Abstract
Investigators now have a wide range of analytical tools to use in measuring metabolites, proteins and transcripts in plant tissues. These tools have the potential to assist genetic studies that seek to phenotype genetic lines for heritable traits that contribute to drought tolerance. To be useful for crop breeding, hundreds or thousands of genetic lines must be assessed. This review considers the utility of assaying certain constituents with roles in drought tolerance for phenotyping genotypes. Abscisic acid (ABA), organic and inorganic osmolytes, compatible solutes, and late embryogenesis abundant proteins, are considered. Confounding effects that require appropriate tissue and timing specificity, and the need for high-throughput and analytical cost efficiency are discussed. With future advances in analytical methods and the value of analyzing constituents that provide information on the underlying mechanisms of drought tolerance, these approaches are expected to contribute to development crops with improved drought tolerance.
Keywords: water stress, compatible solutes, metabolites, osmotic adjustment, abscisic acid, plant breeding
Introduction
Phenotyping involves measurement of observable attributes that reflect the biological functioning of gene variants (alleles) as affected by the environment. In general, phenotyping for crop improvement via breeding requires that hundreds or thousands of genetic lines be assessed. To date, most phenotyping of secondary traits (i.e., those traits in addition to yield, which is often the primary trait) has involved field assessments of easily scored morphological attributes such as plant height, leaf number, flowering date, and leaf senescence. However, investigators have recognized that drought tolerance involves metabolic and regulatory functions, for which measurements of targeted processes are likely to provide valuable information on the underlying biology, and suggest approaches by which it could be modified. Moreover, excellent methods have been developed for assay of such traits, and they have been used in controlled-environment studies to determine the mechanistic basis of drought response. Notwithstanding their positive aspects, many of these methods are too time-consuming, expensive, or technically demanding to be used in large-scale phenotyping. The challenge, then, is to identify those attributes that provide the most meaningful phenotypic information to design sampling methods suitable for use in the field, and design analytical methods that can be efficiently scaled up to the number of samples required in phenotyping projects.
An important prerequisite for the successful phenotyping of secondary traits is to identify key functional attributes that contribute to drought tolerance. Ideally, such identification is based on evidence that there is genetic variation for the trait in the crop of interest, on the trait being correlated with crop performance in drought environments, and on its having sufficient heritability to be used to make progress in a breeding program. Another criterion is that a trait has a clear-cut and rational explanation for its physiological or molecular function in drought tolerance. Some key traits that satisfy this latter criterion include: (i) favorable stomatal behavior; (ii) rooting depth; (iii) osmotic adjustment (OA) and other processes that sustain cell integrity and function; (iv) carbohydrate storage and remobilization; and (v) sustained development (as opposed to abortion) of harvested organs. This article will discuss methods that can contribute to phenotyping of these five traits by analyzing the presence and level of substances that can serve as diagnostic tests to evaluate them. While methods for field-based assessment of these traits are described in other articles in this special issue, the focus of the current article is on laboratory-based analyses of tissue constituents in samples obtained from field-grown plants.
Case Study: Abscisic Acid
Although it is valuable to focus phenotyping efforts on metabolic traits that have a clear-cut rational connection with stress tolerance mechanisms, this goal is often difficult to achieve in practice. A particular constituent may be involved in several metabolic and signaling systems, and its involvement may differ in various tissues and developmental stages. To illustrate the difficulty, the prospects for phenotyping abscisic acid levels as an indicator of stress response will be considered below.
Why phenotype abscisic acid?
Abscisic acid (ABA) plays a central role in plant response to water deficit, and the extent to which a genotype synthesizes and accumulates ABA is a possible indicator of that genotype’s adaptation to drought. It has been demonstrated that ABA has regulatory roles in all five of the traits listed above. While this argues for its importance, it also presents difficulty with respect to the use of ABA analysis as a phenotyping tool, because the multiple effects can confound interpretation.
