Abstract
The main pathway of bacterial sugar phosphorylation utilizes specific phosphoenolpyruvate phosphotransferase system (PTS) enzymes. In addition to the classic PTS system, a PTS-independent secondary system has been described in which nucleotide-dependent sugar kinases are used for monosaccharide phosphorylation. Fructokinase (FK) that phosphorylates d-fructose with ATP as a cofactor has been shown to be a member of this secondary system. Bioinformatics analysis has shown that FK is a member of the “ROK” (bacterial Repressors, uncharacterized Open reading frames, and sugar Kinases) sequence family. In this study, we report the crystal structures of ROK FK from Bacillus subtilis (YdhR) (a) apo and in the presence of (b) ADP and (c) ADP/dfructose. All structures show that YdhR is a homo-dimer with a monomer composed of two similar α/βdomains forming a large cleft between domains that bind ADP and d-fructose. Enzymatic activity assays support YdhR function as an ATP-dependent fructose kinase.
Keywords: Fructokinase, ROK family, metal dependent, ADP and d-fructose binding, reductive methylation
INTRODUCTION
Phosphorylation of monosaccharides is a critical step in carbohydrate metabolism. This fundamental reaction traps sugars inside the cell and targets them for further utilization by specific metabolic pathways. The main pathway of sugar phosphorylation in bacteria utilizes phosphoenolpyruvate phosphotransferase system (PTS) enzymes. In general, the bacterial PTS system has two cytoplasmic proteins: a phosphocarrier-protein and phosphotransferase, and a membrane-bound sugar specific transporter (1–3). The transporter can be part of a large multi-domain protein system or could function as a specific permease. In addition to the PTS system, a PTS-independent secondary phosphotransferase system has been described for a number of bacteria (4). These systems utilize nucleotide-dependent sugar kinases capable of phosphorylating monosaccharides. Many bacterial genomes contain genes for hexokinases, glucokinases, or fructokinases capable of phosphorylating a wide variety of sugar substrates. Hexokinases are known to catalyze the phosphorylation of a broader range of monosaccharides such as glucose, mannose, and fructose, while they are present in high concentrations (5). In contrast, glucokinases and fructokinases (FKs) are much more selective with a relatively high affinity for glucose and fructose, respectively.
Bioinformatic and structural studies of 60 different sugar kinases have revealed three distinct families of kinases: the galactokinase, hexokinase and ribokinase families (4). In addition, Titgemayer and co-workers have noticed that some sugar kinases share a close amino acid sequence relationship to proteins with divergent (or unknown) functions and proposed that they belong to a novel family of proteins called the “ROK family” (6). Based on the amino acid sequence similarity, several sugar kinases, transcriptional repressors, and a large number of uncharacterized proteins, have been grouped into a ROK (bacterial Repressors, uncharacterized Open reading frames, and sugar Kinases) family of enzymes. Currently, there are nearly 5,000 proteins annotated (Pfam PF00480) (7) as members of the ROK family. The members of this family are broadly distributed in nature and are found in bacteria, archaea, eukaryota, plants, and in humans. Also, many ROK sugar kinases phosphorylate a wide range of carbohydrates substrates showing high functional diversity, within the family (6, 8).
To date, only few members of this protein family have been structurally characterized, hence structural coverage of this large and important family of proteins is very limited. The well known members of this family are the xylose operon repressor (XylR)(9, 10), N-acetyl-glucosamine repressor (NagC), glucokinase (EC:2.7.1.2)(11), allokinase (EC:2.7.1.55)(12), ATP-glucomannokinase (13), and the Mlc repressor (14). In order to better structurally characterize this large, important and functionally diverse ROK family of proteins and to understand structure/function relationships the Midwest Center for Structural Genomics (MCSG), a component of the Protein Structure Initiative (PSI), initiated the structural studies of several representatives of this family. Here we present the first crystal structure of ROK fructokinase (YdhR) from B. subtilis. We have determined the 2.10 Å apo structure (YdhR-apo) and 1.66 Å structure of the enzyme with ADP bound (YdhR-ADP) and a 2.45 Å structure with ADP and d-fructose (YdhR-ADP-Fru) molecules bound. The YdhR monomer is composed of two similar α/βdomains and assembles into a homodimer that is observed in solution and in the crystal. Our structures revealed that the YdhR-fold closely resembles that of the actin-like ATPase domain superfamily (CATH 3.30.420.160). The active site is located in a large cleft between domains of the monomer with ADP and d-fructose bound in the middle of the cleft. Fructokinase activity was confirmed by enzymatic assay. Also, point mutation of glycine 59 to alanine showed that this residue plays a critical role in specificity of YdhR to d-fructose. A single zinc ion was found bound to the protein near the active site, where it orients active site His153.
RESULTS AND DISSCUSION
Enzymatic activity
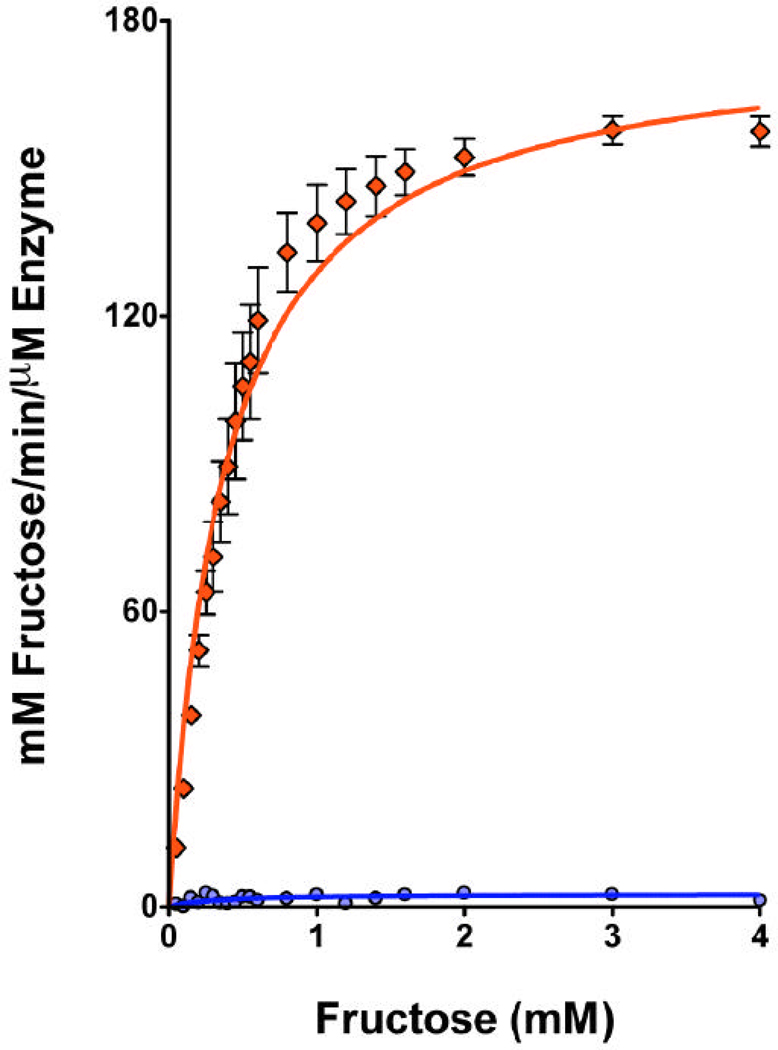
The B. subtilis ROK FK protein (YdhR, Swiss-Prot entry O05510) was predicted to have putative fructokinase function based on genome sequence analysis (15, 16). Searches of the Kyoto Encyclopedia of Genes and Genomes (KeGG) database indicated that YdhR might be involved in fructose and mannose metabolism as well as starch and sucrose metabolism (Pathways bsu00051, bsu00500) (17). Our initial YdhR structural analysis revealed a phosphotransferase-like protein. In order to confirm whether this enzyme is a true fructokinase, the in vitro enzymatic activity of recombinant YdhR was tested. Under standard reaction conditions (see methods), YdhR was shown to catalyze the ATP-dependent phosphorylation of d-fructose to fructose-6-phosphate with the apparent Km and Vmax values of 0.38 mM and 178 U/mg of protein, respectively (Figure 1). Therefore, we conclude that the B. subtilis YdhR is a fructokinase.
Figure 1.
Fructokinase activity of the recombinant B. subtilis YdhR protein expressed in E. coli (red) and G59A mutant (blue) purified and assayed as described in Material and Methods.
Reductive methylation of YdhR improved crystal quality
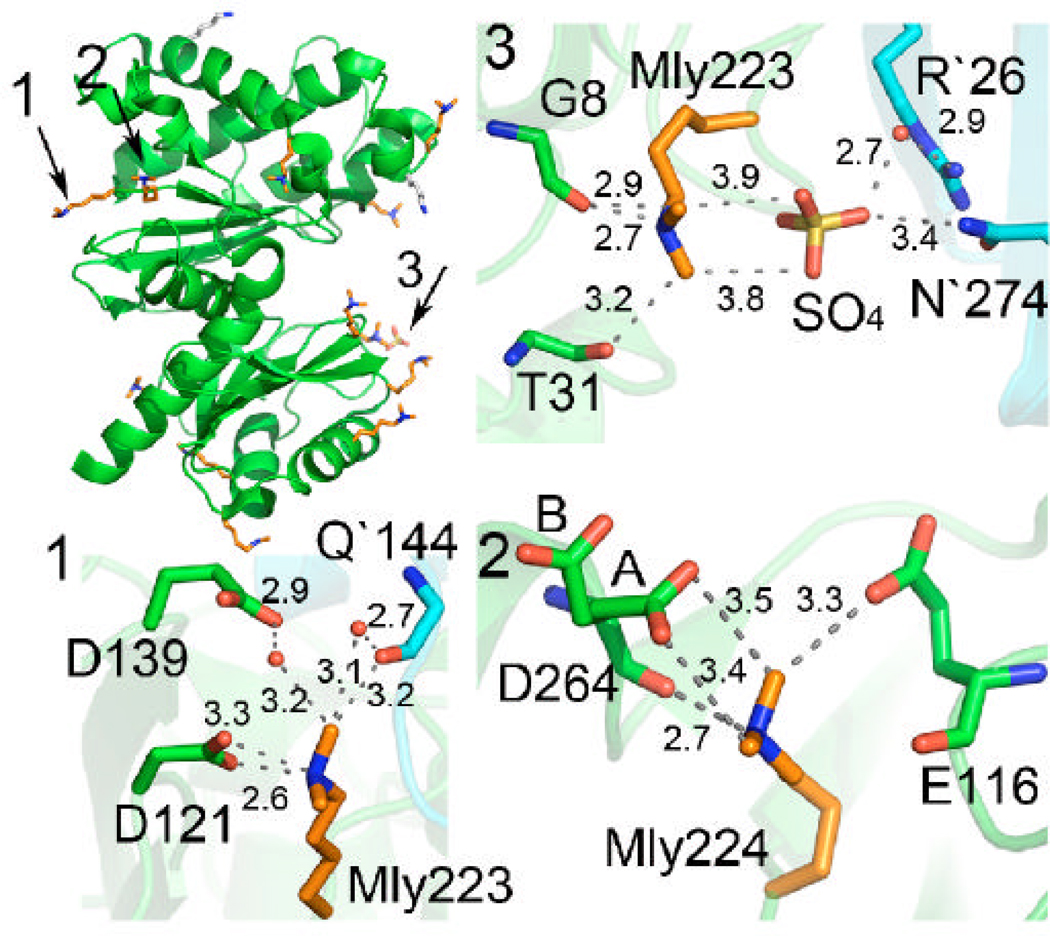
Native YdhR failed to produce crystals suitable for structure solution; therefore, chemical modification of the protein was used. Sequence analysis revealed that YdhR is a good candidate for reductive methylation, based on the number of lysines in the monomer (15 lysines). The protein was methylated according to the protocol described in the methods section (18). Methylated protein was screened for crystallization using the same set of conditions and protocols as for native protein. Diffraction quality crystals were obtained under several conditions and the best crystals were used for data collection (see Materials and Methods). Analysis of the 1.66 Å structure reveals that 12 out of the 15 lysines were doubly methylated and one was singly methylated. Lower resolution structures did not reveal any features in the density maps corresponding to methylated lysine residues, although successful methylation was confirmed by mass spectrometry (17). The methylated lysines are shown to form numerous solvent interactions as well as a few intra- and inter-molecular contacts with the side chains of Asp and Glu residues and the main chain carbonyls. Examples of interactions are shown in Figure 2. The contacts provided by methylated lysine residues appear to be important in obtaining well-diffracting crystals of YdhR.
