Abstract
The Type VI secretion pathway transports proteins across the cell envelope of Gram-negative bacteria. Pseudomonas aeruginosa, an opportunistic Gram-negative bacterial pathogen infecting humans, uses the type VI secretion pathway to export specific effector proteins crucial for its pathogenesis. The HSI-I virulence locus encodes for several proteins that has been proposed to participate in protein transport including the Hcp1 protein, which forms hexameric rings that assemble into nanotubes in vitro. Two Hcp1 paralogues have been identified in the P. aeruginosa genome, Hsp2 and Hcp3. Here, we present the structure of the Hcp3 protein from P. aeruginosa. The overall structure of the monomer resembles Hcp1 despite the lack of amino-acid sequence similarity between the two proteins. The monomers assemble into hexamers similar to Hcp1. However, instead of forming nanotubes in head-to-tail mode like Hcp1, Hcp3 stacks its rings in head-to-head mode forming double-ring structures.
Keywords: Type VI (T6SS) secretion system, Hcp3, Hcp1, X-ray crystallography, Structural genomics
Introduction
Eight distinct protein secretory systems have been described in Gram-negative bacteria (type I (T1SS) to type VIII (T8SS)) [1]. The diversity of these systems has been linked to virulence and efficiency of infections. Typically the genes that code for secretory systems are grouped in virulence loci and are tightly regulated either at the transcriptional or post-transcriptional level by a number of regulatory factors, such as RetS [2], LadS [3] and PpkA [4]. Many of the secretory systems have been shown to be involved in acute and chronic infections. Among the most widespread is the recently discovered Type VI (T6SS) secretion system. The T6SS gene cluster contains a core of 13 highly conserved genes including IcmF, DotU, ClpV homologs of a sub type of the ATPase AAA+ family, IglA/IglB chaperone-like proteins, Hcp, VgrG and several proteins of unknown function [5].
In cystic fibrosis, Pseudomonas aeruginosa secretes Hcp1 (hemolysin-corregulated protein 1) and VgrG (valine-glycine-repeat protein G) using the T6SS [6, 7]. Both of these proteins are encoded by the HSI-I locus. P. aeruginosa also contains 2 loci, HSI-II and HSI-III, which code for Hcp-like proteins as well as other 15–21 proteins homologous to components of the T6SS apparatus (ClpB AAA+ like proteins, IcmF, VgrG, serine-threonine protein kinases and serine-threonine phosphatases [4, 6]). Hcp2 and Hcp3 are believed to be secreted by their own specific T6SS sub-systems [4, 6, 8]. It is also believed that Hcp2 and Hcp3 perform a similar structural function as Hcp1 [4, 6, 9].
The most well characterized Hcp protein is Hcp1 from P. aeruginosa, for which biological properties and the X-ray crystal structure were recently described [6]. The Hcp1 monomer assembles in hexameric rings [6]. In the crystal, Hcp1 hexamers stack head-to-tail and form long nanotubes with a 40 Å wide tunnel [6]. The structure of Hcp1 is similar to that of the gpV tail tube protein of phage lambda [10] and shows an evolutionary relationship to the T4 tail-tube protein gp19 [11]. These phage proteins serve as components of phage genome transfer systems and assemble to form a tail-tube. These similarities have led to the suggestion that Hcp1 forms tubes that can cross membranes and connect different cell compartments. Working together with VgrG it forms a structure on the surface of the cell capable of puncturing the host membrane and facilitating channeling effector proteins across membranes [12, 13].
Here we report the 2.1 Å resolution structure of Hcp3 from P. aeruginosa. Hcp1 and Hcp3 are undeniably structural homologs despite their very low amino acid sequence similarity. The both proteins form hexamers but, in contrast to Hcp1, Hcp3 does not form head-to-tail nanotubes in the crystal. The hexamers of Hcp3 form double rings (dodecamers) in a head-to-head mode, a very different arrangement as compared to Hcp1.
