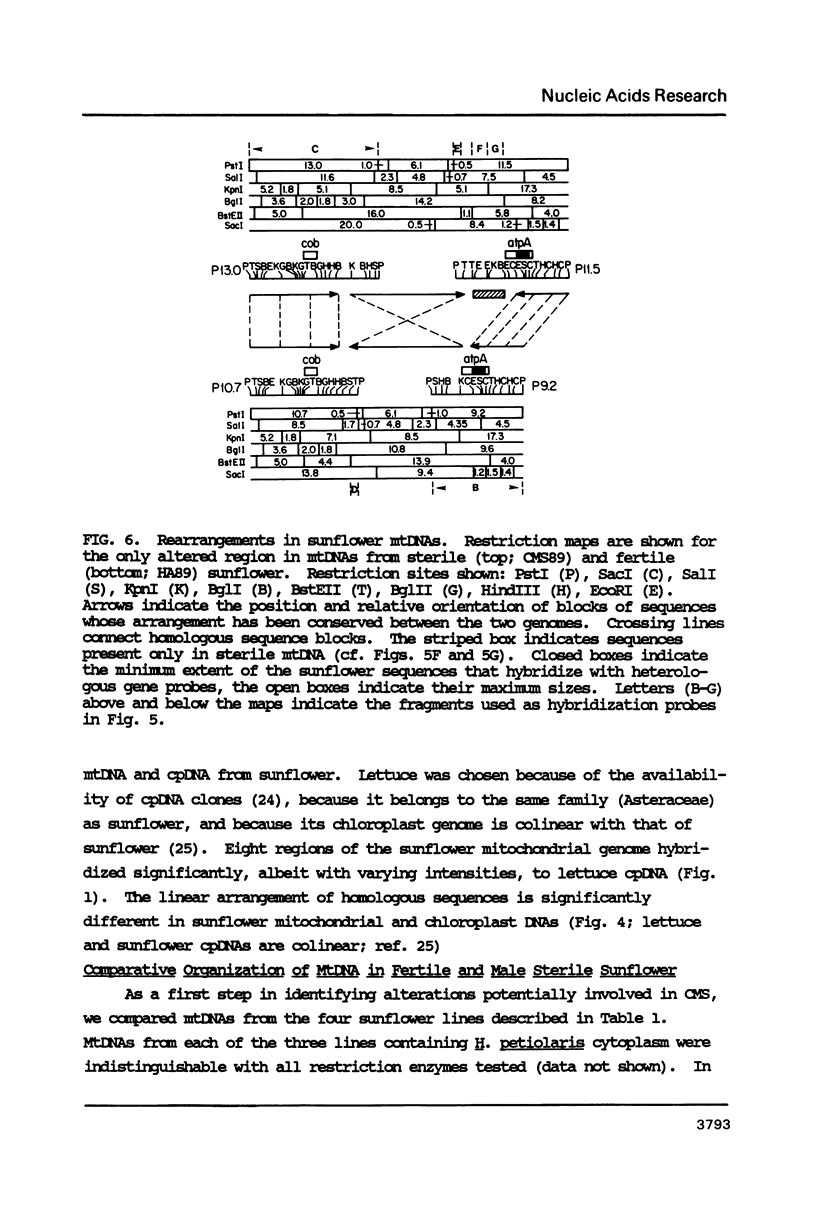
To study the molecular basis of cytoplasmic male sterility (CMS) in sunflower (Helianthus annuus), we compared the physical organization and transcriptional properties of mitochondrial DNAs (mtDNAs) from isonuclear fertile and CMS lines. Mapping studies revealed much greater similarity between the two mtDNAs than in previous comparisons of fertile and CMS lines from other plant species. The two sunflower mtDNAs 1) are nearly identical in size (300 kb and 305 kb); 2) contain the same 12 kb recombination repeat and associated tripartite structure; 3) have the same dispersed distribution of mitochondrial genes and chloroplast DNA-homologous sequences; 4) are greater than 99.9% identical in primary sequence; and 5) are colinear over a contiguous region encompassing 94% of the genome. Detectable alterations are limited to a 17 kb region of the genome and reflect as few as two mutations--a 12 kb inversion and a 5 kb insertion/deletion. One endpoint of both rearrangements is located within or near atpA, which is also the only mitochondrial gene whose transcripts differ between the fertile and CMS lines. Furthermore, a nuclear gene that restores fertility to CMS plants specifically influences the pattern of atpA transcripts. Rearrangements at the atpA locus may, therefore, be responsible for CMS in sunflower.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey-Serres J., Hanson D. K., Fox T. D., Leaver C. J. Mitochondrial genome rearrangement leads to extension and relocation of the cytochrome c oxidase subunit I gene in sorghum. Cell. 1986 Nov 21;47(4):567–576. doi: 10.1016/0092-8674(86)90621-5. [DOI] [PubMed] [Google Scholar]
- Dewey R. E., Levings C. S., 3rd, Timothy D. H. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell. 1986 Feb 14;44(3):439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- Dewey R. E., Siedow J. N., Timothy D. H., Levings C. S., 3rd A 13-kilodalton maize mitochondrial protein in E. coli confers sensitivity to Bipolaris maydis toxin. Science. 1988 Jan 15;239(4837):293–295. doi: 10.1126/science.3276005. [DOI] [PubMed] [Google Scholar]
- Dewey R. E., Timothy D. H., Levings C. S. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5374–5378. doi: 10.1073/pnas.84.15.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Palmer J. D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc Natl Acad Sci U S A. 1987 Aug;84(16):5818–5822. doi: 10.1073/pnas.84.16.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Physicochemical characterization of mitochondrial DNA from pea leaves. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1830–1834. doi: 10.1073/pnas.69.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Fauron C. M. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 1984 Dec 21;12(24):9249–9261. doi: 10.1093/nar/12.24.9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. M., Hodge T. P., Howe C. J., Stern D. B. Maize mitochondrial DNA contains a sequence homologous to the ribulose-1,5-bisphosphate carboxylase large subunit gene of chloroplast DNA. Cell. 1983 Oct;34(3):1007–1014. doi: 10.1016/0092-8674(83)90558-5. [DOI] [PubMed] [Google Scholar]