Figure 2.
Overall fold of YdhR fructokinase (green) with all lysine residues displayed as sticks (methylated lysines are colored orange, unmethylated are shown in grey). Ordered double methylated lysine residues are involved in inter- and intra-molecular interactions with side chains, main chain carboxyls, solvent, and crystallization ions. Close-up views of three examples of interactions are shown on the side (distances are shown in Å).
The YdhR monomer structure
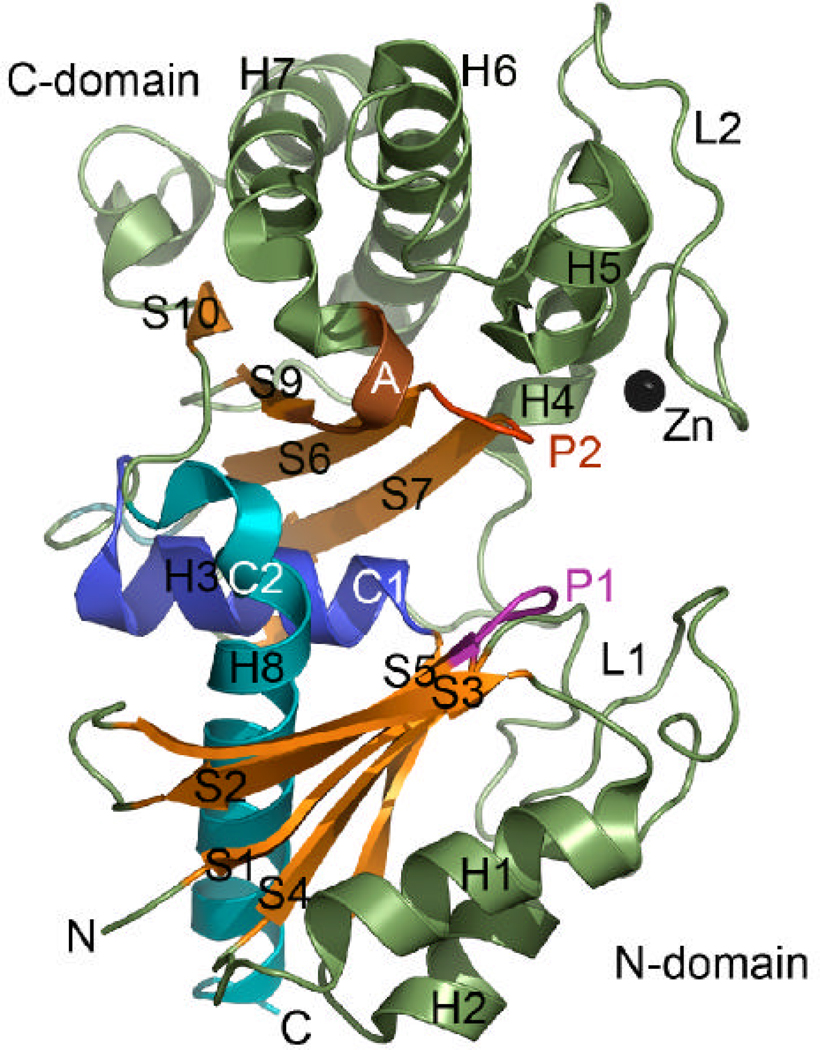
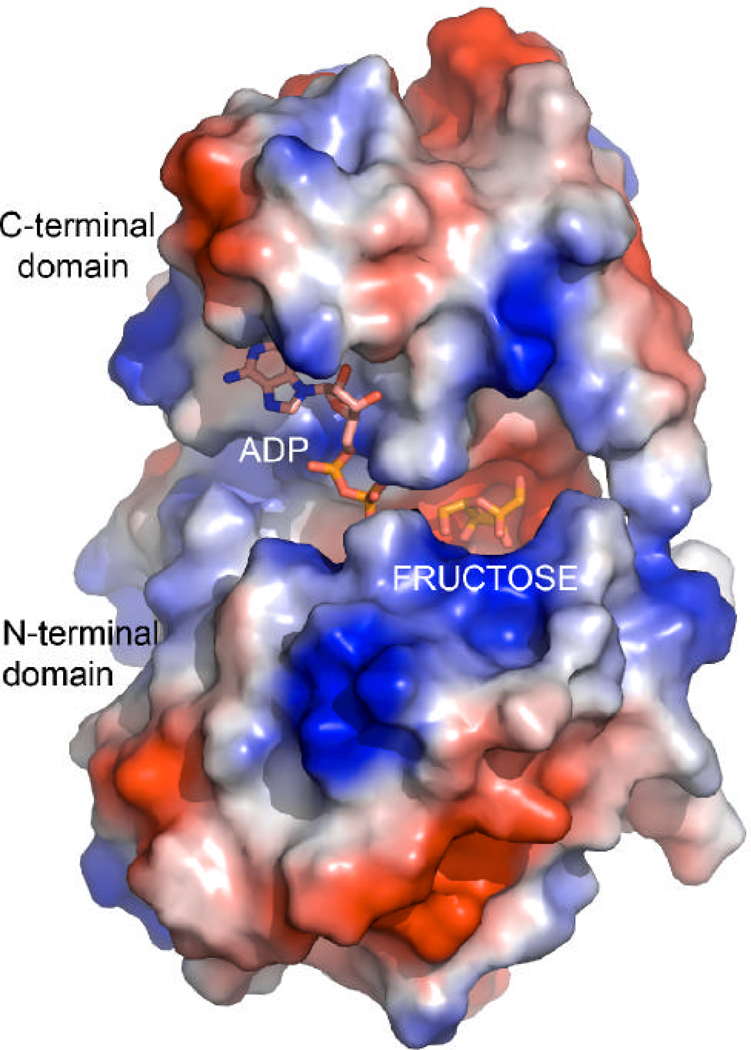
The YdhR crystal structure was determined using the single-wavelength anomalous diffraction (SAD) approach using seleno-methionine (Se-Met) labeled protein. The structure of YdhR described below is based on the high-resolution (1.66 Å) model of YdhR in a complex with ADP. The final model exhibited good crystallographic and geometric statistics and was refined to an Rwork/Rfree of 0.165/0.186 (Table 1). The YdhR monomer is composed of two α/β domains: an N-terminal domain made of two fragments (residues 1–112 and 274–294) and a slightly smaller C-terminal domain (residues 113–273) (Figure 3A). A total of 10 β-strands, 8 α-helices, and 5 310-helices make up the bean-shaped monomer. The N-terminal domain (the lower part of the bean) features a five-stranded β-sheet (S1↑, S2↓, S3↑, S4↑, S5↑) that is sandwiched by two pairs of α-helices, H1 and H2 at the bottom, connected to the β-sheet via an extended 30-residue (loop1, L1) region, and H3 and H8 at the top. The latter two helices are positioned in the center of the monomer and form a bridge between two β-sheets. The C-terminal domain (the upper part of the bean) is divided into two layers. The bottom layer is formed by the five stranded β-sheet (S8↑, S7↓, S6↑, S9↑, S10↑), whereas the top layer is created by a cluster of helices, with helices H6 and H7 forming the core of the cluster. The helical cluster is connected to the β-sheet via a 32-residue loop region (loop 2, L2). In the center of the molecule, there is a large groove (17Å ×18Å ×20Å) (center of the bean (Figure 3A, 3B and 4)), which houses the active site cleft, where ADP, d-fructose and Zn2+ ion bind (see below). The presence of zinc was confirmed by X-ray fluorescence emission measurements of the crystal.
TABLE 1.
| Data collection statistics | YdhR-apo | YdhR+ADP | YdhR+ADP +d-fructose |
|---|---|---|---|
| Space group | P3121 | P3121 | P3121 |
| Unit cell (Å) | a = 112.66 b = 112.66 c = 75.09 |
a = 112.15 b = 112.15 c = 73.72 |
a = 112.33 b = 112.33 c = 73.71 |
| Wavelength (Å) | 0.9793 | 0.9794 | 0.9794 |
| Resolution (Å) | 40-2.10 | 40-1.66 | 40-2.45 |
| Number of observed reflections | 401122 | 381530 | 213353 |
| Number of unique reflections | 31498 | 58984 | 20173 |
| Rmerge (%)a | 6.7(52.0)b | 11.1(65.0)b | 10.1 (66.7)b |
| Completeness (%) | 97.0(98.7) | 98.9(83.2) | 100 (100)b |
| I/σ I | 37.7(4.7)b | 18.6(2.3)b | 22.3 (4.1)b |
| Phasing method | SAD | SAD | MR |
| Phasing resolution range (Å) | 40-2.8 | 40-2.50 | 40-3.0 |
| Number of Se-Met/Pt-ions | -/2 | 6/0 | - |
| Refinement resolution range (Å) | 40-2.1 | 40-1.66 | 40.0-2.45 |
| Rcryst (%) | 18.1 | 16.5 | 15.9 |
| Rfree (%) | 20.3 | 18.6 | 19.6 |
| Number of protein residues | 293 | 293 | 293 |
| ADP/Zn2+/d-fructose/SO42− molecules/glycerol | 0/1/0/-/2 | 1/1/0/1/0 | 1/1/1/1/0 |
| Solvent molecules | 220 | 422 | 82 |
| Bond lengths (Å) | 0.010 | 0.022 | 0.023 |
| Bond angles (deg) | 1.17 | 1.90 | 1.88 |
| B-factors (Å2) | |||
| Protein main chain | 33.0 | 20.0 | 43.7 |
| Protein side chain | 36.1 | 27.3 | 45.9 |
| Zn2+ | 27.5 | 18.5 | 35.8 |
| ADP/FRU | -/- | 26.6/- | 72.2/51.1 |
| Ramachandran Plot (%)d | |||
| Favored | 91.8 | 92.5 | 96.1 |
| Generously allowed | 8.2 | 7.5 | 3.9 |
| Disallowed | 0 | 0 | 0 |
| PDB ID | 1XC3 | 3OHR | 3LM9 |
Rmerge =∑hkl∑i|Ii−<I>|/∑hkl∑i|<I>|, where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
Numbers in parentheses are values for the highest-resolution bin.