Materials and methods
Protein purification
Growth, expression, and purification of the Hcp3 seleno-methionine (SeMet) protein from P. aeruginosa (gene name hcpC, locus tag PA0263) were performed essentially as described by Korolev et al. [14]. Briefly, the expression plasmid was transformed into E. coli BL21-Gold (DE3) strain (Stratagene), which harbors an extra plasmid (pMAGIC) encoding rare tRNA (recognizing AGG and AGA codons for Arg). These E. coli cells were then cultured in M9 minimal media containing 0.4% (w/v) glucose as a carbon source and supplemented with ampicillin (100 g/mL) and kanamycin (50 g/mL). The cultures were incubated at 37°C with shaking until the culture reached an OD600 of 0.8. At this point SeMet, plus an amino acid cocktail (60 mg SeMet; 100 mg of lysine, threonine, and phenylalanine; and 50 mg leucine, isoleucine, and valine) were added. Fifteen minutes later the culture was induced with 0.4 mM IPTG and the cells were allowed to grow for 3 h at 37°C with shaking and allowed to grow overnight at 15°C. Cells were harvested by centrifugation, disrupted by sonication, and the insoluble cellular material was removed by centrifugation. Hcp3 was purified from other contaminating proteins using Ni–NTA affinity chromatography with the addition of 5 mM 2-mercaptoethanol in all buffers. The protein was digested with 0.15 mg TEV protease per 20 mg purified protein for 16 h at 4°C and then passed through a Ni–NTA column to remove both the TEV protease and cleaved histidine tags. The protein was subsequently dialyzed, stored in buffer containing 10 mM HEPES (pH 7.5) and 500 mM NaCl, and quantified using an extinction coefficient of 0.475 M−1 cm−1 for contributions of Trp and Tyr at 280 nm.
Protein crystallization
The initial crystallization condition was determined with a sparse crystallization matrix (Hampton Research Crystal Screen I) at room temperature using the hanging drop-vapor diffusion technique. The optimized crystallization condition consists of 0.1 M Bis–tris buffer pH 7.0 and 3.2 M sodium chloride in the presence of 0.028 mg/ml chymotrypsin. Crystals selected for data collection were flash-cooled in the crystallization buffer supplemented with 16% glycerol.
Data collection, structure determination and refinement
A set of single-wavelength anomalous diffraction (SAD) data was collected from a single SeMet labeled crystal near the selenium absorption peak at 100 K. The data were obtained at the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory using the program SBCcollect. The intensities were integrated and scaled with the HKL3000 suite [15].
The structure was determined by SAD phasing using the HKL3000 suite incorporating the following programs: SHELXC, SHELXD, SHELXE, MLPHARE, and SOLVE/RESOLVE [15]. The initial partial model was autotraced using RESOLVE [16] and more residues were added using ARP/wARP [17]. Manual addition of residues and adjustment using COOT [18] were required to complete the model for refinement. Originally, data were scaled and the structure was determined in the P6322 space group. However, high R-factor values in refinement suggested that the crystal was twinned. Data were re-scaled in the P312 space group and twinning was detected using the PHENIX-xtriage program [19]. Several rounds of manual adjustments using COOT [18] and refinements with detwinning by PHENIX [19] were performed. The stereochemistry of the structure was validated with a WHATCHECK [20] and MOLPROBITY [21] Ramachandran plot. The main chain torsion angles for all residues are in the allowed regions or additional allowed regions. The crystal belongs to the hexagonal space group P312. The asymmetric unit contained 6 protein polypeptides, with 4 SeMet per polypeptide. In addition to water molecules, two molecules of glycerol were modeled in the structure. The final Rwork and Rfree were 21.4% and 25.4%, respectively. A summary of data collection and refinement statistics is given in Tables 1 and 2.
Table 1.
Crystal and data collection statistics
| X-ray wavelength | 0.9792 Å |
| Space group | P312 |
| Unit cell dimensions | a = b = 141.22 Å, c = 105.05 Å, α = β = 90°, γ = 120° |
| Resolutiona | 39.8–2.1 Å (2.14–2.10 Å) |
| No. of unique reflections | 69,851 (3,473) |
| Completeness | 100% (100%) |
| R-merge | 0.120 (0.647) |
| Molecules per asymmetric unit | 6 |
| No. of protein residues | 172 |
| SeMet per asymmetric unit | 24 |
Numbers in parenthesis are shown for the highest resolution shell
Table 2.
Structure refinement statistics
| Resolution range (Å) | 39.8–2.10 |
| Reflections | 69,851 |
| σ cutoff | None |
| R-value (R-work) (%) | 21.4 |
| Free R-value (%) | 25.4 |
| R-value (all) (%) | 21.6 |
| RMS deviations from ideal geometry | |
| Bond length (Å) | 0.008 |
| Angle (degrees) | 1.184 |
| Chiral (Å) | 0.076 |
| No. of atoms | |
| Protein | 6,965 |
| Glycerol | 12 |
| Water | 728 |
| Mean B-factor (Å2) | |
| All atoms | 32.7 |
| Protein atoms | 32.7 |
| Protein main chain | 31.5 |
| Protein side chain | 33.9 |
| Glycerol | 41.5 |
| Water | 33.2 |
| Molprobity Ramachandran plot statistics | |
| Residues in most favored regions (%) | 97.0 |
| Residues in additional allowed regions (%) | 3.0 |
| Residues in disallowed region (%) | 0.0 |
Coordinates
Atomic coordinates and structure factors were deposited into the Protein Data Bank as 3HE1.