- Makaroff C. A., Palmer J. D. Extensive mitochondrial specific transcription of the Brassica campestris mitochondrial genome. Nucleic Acids Res. 1987 Jul 10;15(13):5141–5156. doi: 10.1093/nar/15.13.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Herbon L. A. Tricircular mitochondrial genomes of Brassica and Raphanus: reversal of repeat configurations by inversion. Nucleic Acids Res. 1986 Dec 22;14(24):9755–9764. doi: 10.1093/nar/14.24.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Herbon L. A. Unicircular structure of the Brassica hirta mitochondrial genome. Curr Genet. 1987;11(6-7):565–570. doi: 10.1007/BF00384620. [DOI] [PubMed] [Google Scholar]
- Rottmann W. H., Brears T., Hodge T. P., Lonsdale D. M. A mitochondrial gene is lost via homologous recombination during reversion of CMS T maize to fertility. EMBO J. 1987 Jun;6(6):1541–1546. doi: 10.1002/j.1460-2075.1987.tb02398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C. L., Pring D. R., Lonsdale D. M. Mitochondrial DNA rearrangements associated with fertile revertants of S-type male-sterile maize. Cell. 1985 Nov;43(1):361–368. doi: 10.1016/0092-8674(85)90041-8. [DOI] [PubMed] [Google Scholar]
- Sederoff R. R. Structural variation in mitochondrial DNA. Adv Genet. 1984;22:1–108. doi: 10.1016/s0065-2660(08)60038-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Newton K. J. Isolation of plant mitochondrial RNA. Methods Enzymol. 1986;118:488–496. doi: 10.1016/0076-6879(86)18095-5. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Extensive and widespread homologies between mitochondrial DNA and chloroplast DNA in plants. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1946–1950. doi: 10.1073/pnas.81.7.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Palmer J. D. Tripartite mitochondrial genome of spinach: physical structure, mitochondrial gene mapping, and locations of transposed chloroplast DNA sequences. Nucleic Acids Res. 1986 Jul 25;14(14):5651–5666. doi: 10.1093/nar/14.14.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. L., Anderson R. S., Bendich A. J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell. 1981 Sep;25(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- Wise R. P., Pring D. R., Gengenbach B. G. Mutation to male fertility and toxin insensitivity in Texas (T)-cytoplasm maize is associated with a frameshift in a mitochondrial open reading frame. Proc Natl Acad Sci U S A. 1987 May;84(9):2858–2862. doi: 10.1073/pnas.84.9.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. G., Hanson M. R. A fused mitochondrial gene associated with cytoplasmic male sterility is developmentally regulated. Cell. 1987 Jul 3;50(1):41–49. doi: 10.1016/0092-8674(87)90660-x. [DOI] [PubMed] [Google Scholar]