Calculated with TLS operators
As defined by MOLPROBITY.
Figure 3.
Crystal structure of YdhR.
A) A ribbon representation of the YdhR structure (β-strands are depicted in orange, helices in green, zinc ion in black). Secondary structure elements are indicated with H (helices) and S (strands). The YdhR fold shows homology to that of actin/hexokinase/HSP70 ATPase domains, with the common structural segments labeled and mapped on the three-dimensional structure of YdhR (P1, phosphate 1 region in magenta; P2, phosphate 2 region in red; A, adenosine region in brown; C1, connect 1 helix in blue and C2, connect 2 helix in cyan).
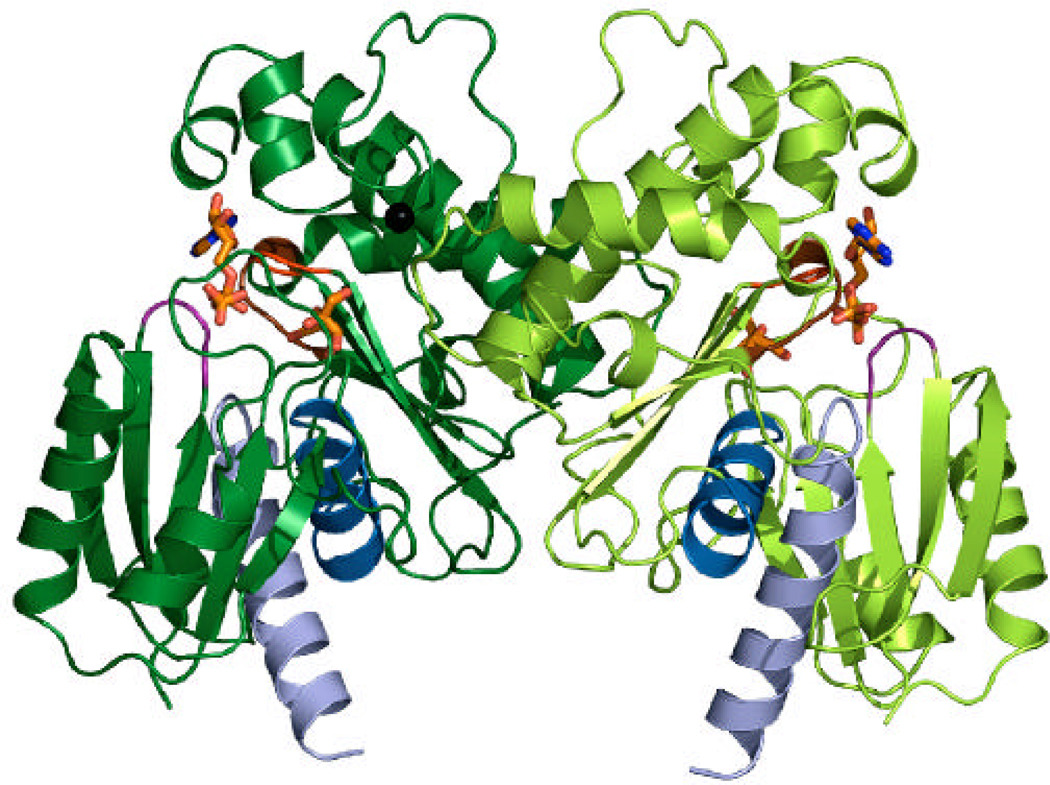
B) A ribbon representation of the YdhR dimer with bound ADP and fructose shown as sticks in orange, blue, and red. Zinc ions are shown as black spheres.
Figure 4.
A surface rendering mapping the local electrostatic potential of the YdhR-ADPFru monomer, showing a deep pocket between domains for binding ADP and d-fructose. Electrostatic figure was calculated using Pymol. Blue and red represent the positive and negative charge potential at the + and − 70?kTe−1 scale, respectively.
The YdhR forms a dimer
The crystals contain one monomer of YdhR in the asymmetric unit. Analysis of the crystal packing revealed that the monomer extensively interacts with a crystallographic two-fold axis related mate and is likely to form a functional dimer (Figure 4). Further analysis showed that the dimeric state is well supported by the strong interaction observed between the helical and loop regions of the monomer C-terminal domains. The YdhR dimer interface is broad and it buries a large solvent accessible surface area of 2166.4 Å2 per monomer. The helical interface is arranged in such a manner that the long helices of one subunit of the dimer complement the shorter helices of the opposite monomer and conversely. This arrangement leads to the formation of a stable V-shaped helical interior which is stabilized by 26 direct hydrogen bonds, several solvent-mediated hydrogen bonds, and four inter-subunit salt bridges formed between R141(A)/E139(B), R157(A)/D173(B), and their corresponding symmetry mates. Noteworthy, is the interaction of the loop located between helices H7 and G3 of the one subunit (residues 249–255), with the highly conserved residues of the N- (residues 58–60) and the C-terminal (residues 168–172) loops of the opposite subunit. Close interaction of these loops leads to the enclosure of one side of the central groove of each dimer subunit and to the formation of the well-defined substrate pockets (Figure 4). Interactions involving swapping side chains between monomers are quite extensive and include the side chains of residue Y252 that cross over to the other subunit pocket. The YdhR dimeric state in solution has been confirmed by size exclusion chromatography experiments (data not shown), and is consistent with the data reported earlier (14) for fructokinases from other organisms (19, 20).
Structural homologs of B. subtilis YdhR
Structural comparison searches revealed that YdhR is a member of the actin-like ATPase domain superfamily (CATH 3.30.420.160). Members of this functionally diverse protein superfamily share common structural motifs characterized by five conserved protein segments termed phosphate1 (P1), phosphate2 (P2), adenosine (A), connect1 (C1), and connect2 (C2) (Figure 3A) (21). The first three segments indicate regions that are involved in the nucleotide binding, whereas the last two segments describe helices connecting domains and leading to the formation of the inter-domain hinge. The sequence analysis of YdhR reveals the presence of two phosphate-binding loops with a conserved GT sequence [G9, T10, (P1); G129, T130, (P2)]. The presence of the two conserved loops with a GT sequence motif further classifies this protein into the subclass of prokaryotic and eukaryotic sugar kinases within the ATPase domain superfamily. Structural comparison of YdhR with structures deposited in the Protein Data Bank (PDB) using the DALI program (22) show that the closest structural relatives are glucokinase (PDB code 2QM1, Z-score 29.5, RMSD 2.3 Å, sequence identity 26%), E. coli N-acetylmannosamine kinase (PDB code 2AA4, Z-score 29.0, RMSD 2.5 Å, sequence identity 25%), T. maritima N-acetylglucosamine kinase, (PDB code 2HOE, Zscore 28.2, RMSD 2.5 Å, sequence identity 21%), Arthrobacter sp. inorganic polyphosphate/ATP-glucomannokinase (PDB code 1WOQ, Z-score 25, RMSD 2.5 Å, sequence identity 25%) (13), and Trypanosoma cruzi ATP-dependent glucokinase (PDB code 2Q2R, Z-score 21.0, RMSD 3.1 Å, sequence identity 16%) (23). Other significant homologs include glucose and hexokinases (PDB IDs 1SZ2, 1Q18, 3B8A, 1BG3, 3IMX which show Z-scores in the range of 19–24 and RMSD at around 3Å with sequence identity ranging from 14 to 20 %) (24–27).
YdhR-FK ROK family members
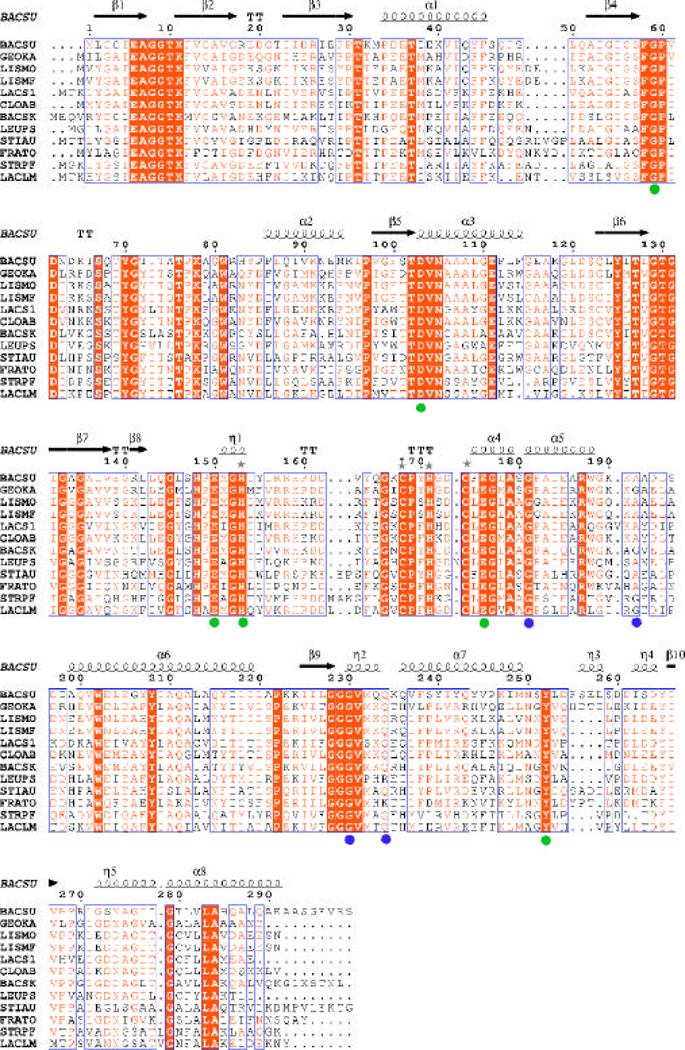
The common sequence signature of the ROK family proteins comprises of ~180 residues often distributed between two domains with the majority of residues grouped into a highly conserved core domain. ROK family members could be recognized by the presence of the sequence motif [LIVM]-X2-G-[LIVMFCT]-G-X-[GA]-[LIVMFA]-X3-{V}-X4-G-X(3,5)-[GATP]-{G}-x-G-[RKH]. Another ROK family motif includes the presence of the zinc binding motif: motif 1 (GHX(9–11)CXCGX2G(C/H)XE) or motif 2 (GHX(11–17)CX2HX2CXE), where X represents any residue (14, 28, 29). In YdhR, these sequence fingerprints are located in the middle of the protein sequence (residues 128–177). The N-terminal part of the fingerprint sequence (residues 129–135) represents a glycine-rich loop involved in the interaction of the polyphosphate moiety of the nucleotide (see ADP-binding) (Figure 5). The central part specifies the loop region interacting with the sugar-substrate (residues 146–153). The conserved cysteine motif 2 is situated between the residues 154–177 and binds zinc ion (Figure 5). The ROK family is an example of a fold that has been utilized to serve several different functions.