Results and discussion
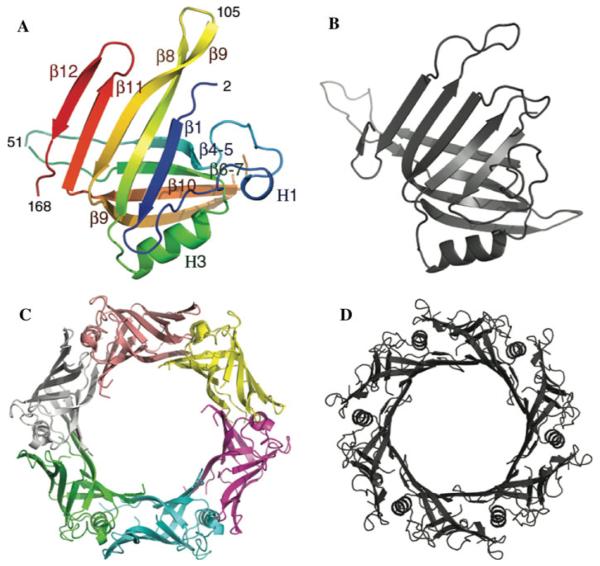
The overall structure of P. aeruginosa Hcp3 closely resembles the P. aeruginosa Hcp1 structure [6] despite low amino acid sequence similarity (Figs. 1, 2). Only a stretch of 109 residues (45–146) of Hcp3 shows 19.3% sequence identity to the 57–165 residues of Hcp1 protein (based on FASTA UVa server). The main body of the Hcp3 monomer contains two β-sheets consisting of 4 and 5 β-strands each. The β-sheets interact with each other and form a tight β-barrel. One side of the β-barrel is flanked by a 12-residue α-helix (H3) and two one-turn α-helices while the other side is open to accept the α-helix from an adjacent subunit. The α-helix H3 is nearly parallel to the sixfold axis of the hexamer (Fig. 2c, d). It is very hydrophobic with the sequence LLLAAL and provides a binding surface for β9 and β10 strands from the same subunit and β11 and β12 strands from an adjacent subunit. Therefore, α-helices H3 serve as locking pins holding the hexamer together. The structure of the Hcp3 monomer is very similar to Hcp1 and can be aligned with an RMSD for Cα atoms of 2.1 Å.
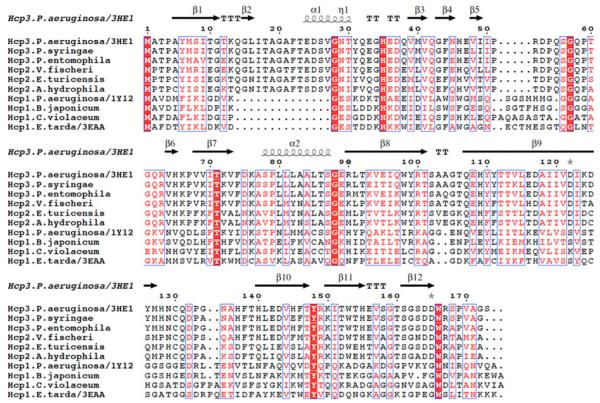
Fig. 1.
The sequence alignment of Hcp1 and Hcp2 proteins compared to P. aeruginosa Hcp3 sequence and its secondary structures
Fig. 2.
The crystal structures of Hcp3 (in color) and Hcp1 (in grey) proteins from P. aeruginosa. a, b Structures of Hcp3 and Hcp1 monomers, respectively, c, d Structures of Hcp3 and Hcp1 hexamers, respectively
The Hcp3 crystal asymmetric unit contains 6 protein monomers grouped in 3 dimers related by an approximate threefold axis. Each of the dimers contributes to a different hexameric unit in the protein crystal. The inside channel of the hexameric rings has a diameter of ~40 Å (Fig. 2c) and closely resembles Hcp1 [6]. The inner surface of the ring is formed by a continuous β-sheet created from 6 individual β-sheets consisting of β4, β5, β6, β7, β9, and β10 strands from each monomer. The β9 strand of each monomer interacts with the β4 and β5 strands of an adjacent subunit inside the ring. Similarly to Hcp1, this inner surface is partly hydrophilic and partly hydrophobic. The outer surface of the ring has a hexagonal shape with a diameter of 90 Å (Fig. 2c) and shows one strongly negatively chargedside of the ring (data not shown). The thickness of the hexameric ring is about 30Å.
