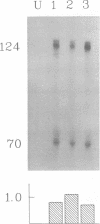
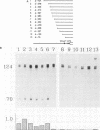
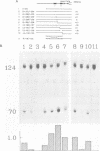
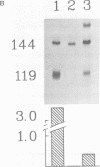
Abstract
The DNA sequences required for efficient initiation of transcription of the Xenopus transcription factor IIIA (TFIIIA) gene were determined by microinjecting a series of deletion and linker substitution mutants into Xenopus oocyte nuclei. An upstream activating sequence, which resides between residues -283 and -238, and perhaps a second sequence between -167 and -122 preceding the transcription start site, together stimulate transcription about 30-fold. The distance between the two sequences and the distance from them to the initiation site can vary by at least 13 base pairs without loss of activity. The TFIIIA upstream sequences can stimulate transcription of other genes, for example, they stimulate transcription from the herpes thymidine kinase promoter about 30-fold.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BROWN D. D., LITTNA E. RNA SYNTHESIS DURING THE DEVELOPMENT OF XENOPUS LAEVIS, THE SOUTH AFRICAN CLAWED TOAD. J Mol Biol. 1964 May;8:669–687. doi: 10.1016/s0022-2836(64)80116-9. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Schlissel M. S. A positive transcription factor controls the differential expression of two 5S RNA genes. Cell. 1985 Oct;42(3):759–767. doi: 10.1016/0092-8674(85)90272-7. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Sugimoto K. The structure and evolution of ribosomal and 5S DNAs in Xenopus laevis and Xenopus mulleri. Cold Spring Harb Symp Quant Biol. 1974;38:501–505. doi: 10.1101/sqb.1974.038.01.054. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Southern E. M. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973 Jan 3;241(105):7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Gilbert D. M. Temporal order of replication of Xenopus laevis 5S ribosomal RNA genes in somatic cells. Proc Natl Acad Sci U S A. 1986 May;83(9):2924–2928. doi: 10.1073/pnas.83.9.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinta D. R., Korn L. J. Differential order of replication of Xenopus laevis 5S RNA genes. Mol Cell Biol. 1986 Jul;6(7):2536–2542. doi: 10.1128/mcb.6.7.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinta D. R., Tso J. Y., Narayanswami S., Hamkalo B. A., Korn L. J. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5150–5154. doi: 10.1073/pnas.83.14.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. H., Wu R. Exonuclease III: use for DNA sequence analysis and in specific deletions of nucleotides. Methods Enzymol. 1983;100:60–96. doi: 10.1016/0076-6879(83)00046-4. [DOI] [PubMed] [Google Scholar]
- Korn L. J. Transcription of Xenopus 5S ribosomal RNA genes. Nature. 1982 Jan 14;295(5845):101–105. doi: 10.1038/295101a0. [DOI] [PubMed] [Google Scholar]
- Ladiges W. C., Raff R. F., Brown S., Deeg H. J., Storb R. The canine major histocompatibility complex. Supertypic specificities defined by the primed lymphocyte test (PLT). Immunogenetics. 1984;19(4):359–365. doi: 10.1007/BF00345410. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- McKnight S. L. Constitutive transcriptional control signals of the herpes simplex virus tk gene. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):945–958. doi: 10.1101/sqb.1983.047.01.108. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Wormington W. M., Brown D. D. Related 5S RNA transcription factors in Xenopus oocytes and somatic cells. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1760–1764. doi: 10.1073/pnas.78.3.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Shastry B. S., Honda B. M., Roeder R. G. Altered levels of a 5 S gene-specific transcription factor (TFIIIA) during oogenesis and embryonic development of Xenopus laevis. J Biol Chem. 1984 Sep 25;259(18):11373–11382. [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Taylor W., Jackson I. J., Siegel N., Kumar A., Brown D. D. The developmental expression of the gene for TFIIIA in Xenopus laevis. Nucleic Acids Res. 1986 Aug 11;14(15):6185–6195. doi: 10.1093/nar/14.15.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso J. Y., Sun X. H., Wu R. Structure of two unlinked Drosophila melanogaster glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985 Jul 5;260(13):8220–8228. [PubMed] [Google Scholar]
- Tso J. Y., Van Den Berg D. J., Korn L. J. Structure of the gene for Xenopus transcription factor TFIIIA. Nucleic Acids Res. 1986 Mar 11;14(5):2187–2200. doi: 10.1093/nar/14.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Schlissel M., Brown D. D. Developmental regulation of Xenopus 5S RNA genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):879–884. doi: 10.1101/sqb.1983.047.01.101. [DOI] [PubMed] [Google Scholar]