Figure 5.
A multiple sequence alignment of YdhR fructokinase homologs. Sequence identities are highlighted in red and similarities are shown as red letters. The corresponding secondary structures of YdhR from B. subtilis are shown on the top (black), whereas on the bottom, green and blue dots represent residues interacting with d-fructose, and ADP, respectively. Grey stars highlight residues coordinating the zinc ion. Helices (H, a-helix; G, 310 helix) appear as small squiggles, beta strands (S, β-strand) as arrows. The following abbreviations were used with Unit-Prot accession numbers indicated in parentheses: BACSU, B. subtilis (o05510); GEOKA, Geobacillus kaustophilus (Q5KUZ7); LISMO, Listeria monocytogenes str. 4b H7858 (Q4EFF6); LISMF, Listeria monocytogenes serotype 4b (strain F2365) (Q722A5); LACS1, Lactobacillus salivarius (Q1WS64); CLOAB, Clostridium acetobutylicum (Q97IW7); BACSK, Bacillus clausii (Q5WD01);LEUPS, Leuconostoc pseudomesenteroides (Q8KMM1); STIAU, Stigmatella aurantiaca DW4/3-1 (Q096C7); FRATO, Francisella tularensis (Q0BKX6); STRPF, Streptococcus pyogenes (Q1J538); LACLM, Lactococcus lactis (A2RJZ3)(66). Figure prepared with ESPript/ENDscript webserver (66).
Zinc binding
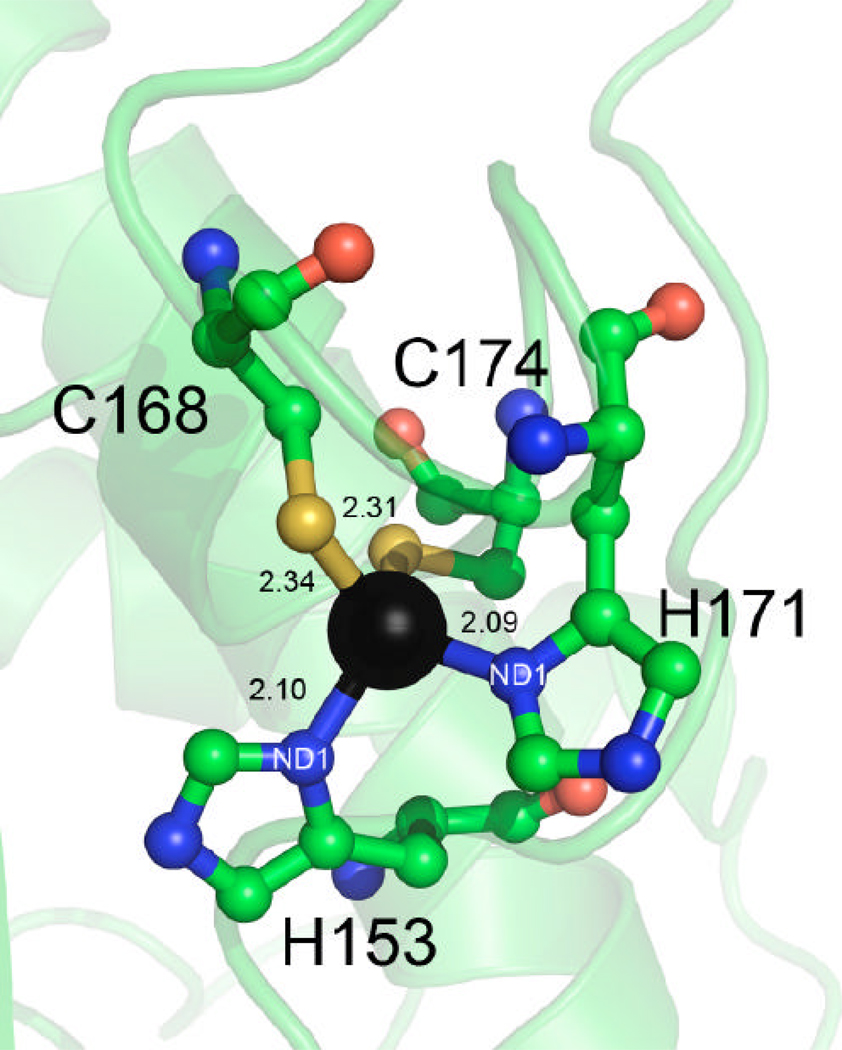
Zinc ions are present in all YdhR structures, which we have determined. In each structure, a single zinc ion is positioned at the loop region (L2) of the C-terminal domain (motif 2). It is situated in close proximity to the large inter-subunit cleft, in close distance to the d-fructose binding pocket. The zinc ion adopts a distorted tetrahedral geometry and is coordinated by a pair of conserved cysteine and a pair of histidine residues (Figures 5 and 6A). Based on the high resolution 1.66 Å YdhR-ADP structure, the Zn-S distances are 2.34 Å for C168 and 2.31 Å for C174, whereas Zn-Nδ distances are shorter, 2.09 Å and 2.10 Å for H153 and His171, respectively. Analysis of the YdhR-ADP-Fru structure indicates that His153 (N∍ of the side chain imidazole) further stabilizes the d-fructose by interaction with the C1 hydroxyl group (Figure 7). Therefore it appears that the zinc ion imposes a proper orientation of the H153 side chain in order to position the substrate in the active center. Nearby, E150, which is centrally located at the long loop, also interacts with the d-fructose (see fructose binding), underscoring the importance of the zinc-binding region. The functional importance of the cysteine residues involved in zinc binding was confirmed by the mutagenesis studies of the corresponding cysteine residues in other enzymes. It was shown that replacement of equivalent cysteines to alanines in B. subtilis glucokinase and the ROK Mlc regulator in E. coli, resulted in an inactive glucokinase enzyme and dramatically impaired Mlc repressor function (14, 30).
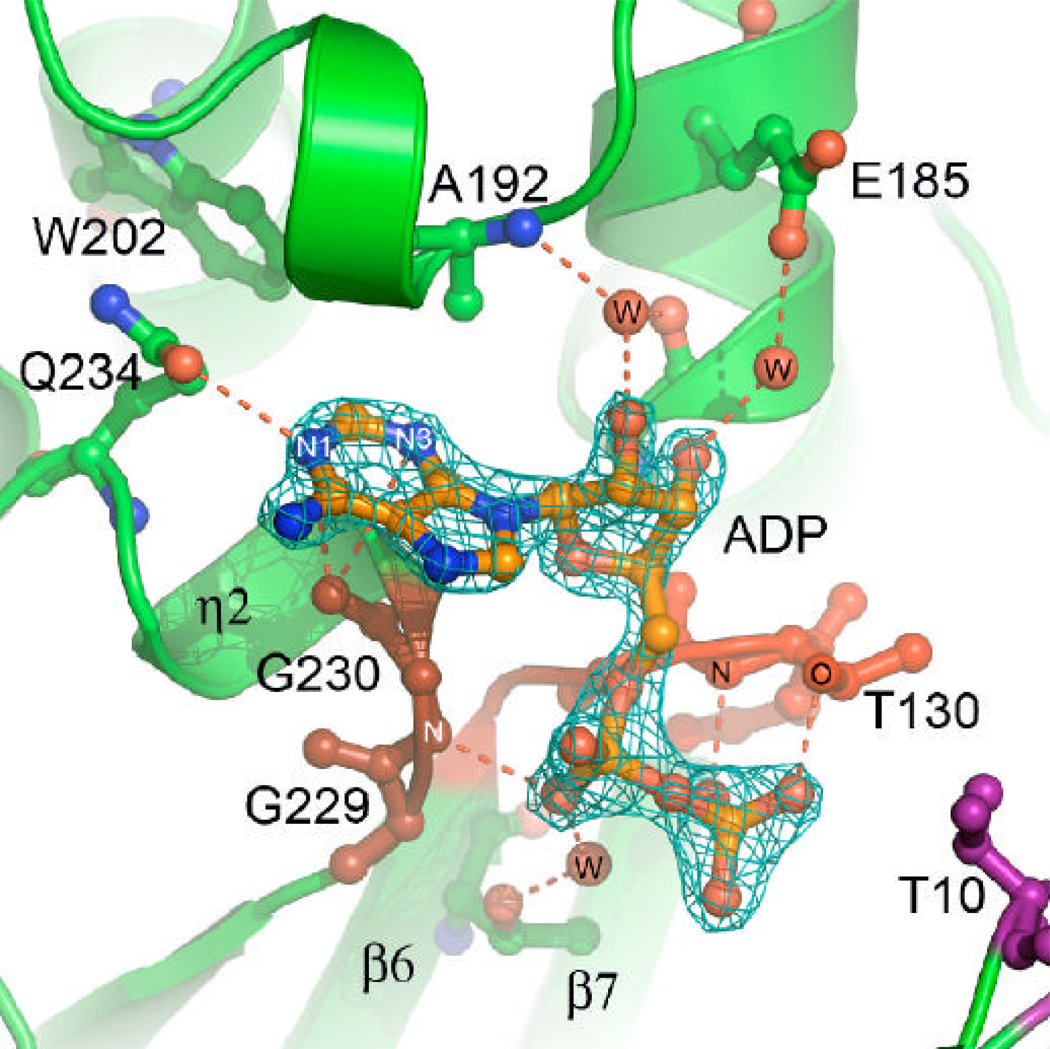
Figure 6.
ADP and zinc binding.
A) A close-up view of the Zn+2 ion coordination environment (distances are shown in Å).
B) 2Fo–Fc electron density map contoured at 3s around the molecule of ADP. Critical direct and water-mediated protein-ADP interactions are represented by the dashed lines.
Figure 7.