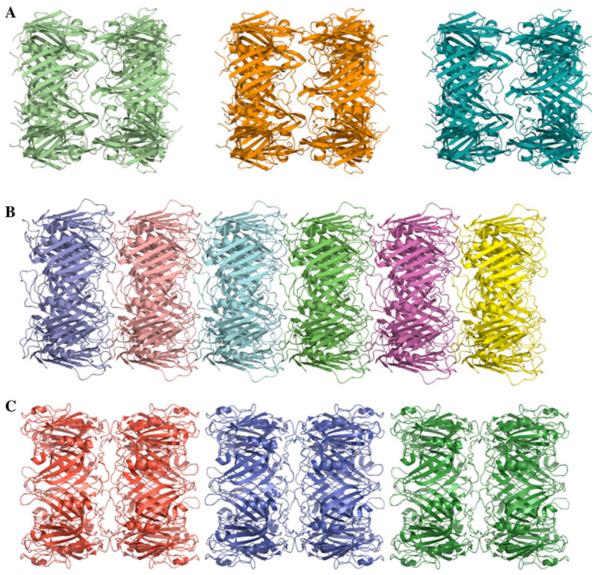
In the crystal, Hcp3 hexamers interact with the next hexameric unit in head-to-head mode to form dodecamers (Fig. 3a). The thickness of the double ring is about 70 Å. The main contacts between the rings result from the interaction between loops 31–33 and 104–106. The hexamer interactions in Hcp3 crystals are very different from that found in the P. aeruginosa Hcp1 structure, where hexamers are connected in head-to-tail mode and form long nanotubes (Fig. 3b). The inter-ring contacts for the Hcp1 structure are more extensive and involve 4 loops. Two of these loops are in a structurally similar position to those of Hcp3 (residues 17–19 and 89–92) and two loops are very different (residues 42–45 and 157–158). Hcp3 does not form continuous nanotubes in the crystal. Recently, a structure of the EvpC protein from Edwardsiella tarda was reported [22]. The EvpC shows sequence similarity to Hcp1 (Fig. 1). This structure is also a hexameric assembly with rings interacting in head-to-head mode (Fig. 3c) similar to the Hcp3 structure. However, the double rings are packed more densely than in the case of Hcp3 and form nanotubes in the crystal in alternating head-to-head and tail-to-tail modes. The inter-ring contacts are generated by two loops (residues 44–51 and 158–160) that are in a structurally similar position to Hcp1 loops (residues 42–45 and 157–158). It is quite striking that similar head-to-head interactions in Hcp3 and EvpC can be accomplished by two different sets of structural elements.
Fig. 3.
Crystal packing of Hcp3 and Hcp1 proteins. a “Isolated” double-rings of P. aeruginosa Hcp3. b Nanotubes of P. aeruginosa Hcp1 protein. c “Close” double-rings of E. tarda Hcp1 (EvpC) protein
It is possible that these different modes of inter-ring interactions in Hcp1 and Hcp3 crystals result from different crystal packing. Both crystals have honey-bee cluster type packing in planes perpendicular to their ring axes, apparently because of the hexagonal shape of flat protein rings. Such dense packing may force ring stacking and form Hcp1 nanotubes with head-to-tail mode of packing or in alternating head-to-head and tail-to-tail modes as found in EvpC (Hcp1). In the case of Hcp3, the crystal packing resulted in head-to-head interactions generating dodecamers. The formation of Hcp1 head-to-tail nanotubes has been observed by electron microscopy and was shown to be reversible [6, 12]. This still needs to be shown for alternating head-to-head and tail-to-tail nanotubes.
Currently available data suggest that Hcp hexamers show very distinctive properties. These hexameric discs contain several loop regions located on their flat surfaces that are capable of promoting inter-ring stacking of these discs in head-to-tail, head-to-head and tail-to-tail modes. In the case of Hcp1, it leads to the formation of head-to-tail nanotubes and in the case of EvpC, it results in the formation of alternating head-to-head and tail-to-tail nanotubes. Although this is not observed in the crystals of Hcp3, we cannot exclude the possibility of the nanotube formation similar to that of EvpC under conditions different from those used by us for protein crystallization. One can easily imagine that double hexamer Hcp3 rings can stack on top of each other forming a nanotube. This could be achieved because of the flexibility of 52–60 and 129–137 Hcp3 loops.