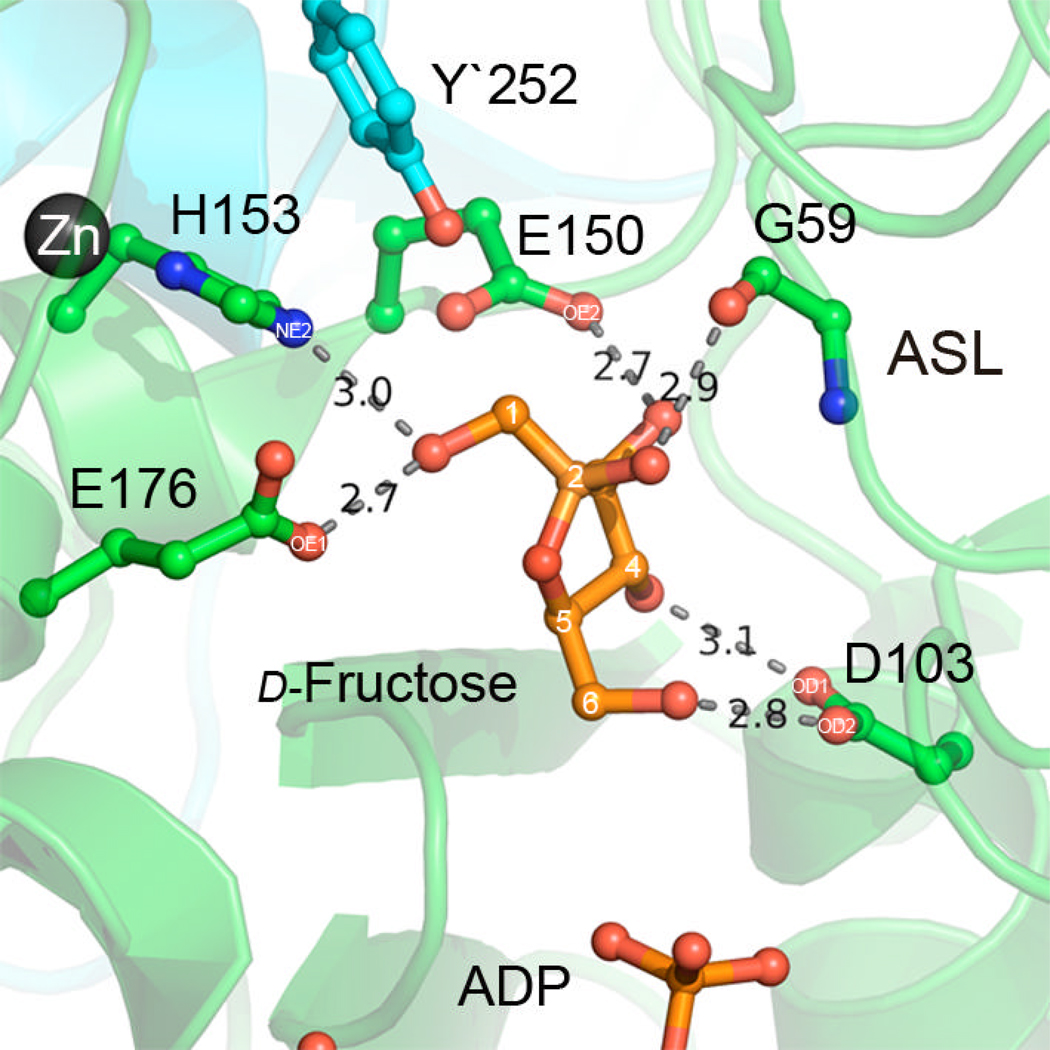
A close-up view of the d-fructose binding pocket. The interacting residues are shown as ball-and-stick. The hydrogen bonds are shown as red dashed lines.
ADP binding
The structure of YdhR in complex with ADP was determined and refined at 1.66 Å resolution. Close inspection of the Fo – Fc and 2Fo-Fc difference maps revealed excellent electron density for the ligand, permitting unambiguous placement of the ADP molecule (Figure 5B). The nucleotide is bound to the protein in an extended conformation at the C-terminal part of the groove (Figure 6B). The adenine ring of ADP binds in a small sub-cavity made by the loop connecting helices H5 and H6 (residues 189–198) on one side, and a highly conserved hydrophobic loop (residues 228–232) connecting β-strand S9 and helix G1 on the other side. There are a few hydrogen bond interactions, including the adenine N1 atom and side chain atom OE1 of the semi-conserved Q234 and weaker hydrogen bond (~3.4 Å) interactions between the carbonyl oxygen of G230 and N1 or N3 of the adenine ring.
The sugar moiety of the ADP, which displays C2'-endo geometry, is anchored by a limited number of water mediated interactions. The 2`-hydroxyl of the ribose ring of the ADP is involved in an interaction with a well-ordered water molecule stabilized by the interaction between an adjacent backbone amide group of A192 and peptide bond carbonyl of the conserved G181. The 3`-hydroxyl interacts with a water molecule in close proximity, which in turn interacts with the OE1 atom of the E185. The alpha and beta phosphate groups of the ADP cofactor bind on the edge of the C-terminal β-sheet at the loop region connecting S9 and G2 and the loop region connecting S6 and S7, respectively (Figures 3A and B, 4, 6B).
In contrast to the adenyl and ribose moieties, the pyrophosphate is well exposed and involved in extensive solvent interactions. There are also a few important direct interactions. The alpha phosphate is stabilized by the direct hydrogen bond interaction between O2 and G230. The beta phosphate is stabilized by the interactions of O2 and the hydroxyl group of T130 (phosphate 2 motif) as well as the O3 and backbone N of T130 (Figure 6B).
Fructose binding
YdhR was also co-crystallized with ADP and d-fructose to reveal how products/substrates are bound to the enzyme active site simultaneously. The structure of the YdhR-ADP-Fru complex was determined by MR and refined to a resolution of 2.45 Å and an R factor of 0.159 (Rfree = 0.196). Close examination of the Fo- Fc and 2Fo - Fc difference electron density maps in the cleft region revealed the unambiguous electron density consistent with ADP and d-fructose molecules. The d-fructose molecule binds in the smaller sub-cavity in the bottom of the cleft area between the edge of the N- and C-terminal β-sheets and helix H3. In YdhR-apo structure, two glycerol molecules were observed to bind there. The sub-cavity is mainly negatively charged and is gated from one side by the bound nucleotide and two structurally conserved loops (the loop connecting β-strands S1, S2 (P1), S6 and S7(P2)).
The fructose molecule binds in a chair conformation with the anomeric carbon in the β-position. The substrate is bound within the active center via an extensive network of interactions with highly conserved residues (Figure 7). The hydroxyl group at the C1 position of d-fructose forms hydrogen bonds with E176 OE1 (2.68 Å), H153 NE2 (3.03 Å). The orientation of the H153 side chain is stabilized by the zinc ion, underlying the importance of the metal ion. The sugar is further coordinated by the interaction of the C3 hydroxyl group with E150 NE2 (2.69 Å). The N-terminal domain stabilizes the sugar with two interactions: the peptide bond carbonyl of G59 and the hydroxyl group of C2 of d-fructose (2.86 Å); and the bidentate interaction of D103 with the hydroxyl groups of the sugar (OD2-FO6H, 2.86 Å and OD1- FO4H, 3.10 Å)
Comparison of YdhR-apo, YdhR-ADP and YdhR-ADP+d-Fructose structures
Superimposition of all three structures shows that they are essentially identical. Structure of YdhR-apo superimposes over YdhR-ADP-Fru with r.m.s.d of 0.27 Å for 294 C-α atoms, while superimposition of YdhR-apo over YdhR-ADP results in r.m.s.d of 0.25 Å for 294 C-α atoms as calculated using SSM server (31). It suggests that the enzyme adopts the active conformation and does not require any large conformational changes. A comparison of the apo-structure with YdhR-ADP shows only minor movements that could be contributed to the binding of the ligands. One of the notable changes is movement of Ala192 in the nucleotide-binding pocket. The alanine residue moves ~0.6 Å in order to interact via water with the ADP molecule. Another noteworthy change is the ~0.5 Å movement of G59 upon binding of the fructose.
Structural insights into d-fructose specificity of YdhR
In an attempt to gain structural insights into the specificity of YdhR ROK fructokinase towards d-fructose (no activity towards glucose is observed, see Material and Methods), we compared the architecture of the sugar binding pocket of YdhR fructokinase with close structural homolog, ROK ATP-glucomannokinase (GMK) (13, 32). GMK, which specifically binds glucose, is the only other structure of the ROK family determined in the presence of a substrate.
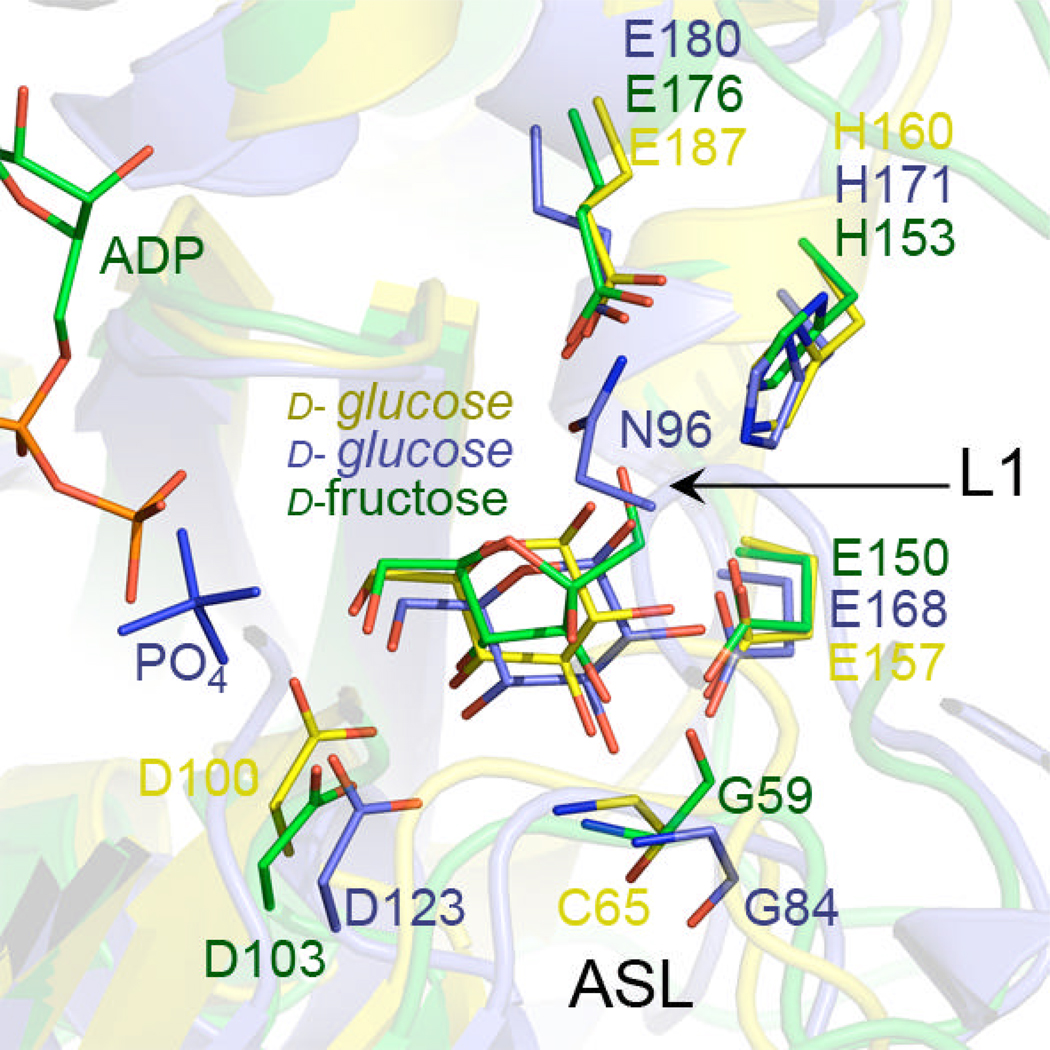
Comparison of the active site pockets of YdhR vs. GMK shows that the majority of residues involved in substrate binding are structurally conserved between the enzymes (D103, E150, E153, E176 numbering according to YdhR). In YdhR, D103 corresponds to D123 in GMK, E176 corresponds to E180 in GMK, H153 corresponds to H171 in GMK, and E150 corresponds to E168 in GMK (Figures 7 and 8). The side chains of these residues assume similar conformations in these enzymes when respective carbohydrate substrates are present. Also, both fructose and glucose substrates present in cyclic form bind in relatively similar positions.
Figure 8.
Superimposition of the active site pockets of YdhR fructokinase (green) and GMK (1woq, blue) and non-ROK glucokinase (1SZ2, yellow) shows similar arrangement of the conserved active site residues. The most notable difference are visible at the active site loop (ASL) at the position of Gly59 and corresponding Gly 84 (GMK, blue) and Cys65(non-ROK glucokinase, for clarity only main chain is shown). Superimposition was done using SSM server and Pymol.
Since fructose is an isomer of glucose, the mechanism by which YdhR distinguishes and selects fructose presents an important question. An overlay of the enzymes reveals that one of the main differences between YdhR and GMK structures is observed in the positioning of the N-domain loop overlaying active site (L1) (Figure 8). In GMK, the loop equivalent to YdhR L1 overhangs over the substrate pocket and encloses the active site. Also, the middle of the loop contains the N96 residue, which is involved in binding and stabilization of d-glucose. In contrast, YdhR L1 does not interact with the substrate, and the configuration seems to be more open, making the active site more exposed. The other differences between the enzymes could be observed at the so-called “active site part” of loop L1 (8) (Figure 8). Although the active site loops assume similar positions in the enzymes, the main difference is observed in the location of the main chain carbonyl group of YdhR G59. This residue, conserved among many ROK members, forms the cis peptide bond with P60 in all three structures of YdhR that we determined. The position of the G59 carbonyl oxygen allows interaction with the C2 hydroxide of d-fructose. In the GMK structure, G84 is in the regular trans conformation and is facing in the opposite direction (Figure 8B). This results in a ~5Å difference in the position of the carbonyl group between YdhR and GMK, which is a considerable difference. We propose that this conformational “switch” provides d-fructose specific contact with the C2 hydroxyl. The presence of glycine in this position seems critical since no other amino acid allows such conformational flexibility. A point mutation of the G52 to alanine in YdhR shows practically no enzymatic activity towards fructose, therefore verifying its critical role in substrate recognition and binding (Figure 1).
Another difference between YdhR and GMK is the presence of the active site of Y`252, which crosses over from the other dimer subunit. Even though the Y`252 seems too far away to interact with fructose (~3.74 Å apart from hydroxyl groups of C1 of d-fructose), it contributes to the active site pocket and encloses it from the side. In GMK, however, no residues from the other subunit of the dimer contribute to the active site pocket. This identifies an important difference between architecture of YdhR and GMK dimers and corroborates a different architecture of the active site pockets. In GMK, the loop1 (L1) encloses the active site pocket, while in YdhR, the active site pocket enclosure requires residues from both subunits.
Proposed catalytic mechanism
Superimposition of non-ROK E. coli glucokinase (33) with YdhR fructokinase also reveals almost identical location and conservation of the residues in the active sites (Figure 8). For example, in both enzymes, the aspartic residue (D103 in B. subtilis YdhR and D100 in E. coli glucokinase) could act as a general base in catalysis, as it is positioned in a similar arrangement interacting with the C4 and C6 hydroxides of their respective substrates. Based on the structural and mutagenesis studies of several representatives of the glucokinase and hexokinase subfamily of enzymes, the phosphorylation of sugar substrates proceeds by a nucleophilic substitution via SN2 mechanism (33, 34). Analogously, based on the high conservation of the structure and the active site residues, it is highly likely that the phosphorylation of the fructose by YdhR also follows the same mechanism.
In the proposed SN2 mechanism, the O6 atom of the d-fructose substrate nucleophile attacks the electropositive P atom of the γ-phosphoryl group of ATP (33). Based on the YdhR-ADP-Fru structure it is straightforward to envision that ATP is likely to bind in a similar way as the ADP molecule. Modeling the ATP at the ADP molecule positions the terminal phosphoryl group of trinucleotide near the O6 atom within a distance of ~5Å. Correctness of this modeling could be confirmed by the fact that γ-phosphate aligns closely with the position of one of the phosphates present in the glucose-phosphate complex of ROK ATP/GMK (13,32). This distance would be suitable for in-line nucleophilic attack (34–36). Located in close proximity (2.9 Å) to the d-fructose is the D103 side chain (Figure 7 and 8). Its carboxylate hydrogen bonds to the d-fructose oxygen O6 and apparently acts as a base. It is likely to deprotonate the alcohol group of the C6 atom of the d-fructose and creates a nucleophilic oxygen atom. The general base role of D103 is in agreement with the role proposed and confirmed by the mutagenesis studies for the corresponding D100 and D657 in E. coli glucokinase and human hexokinase, respectively (37–39). It is hard to exactly predict, which residue plays the role of general acid, activates the terminal ATP phosphoryl group, and plays an important role in transition state stabilization. One of the possible candidates could be a K11 residue, which is located 7.6 Å apart from the β-phosphate of ADP. Simple torsion rotation movements put it ~3 Å from the β-phosphate, confirming this option. This possibility is supported by the observation that unmethylated fructokinase showed 50 times lower Km and 1.8 times higher Vmax (data not shown). The K11 residue is doubly methylated in the crystal. Another possibility could be that a proton is donated directly from the solvent. The other likely scenario may involve a magnesium-bound water molecule that functions as a general acid, as it had been proposed earlier in the case of human hexokinase I (37, 39). This could also explain the essential role of divalent cations in the catalysis. Although YdhR-ADP-Fru and YdhR-ADP were crystallized in the presence of magnesium ions, we were unable to observe bound magnesium in the structure, indicating that it may be coordinated by the β- and γ-phosphates of the ATP molecule and that the β-phosphate of ADP alone is not adequate to provide sufficient coordination to bind a magnesium ion.
Conclusions
Here we present the first structural and functional analysis of YdhR fructokinase, a member of the ROK family. We have determined the structure of an apo, ADP- and ADP-d-fructose bound YdhR enzyme and confirmed its fructokinase function. This is only the second extensive structural study of an enzyme in this large and versatile family, and only the second structure of an ROK enzyme solved in the presence of its substrate. Superimposition of three structures of YdhR does not reveal any significant structural changes upon binding of the substrate (YdhR+ADP+Fru) or release of product (YdhR+ADP). This suggests that ROK YdhR fructokinase is structurally arranged for the catalytic events and does not require large conformational changes to allow substrates to bind. In contrast to YdhR, other sugar binding enzymes like hexokinase or glucokinase (27, 33) have been shown to undergo conformational changes upon substrate binding. It has been reported that conformational changes are prerequisites for ADP binding in E. coli glucokinase, suggesting that the enzyme in its open conformation may have low affinity for ADP (27, 33). This is clearly not the case for YdhR; the active site is quite open and the crystals of the YdhR/ADP complex are very well ordered and diffract to the highest resolution. This further suggests that YdhR is well ordered while binding ADP and does not follow the induced fit model of glucokinases and hexokinases.
Structural comparison of YdhR with other proteins from PDB reveals that YdhR is closely related with other ROK family members, such as acetylmannosamine kinase, N-acetylglucosamine kinase, and ATP glucomannokinase, as well as with non-ROK glucokinases. It has been previously suggested that ROK and non-ROK glucokinases diverged from a common ancestor, explaining close structural similarity of these proteins (32). While this is not surprising, the fact that many ROK enzymes are functionally specialized to use versatile carbohydrate substrates other than glucose, while keeping almost unchanged active site architecture, remains puzzling. Certainly, preservation of the same residues and maintaining the same arrangement is clearly advantageous from the perspective of preserving the same mechanism. However, understanding how these enzymes, by using nearly identical active sites, control specificity towards particular substrates requires careful studies of each representative. Our studies of YdhR fructokinase shed some light on how this enzyme specifically recognizes d-fructose. Superimposition of the structures of YdhR and a few other sugar kinases revealed that the so-called active site loop plays a critical role in substrate specificity. In particular, two residues, glycine 59 and proline 60, form the cis peptide bond that interacts with the C2 hydroxide oxygen of d-fructose. It appears that this interaction is critical in binding fructose, as the point mutation of the glycine to alanine nearly completely abolishes the fructokinase activity (Km = 0.3 mM and Vmax = 2.7 U/mg of protein). Similarly, biochemical and mutagenesis studies of another ROK family member, E. coli K-12 d-allose kinase (AlsK) (40), showed that the corresponding active site loop residues play a critical role in AlsK specificity towards its natural substrate, d-allose. Sequence analysis of ROK family enzymes revealed that the majority of ROK family members have a conserved glycine residue (equivalent to YdhR glycine 59), except for AlsK, which instead has an alanine residue (40). Mutation of the AlsK alanine residue back to glycine leads to a more promiscuous enzyme, decreasing its selectivity towards non-native carbohydrates such as D-glucose, D-altrose, 2`-deoxy-D-glucose and D-mannose (40). However, enzyme activity towards its natural substrate D-allose remained unchanged.
Both studies consistently point towards the important role of equivalent position residues (cis-glycine in YdhR and the L-alanine in AlsK) in substrate specificity, indicating that small changes in the active site play a large role in controlling specificity of substrates. Towards this end, in another study, replacement of the active site residue W169 in α-glucosidase enzyme to tyrosine reversed this enzyme enzymatic preference, indicating it as a selectivity determinant between isomaltose and maltose substrates(41).
The above examples strongly suggest that a change of one residue may lead to gaining alternate function or loosing activity towards the natural substrate. It appears that carbohydrate-binding enzymes, especially ROK enzymes, seem to use this approach quite effectively. Further biochemical and structural studies of other representatives of the ROK family are required to show if this is a common scheme controlling specificity of these enzymes.
MATERIAL AND METHODS
Gene cloning and protein expression
The ORF of YdhR (gi|3914959) was amplified by PCR from B. subtilis MC58 genomic DNA (ATCC) with KOD DNA polymerase using conditions and reagents provided by the vendor (Novagen, Madison, WI). The gene was cloned into the pMCSG7 vector by using a modified ligation-independent cloning protocol (42). This process generated an expression clone of a fusion protein with an N-terminal His6-tag and a TEV protease recognition site (ENLYFQ↓S). The fusion protein was expressed in an E. coli BL21-derivative that harbored a plasmid pMAGIC encoding three rare E. coli tRNAs (Arg (AGG/AGA) and Ile (ATA)). This construct provided an N-terminal His6-tag separated from the gene by a TEV protease recognition sequence. A Se-Met derivative of the expressed protein was prepared as described earlier (43). The transformed BL21 cells were grown at 37°C in M9 medium supplemented with 0.4% (w/v) glucose, 8.5mM NaCl, 0.1mM CaCl2, 2mM MgSO4, and 1% (w/v) thiamine. At an A595 of 1.0–1.5, 0.01% (w/v) leucine, isoleucine, lysine, phenylalanine, threonine, and valine were added to inhibit the metabolic pathway of methionine and encourage Se-Met incorporation. Se-Met (90mg) was added to one liter of culture and protein expression was induced with 1mM isopropyl-β-d-thiogalactoside (IPTG). The cells were then incubated at 20°C overnight. The harvested cells were re-suspended in lysis buffer (500mM NaCl, 5% (v/v) glycerol, 50mM Hepes pH 8.0, 10mM imidazole, 10mM 2-mercaptoethanol) and stored at − 20 °C.