Sequence comparisons of Hcp1, 2 and 3 proteins using BLAST server revealed that Hcp proteins fall into two rather than three sequence subgroups, with Hcp1 forming one subgroup and Hcp2 and Hcp3 belonging to a second subgroup (Fig. 1). The largest difference between these subgroups is a deletion of 14 residues in Hcp1 (region 14–27 between β1 and α1), an insertion of four residues between β5 and β6, and 2–3 additional residues on the C-terminus. There are very few residues that are conserved among Hcp proteins; these include three Gly (28, 57, 88), His35, Thr71, Tyr148 and Trp166 (Fig. 1). With the exception of Thr71, which lines up the inner surface of the channel, all other conserved residues are on the outside surface of the protein or are not solvent accessible. There are some residues conserved only in either Hcp1 (for example A2, D4, D32, G127) or in Hcp2/3 (for example T3, P4, P54, W98, C130, W153) subgroups, but there is no correlation between these conserved residues and a mode of inter-ring interaction. It is also unlikely the mixed Hsp1/Hcp2/Hcp3 nanotubes can form. The sequence diversity among different Hcp proteins suggests that the secretion apparatus may have some specificity and that Hcp proteins may be optimized to perform a particular secretion function. In P. aeruginosa there are three HSI loci each encoding Hcp proteins as well as other components of the T6SS apparatus. Each of these loci may be responsible for secretion of different proteins and function in response to different signals. The exact role of each component of the T6SS apparatus, the secretion specificity and the identification of proteins that are being secreted by each loci await further studies.
Acknowledgments
We thank all members of the Structural Biology Center at Argonne National Laboratory and at the Ontario Centre for Structural Proteomics for their help in conducting these experiments. This work was supported by National Institutes of Health grant GM074942 and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357 and by the US National Science Foundation grant MCB 0231319 (to R.H.W.), and by Genome Canada (through the Ontario Genomics Institute).
Abbreviations
- T6SS
Type VI secretion system
- Hcp1
Hemolysin-corregulated protein
- PDB
Protein Data Bank
- SeMet
Seleno-methionine
- SAD
Single-wavelength anomalous diffraction
- RMSD
Root mean square deviation
Footnotes
The submitted article has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Contributor Information
Jerzy Osipiuk, Argonne National Laboratory, Biosciences Division, Midwest Center for Structural Genomics and Structural Biology Center, 9700 S. Cass Ave., Argonne, IL 60439, USA.
Xiaohui Xu, University of Toronto, Structural Genomics Consortium, Toronto, Canada.
Hong Cui, University of Toronto, Structural Genomics Consortium, Toronto, Canada.
Alexei Savchenko, University of Toronto, Structural Genomics Consortium, Toronto, Canada.
Aled Edwards, University of Toronto, Structural Genomics Consortium, Toronto, Canada; Clinical Genomics Centre/Proteomics, University Health Network, 200 Elizabeth St., Toronto, ON M5G 1L7, Canada.
Andrzej Joachimiak, Argonne National Laboratory, Biosciences Division, Midwest Center for Structural Genomics and Structural Biology Center, 9700 S. Cass Ave., Argonne, IL 60439, USA; Department of Biochemistry and Molecular Biology, University of Chicago, 920 E. 58th St., Chicago, IL 60637, USA.
References
- 1.Desvaux M, et al. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 2009;17(4):139–145. doi: 10.1016/j.tim.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Goodman AL, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7(5):745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Ventre I, et al. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA. 2006;103(1):171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mougous JD, et al. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9(7):797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 5.Boyer F, et al. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312(5779):1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009;12(1):11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Schell MA, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64(6):1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 10.Pell LG, et al. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106(11):4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballister ER, et al. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci USA. 2008;105(10):3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filloux A. The type VI secretion system: a tubular story. EMBO J. 2009;28(4):309–310. doi: 10.1038/emboj.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korolev S, et al. Autotracing of Escherichia coli acetate CoA-transferase alpha-subunit structure using 3.4 A MAD and 1.9 A native data. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 12):2116–2121. doi: 10.1107/s0907444902017055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minor W, et al. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 16.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 17.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 20.Hooft RW, et al. Errors in protein structures. Nature. 1996;381(6580):272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 21.Davis IW, et al. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jobichen C, et al. Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS One. 2010;5(9):e12910. doi: 10.1371/journal.pone.0012910. [DOI] [PMC free article] [PubMed] [Google Scholar]