Protein purification
Both native and Se-Met proteins were purified using the procedure described in Kim et. al. (44). The harvested cells were re-suspended in lysis buffer supplemented with 1mg/mL lysozyme and 100ml of protease inhibitor (Sigma, P8849) per 2g of wet cells. This mixture was kept on ice for 20 min and then sonicated. The lysate was clarified by centrifugation at 36,000g for 1 hour and filtered with a 0.44µm membrane. The clarified lysate was applied to a 5mL HiTrap Ni-NTA column (GE Health Systems) on an AKTAexpress system (GE Health Systems). The His6-tagged protein was released with elution buffer (500mM NaCl, 5% glycerol, 50mM Hepes pH 8.0, 250mM imidazole, 10mM 2-mercaptoethanol) and the fusion tag was removed with treatment of recombinant His6-tagged TEV protease (a gift from Dr D. Waugh, NCI). Ni-NTA affinity chromatography was performed to remove the His6-tag, uncut protein, and His6-tagged TEV protease. Protein was concentrated using a Centricon centrifugal filter device with a 5000 Mr cutoff (Millipore), flash frozen, and stored in liquid nitrogen. The YdhR protein was dialyzed against crystallization buffer containing 200mM NaCl, 20mM Hepes pH 8.0, 2mM DTT and then concentrated to 81mg/ml for crystallization.
Point mutation
The G59A point mutation was prepared using PIPE cloning (Polymerase Incomplete Primer Extension) method applied to high-throughput cloning and site-directed mutagenesis (45). The efficiency of cohesive ends creation was enhanced by T4 polymerase treatment of amplified plasmid. Briefly, plasmid carrying the YdhR gene was PCR amplified by Kod Hot Start polymerase in the presence of 1M betaine and the following primers: 5’TCCTTTGCACCCGTTGATAACGATAAAACCAGTC3’, 5’CAACGGGTGCAAAGGAGCCGATGCCGATC3’. Unpurified PCR product was digested with T4 polymerase without any dNTPs (46). The T4 treated mixture was transformed to E. coli BL21 Magic strain. Extracted plasmid was amplified using the following primers: T7 short 5'-TAATACGACTCACTATAGGG-3' and T7 term 5'-GCTAGTTATTGCTCAGCGG-3'. The purified PCR product was sequenced at the University of Chicago Cancer Research DNA Sequencing Facility.
Enzymatic activity
Determination of the fructose and glucose kinase activity was measured according to the coupled assay described by Martinez-Barajas et al. (47). The fructose kinase activity was measured using UV–VIS spectrophotometry (BMG FLUOstar Galaxy v4.30-0) at 340 nm by detecting the reduction of NAD+ (? 340=6.22cm−1 mM−1) at 25°C in an assay buffer (25mM Tris-HCl pH 8.0, 50mM KCl, 0.5mM NAD+, 1.5mM ATP, 1.5mM MgCl2, 0.1 to 6.0mM fructose, 1.2 unit/ml glucose-6-phosphate dehydrogense, and 0.8 unit/ml phosphoglucose isomerase). The assay is initiated when 0.12µM of unmethylated enzyme is added and the absorbance at 340 nm is measured after 15 min. Enzyme activity is expressed as mM of substrate converted per min per µM of enzyme.
The activity of glucose kinase was measured in a glucose-6-phosphate dehydrogenase coupled reaction (47). The reduction of NADP (? 340=6.22cm−1 mM−1) was monitored by spectrometer at 340mm in assay buffer (100mM Tris-HCl pH 7.5, 10mM d-glucose, 5mM MgCl2, 0.5mM NADP, 1mM ATP, and 0.7 unit/ml glucose-6-phosphate dehydrogenase). The assay is initiated when 0.1µM of enzyme was added. The absorbance at 340 nm is measured for up to 4 hours. For determination of the Km and Vmax, the assays contained substrates at concentrations of 0.1–4.0mM. Kinetic parameters were determined by a nonlinear curve fitting the Lineweaver-Burk plot using GraphPad Prism software (version 4.00 for Windows, GraphPad Software, San Diego, CA).
Size exclusion chromatography
The molecular weight of native YdhR protein in solution was determined by size exclusion chromatography using a Superdex-200 10/30 column (Pharmacia) according to the method described previously(48). The column was calibrated with chymotrypsinogen A (25kDa), ovalbumin (40kDa), albumin (67kDa), aldolase (158kDa), and blue dextran (2,000kDa). The chromatography was carried out at 4°C at a flow rate of 1 mL/min. The calibration curve of Kav versus log molecular weight was prepared using the equation Kav = Ve–Vo/Vt –Vo, where Ve = elution volume for the protein, Vo = column void volume, and Vt = total bed volume.
Reductive methylation
Reductive methylation of YdhR was performed using a modified version of a reductive alkylation protocol reported previously (49, 50) using dimethylamine-borane complex as a reducing agent (ABC, Sigma-Aldrich). All reagents were freshly prepared and solutions were kept on ice. The YdhR protein was concentrated to a volume of 5 ml at concentrations of 5–10 mg/ml. 40 µl of 1M formaldehyde (per 1 ml of protein solution) was added and mixed gently, followed immediately by 20 µl of 1M ABC (per 1 ml of protein solution) and again gently mixed. The solution was incubated at 4°C for 2 hours and the procedure was repeated. At the end of the incubation, an additional 10 µl of ABC (per 1 ml of protein solution) was added. The solution was incubated at 4°C overnight. The following day, 1 mg of glycine and 5mM DTT were added to quench the reaction and the solution was left on ice for 2 hours. A gel filtration step was applied and the pure protein was concentrated using a Centricon Plus-20 centrifugal concentrator (Millipore).
Crystallization
YdhR-apo
Native crystals of methylated YdhR protein (~80 mg/ml) crystals were obtained from a reservoir of 0.1M Tris-HCl pH 8.5, 1.5M ammonium sulfate. Crystals were then transferred to hanging drops to which K2PtCl4 was added to a concentration of 10mM and soaked for approximately one hour. Crystals were transferred briefly to a cryo-solution (0.1M Tris-HCl pH 8.5, 1.5M ammonium sulfate, 25% (w/v) glycerol) with nylon cryo-loops and then flash frozen in liquid nitrogen.
YdhR-ADP and YdhR-ADP-d-fructose
The best crystals were obtained from methylated protein in a condition containing 0.1M Tris-HCl pH 8.5 and 1.5M ammonium sulfate. In order to obtain the YdhR-ADP complex crystals, 20mM MgCl2 and 40mM ADP were added to the YdhR protein and incubated on ice for 1 hour prior to crystallization. A similar approach was used to obtain the YdhR-ADP-FruRU complex crystals using 20mM MgCl2, 40mM d-fructose, and40 mM ADP. A grid screen containing four different concentrations of ammonium sulfate salt (1 M, 1.5 M, 2.0 M, 2.5 M) versus various Tris buffers (pHs 6, 6.5, 7, 7.5, 8, and 8.5) were set up by mixing 1µl of protein with 1µl of the corresponding well solution. The best quality crystals were grown in 1M ammonium sulfate and 0.1M Bis-Tris pH 8.0. The crystals reached a size of 0.7 × 0.6 × 0.5 µm within 1 day. For data collection, 3M ammonium sulfate supplemented with 10% glycerol was used as a cryo-protectant. The crystals were picked up with a nylon loop and flash-cooled in liquid nitrogen.
Data collection
YdhR-apo
The platinum-derivatized crystal belonged to space group P3121 and diffracted to 2.10 Å. A single-wavelength anomalous diffraction (SAD) dataset was collected at 0.9793 Å keV at the Structural Biology Center beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory. Emission peaks were detected for Pt and Zn in an X-ray emission spectrum of the crystal at the data-collection energy. The crystal temperature was maintained at 100K and inverse beam geometry was utilized to maximize Bijvoet pair coverage. Images were recorded on a custom made CCD area detector, SBC2 (51). Data collection strategy, integration, and scaling were performed with the HKL2000 program package (46–52). A summary of the crystallographic data can be found in Table 1.
YdhR-ADP, YdhR-ADP-d-fructose
A SAD experiment was carried out on the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source using the Se-Met YdhR-ADP crystals (51). Diffraction data was collected from a Se-Met-substituted crystal of YdhR-ADP using inverse-beam geometry near the selenium absorption edge (0.9794 Å) to 1.66 Å. Diffraction data from a native YdhR-ADP-Fru crystal was collected to 2.45 Å. In both cases, before data collection, an X-ray florescence spectrum was recorded from each sample, which identified the presence of Se and Zn in the protein crystals (data not shown). The SBC-Collect program was used for data collection and visualization. All data was integrated and scaled with the HKL3000 software package (52–58). Data collection and processing statistics are shown in Table 1.
Structure determination
YdhR-apo
Three heavy atom positions were found by the substructure solution engine HySS of the Phenix software suite (59). These sites were optimized by iterative residual analysis and used for phasing, density modification, and model-building with the autoSHARP program suite(60). Manual rebuilding with the program COOT (61) completed the model. Restrained refinement was performed with REFMAC5 against data to 2.10 Å and Hendrickson-Lattmann coefficients as calculated by autoSHARP prior to density modification. The final model contained one YdhR molecule, two Pt, one Zn, 221 water molecules and two glycerol molecules. There was no interpretable electron density for part of the cloning tag, (SNA), and C-terminal residues 295–299. All residues fell within acceptable regions of a MolProbity (65) Ramchandran plot, with 97.0% of all residues in favored regions. Refinement details are shown in Table 1.
YdhR-ADP, YdhR-ADP-d-fructose
For the YdhR-ADP complex crystal, the initial phases were determined by SAD phasing, density modification, and the initial protein model was built using the HKL3000 software package (52–58). The initial rounds of rebuilding and refinement were performed using COOT and REFMAC (61, 62). The structure of YdhR-ADP-d-fructose was determined by molecular replacement with MOLREP (63) (R-factor of 0.35, correlation coefficient of 0.70) using the model of YdhR-ADP. A few rounds of rebuilding and refinement were performed using COOT and REFMAC (61, 62). The final models were refined against all reflections except for 5% randomly selected reflections, which were used for monitoring Rfree. The final rounds of refinement were carried out using TLS refinement. The TLS groups were calculated using the TLSMD server (64). The quality of both structures was checked with the validation tools included in the programs COOT and Molprobity (65). The final refinement statistics are presented in Table 1. All residues fell within acceptable regions of the Ramachandran plot, with 97.0% of all residues in favored regions. Refinement details are summarized in Table 1.
Coordinates
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, with the following PDB ID numbers: YdhR-apo, PDB ID 1XC3; YdhR-ADP, PDB ID 3OHR; and YdhR-ADP-FruRU, PDB ID 3LM9.
Highlight.
Fructokinase YdhR phosphorylates d-fructose using ATP as a cofactor
YdhR is a member of the “ROK” sequence family
YdhR is a homo-dimer with a monomer composed of two similar α/β domains
Structures of an apo, ADP- and ADP-d-fructose form of YdhR were determined
cis-Gly59 is important for specific binding of d-fructose.
Acknowledgements
We wish to thank all members of the Structural Biology Center at Argonne National Laboratory for their help in conducting these experiments. This work was supported by National Institutes of Health grant GM074942 and by the U. S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
REFERENCES
- 1.Titgemeyer F, Walkenhorst J, Reizer J, Stuiver MH, Cui X, Saier MH., Jr Identification and characterization of phosphoenolpyruvate:fructose phosphotransferase systems in three Streptomyces species. Microbiology (Reading, England) 1995;141(Pt 1):51–58. doi: 10.1099/00221287-141-1-51. [DOI] [PubMed] [Google Scholar]
- 2.Wilbrandt W, Rosenberg T. The concept of carrier transport and its corollaries in pharmacology. Pharmacol Rev. 1961;13:109–183. [PubMed] [Google Scholar]
- 3.Postma PW, Lengeler JW. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985;49:232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Middleton RJ. Hexokinases and glucokinases. Biochem Soc Trans. 1990;18:180–183. doi: 10.1042/bst0180180. [DOI] [PubMed] [Google Scholar]
- 6.Titgemeyer F, Reizer J, Reizer A, Saier MH., Jr Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology (Reading, England) 1994;140(Pt 9):2349–2354. doi: 10.1099/13500872-140-9-2349. [DOI] [PubMed] [Google Scholar]
- 7.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conejo MS, Thompson SM, Miller BG. Evolutionary bases of carbohydrate recognition and substrate discrimination in the ROK protein family. J Mol Evol. 70:545–556. doi: 10.1007/s00239-010-9351-1. [DOI] [PubMed] [Google Scholar]
- 9.Dahl MK, Schmiedel D, Hillen W. Glucose and glucose-6-phosphate interaction with Xyl repressor proteins from Bacillus spp. may contribute to regulation of xylose utilization. Journal of bacteriology. 1995;177:5467–5472. doi: 10.1128/jb.177.19.5467-5472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodionov DA, Mironov AA, Gelfand MS. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiology Letters. 2001;205:305–314. doi: 10.1111/j.1574-6968.2001.tb10965.x. [DOI] [PubMed] [Google Scholar]
- 11.Hansen T, Schonheit P. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol Lett. 2003;226:405–411. doi: 10.1016/S0378-1097(03)00642-6. [DOI] [PubMed] [Google Scholar]
- 12.Gibbins LN, Simpson FJ. The purification and properties of D-allose-6-kinase from Aerobacter aerogenes. Can. J. Biochem. 1963:769–779. [Google Scholar]
- 13.Mukai T, Kawai S, Mori S, Mikami B, Murata K. Crystal structure of bacterial inorganic polyphosphate/ATP-glucomannokinase. Insights into kinase evolution. The Journal of biological chemistry. 2004;279:50591–50600. doi: 10.1074/jbc.M408126200. [DOI] [PubMed] [Google Scholar]
- 14.Schiefner A, Gerber K, Seitz S, Welte W, Diederichs K, Boos W. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. The Journal of biological chemistry. 2005;280:29073–29079. doi: 10.1074/jbc.M504215200. [DOI] [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Sadaie Y, Yata K, Fujita M, Sagai H, Itaya M, Kasahara Y, Ogasawara N. Nucleotide sequence and analysis of the phoB-rrnE-groESL region of the Bacillus subtilis chromosome. Microbiology (Reading, England) 1997;143(Pt 6):1861–1866. doi: 10.1099/00221287-143-6-1861. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, Fan Y, Bigelow L, Maltseva N, Wu R, Borovilos M, Duggan E, Zhou M, Binkowski TA, Zhang RG, Joachimiak A. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5:853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pego JV, Smeekens SC. Plant fructokinases: a sweet family get-together. Trends Plant Sci. 2000;5:531–536. doi: 10.1016/s1360-1385(00)01783-0. [DOI] [PubMed] [Google Scholar]
- 20.Qu Q, Lee SJ, Boos W. Molecular and biochemical characterization of a fructose-6-phosphate-forming and ATP-dependent fructokinase of the hyperthermophilic archaeon Thermococcus litoralis. Extremophiles. 2004;8:301–308. doi: 10.1007/s00792-004-0392-5. [DOI] [PubMed] [Google Scholar]
- 21.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 23.Cordeiro AT, Caceres AJ, Vertommen D, Concepcion JL, Michels PA, Versees W. The crystal structure of Trypanosoma cruzi glucokinase reveals features determining oligomerization and anomer specificity of hexose-phosphorylating enzymes. J Mol Biol. 2007;372:1215–1226. doi: 10.1016/j.jmb.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 24.Anderson CM, Stenkamp RE, Steitz TA. Sequencing a protein by x-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 A resolution. J Mol Biol. 1978;123:15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- 25.Rosano C, Sabini E, Rizzi M, Deriu D, Murshudov G, Bianchi M, Serafini G, Magnani M, Bolognesi M. Binding of non-catalytic ATP to human hexokinase I highlights the structural components for enzyme-membrane association control. Structure. 1999;7:1427–1437. doi: 10.1016/s0969-2126(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 26.Weihofen WA, Berger M, Chen H, Saenger W, Hinderlich S. Structures of human N-Acetylglucosamine kinase in two complexes with N-Acetylglucosamine and with ADP/glucose: insights into substrate specificity and regulation. J Mol Biol. 2006;364:388–399. doi: 10.1016/j.jmb.2006.08.085. [DOI] [PubMed] [Google Scholar]
- 27.Nishimasu H, Fushinobu S, Shoun H, Wakagi T. Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii. The Journal of biological chemistry. 2007;282:9923–9931. doi: 10.1074/jbc.M610678200. [DOI] [PubMed] [Google Scholar]
- 28.Concha MI, Leon G. Cloning, functional expression and partial characterization of the glucose kinase from Renibacterium salmoninarum. FEMS Microbiol Lett. 2000;186:97–101. doi: 10.1111/j.1574-6968.2000.tb09088.x. [DOI] [PubMed] [Google Scholar]
- 29.Park SY, Kim HK, Yoo SK, Oh TK, Lee JK. Characterization of glk, a gene coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiol Lett. 2000;188:209–215. doi: 10.1111/j.1574-6968.2000.tb09195.x. [DOI] [PubMed] [Google Scholar]
- 30.Mesak LR, Mesak FM, Dahl MK. Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages. BMC Microbiol. 2004;4:6. doi: 10.1186/1471-2180-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang DT, Chen CY, Chung WC, Oyang YJ, Juan HF, Huang HC. ProteMiner-SSM: a web server for efficient analysis of similar protein tertiary substructures. Nucleic Acids Res. 2004;32:W76–W82. doi: 10.1093/nar/gkh425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai S, Mukai T, Mori S, Mikami B, Murata K. Hypothesis: structures, evolution, and ancestor of glucose kinases in the hexokinase family. Journal of bioscience and bioengineering. 2005;99:320–330. doi: 10.1263/jbb.99.320. [DOI] [PubMed] [Google Scholar]
- 33.Lunin VV, Li Y, Schrag JD, Iannuzzi P, Cygler M, Matte A. Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose. Journal of bacteriology. 2004;186:6915–6927. doi: 10.1128/JB.186.20.6915-6927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matte A, Tari LW, Delbaere LT. How do kinases transfer phosphoryl groups? Structure. 1998;6:413–419. doi: 10.1016/s0969-2126(98)00043-4. [DOI] [PubMed] [Google Scholar]
- 35.Flaherty KM, Wilbanks SM, DeLuca-Flaherty C, McKay DB. Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment. The Journal of biological chemistry. 1994;269:12899–12907. [PubMed] [Google Scholar]
- 36.Muller CW, Schulz GE. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. J Mol Biol. 1992;224:159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- 37.Zeng C, Aleshin AE, Hardie JB, Harrison RW, Fromm HJ. ATP-binding site of human brain hexokinase as studied by molecular modeling and site-directed mutagenesis. Biochemistry. 1996;35:13157–13164. doi: 10.1021/bi960750e. [DOI] [PubMed] [Google Scholar]
- 38.Aleshin AE, Zeng C, Bourenkov GP, Bartunik HD, Fromm HJ, Honzatko RB. The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate. Structure. 1998;6:39–50. doi: 10.1016/s0969-2126(98)00006-9. [DOI] [PubMed] [Google Scholar]
- 39.Mulichak AM, Wilson JE, Padmanabhan K, Garavito RM. The structure of mammalian hexokinase-1. Nat Struct Biol. 1998;5:555–560. doi: 10.1038/811. [DOI] [PubMed] [Google Scholar]
- 40.Larion M, Moore LB, Thompson SM, Miller BG. Divergent evolution of function in the ROK sugar kinase superfamily: role of enzyme loops in substrate specificity. Biochemistry. 2007;46:13564–13572. doi: 10.1021/bi700924d. [DOI] [PubMed] [Google Scholar]
- 41.Tan K, Tesar C, Wilton R, Keigher L, Babnigg G, Joachimiak A. Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch. FASEB J. 24:3939–3949. doi: 10.1096/fj.10-156257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 43.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol Crystallogr. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 46.Dieckman L, Gu M, Stols L, Donnelly MI, Collart FR. High throughput methods for gene cloning and expression. Protein Expr Purif. 2002;25:1–7. doi: 10.1006/prep.2001.1602. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Barajas E, Krohn BM, Stark DM, Randall DD. Purification and characterization of recombinant tomato fruit (Lycopersicon esculentum Mill.) fructokinase expressed in Escherichia coli. Protein Expr Purif. 1997;11:41–46. doi: 10.1006/prep.1997.0762. [DOI] [PubMed] [Google Scholar]
- 48.Nocek B, Chang C, Li H, Lezondra L, Holzle D, Collart F, Joachimiak A. Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J Mol Biol. 2005;354:91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- 50.Rypniewski WR, Holden HM, Rayment I. Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme: an X-ray analysis at 1.8-A resolution. Biochemistry. 1993;32:9851–9858. doi: 10.1021/bi00088a041. [DOI] [PubMed] [Google Scholar]
- 51.Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallographica Section D-Biological Crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 53.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron-Density Maps and the Location of Errors in These Models. Acta Crystallographica Section A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 55.Cowtan KD, Main P. Improvement of Macromolecular Electron-Density Maps by the Simultaneous Application of Real and Reciprocal Space Constraints. Acta Crystallographica Section D-Biological Crystallography. 1993;49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- 56.Terwilliger TC. Improving macromolecular atomic models at moderate resolution by automated iterative model building, statistical density modification and refinement. Acta Crystallographica Section D-Biological Crystallography. 2003;59:1174–1182. doi: 10.1107/S0907444903009922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 64.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallographica Section D-Biological Crystallography. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 65.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]