A situation where higher leaf ABA could be advantageous is in environments where water-use efficiency (WUE) or postponement of dehydration is desirable. A recent example related to this idea involved production of transgenic canola (Brassica napus) with an enhanced response to ABA signaling (Wang et al., 2005). Investigators anti-sensed farnesyl transferase, a negative regulator of ABA signaling. They used a drought-responsive promoter (RD29A), thereby confining the expression to drought episodes. Studies showed a slightly earlier stomatal closure response during the onset of water deficit and, in field trials, yields appeared to be slightly higher in drought environments. In addition to ABA sensitivity, genotypes that generally have higher activity for ABA synthesis in leaves might maintain lower stomatal conductance, closing their stomata at an earlier stage of soil water depletion, or closing them sooner after daybreak, thereby keeping the tissues at a high water potential and avoiding damage from dehydration. Maintaining a low stomatal conductance can also increase WUE, because it keeps the CO2 concentration in the leaf internal airspace (Ci) below the CO2-saturating (asymptote) portion of the photosynthesis versus Ci function. Elevated ABA in the afternoon can restrict stomatal opening when temperature and leaf-air vapor pressure deficit (VPD) are highest, and when a genotype couples this behavior with low ABA and partial stomatal opening in the morning, WUE is enhanced. While analysis of ABA is a way to predict stomatal status, other methods, might be better suited to this purpose. Notably, stable isotope assay of 13C discrimination, or field sampling of stomatal diffusive conductance is more direct and, in the case of 13C, are more integrative of Ci throughout the season (Condon et al., 2004, 2007).
However, the basis for a genotype’s tendency to accumulate leaf ABA may not be as straightforward as a genetic tendency for high ABA synthetic activity. A genotype with a shallow root system or low hydraulic conductance, which restricts the supply of water to the leaves, can also cause high ABA accumulation. Evidence that measured ABA levels can be related to root architecture is provided by presence of a quantitative trait locus (QTL) in maize, Root-ABA1, which was initially identified as a QTL that affected leaf ABA (Lebreton et al., 1995; Giuliani et al., 2005; Landi et al., 2007). The QTL was identified in two different maize populations, and in several different environments and locations. Yet, in both cases, root architecture was related to leaf ABA accumulation. This QTL, in Bin 2.04 of chromosome 2, was initially identified by QTL mapping of leaf ABA in populations scored under water limited environments. The QTL allele for high ABA also confers decreased stomatal conductance, as expected, given the role of ABA in stomatal closure (Lebreton et al., 1995; Giuliani et al., 2005). In addition, greenhouse studies of backcross-derived lines have shown that the high ABA QTL allele is associated with increased root dry weight, root diameter, and root:shoot ratio, and with decreased root lodging (Giuliani et al., 2005). While it is not yet known whether the observed phenomenon is related to the effect of ABA in stimulating seedling root growth (Sharp et al., 1994; Ober and Sharp, 2007), an alternative interpretation is that the QTL allele alters root architecture to a shallower root system that rapidly depletes surface moisture. This would be consistent with the observation in these studies that the QTL allele for high ABA is associated with lower grain yield (Landi et al., 2007) and with other genetic studies that have shown a correlation between high leaf ABA and lower grain yield in water deficit environments (Mugo et al., 1998; Setter et al., 2011). Thus, to gain valuable phenotypic information, data on leaf ABA levels need to be interpreted in relation to other information, such as the depth of soil water availability and stomatal behavior. An ideal genotype may be one with relatively low levels of leaf ABA, indicating good root depth and water transport properties, but with desirable stomatal behavior, indicating high stomatal sensitivity to ABA.
Interpreting ABA phenotypic data is tissue-dependent
ABA levels in organs other than leaves also require interpretation to be valuable for phenotyping. For example, as a stress-signaling hormone, ABA increases the expression of numerous gene products that have putative roles in stabilizing proteins and membrane systems so that they are better able to tolerate desiccation stress. However, ABA is also associated with the arrested development of sink organs, including soybean seeds (Liu et al., 2004), rice spikelets (Ji et al., 2011), maize kernels (Ober et al., 1991; Setter et al., 2001), and wheat grains (Westgate et al., 1996; Ji et al., 2011). Consistent with this, among lines of maize, a negative correlation has been found between ear ABA levels and prepollination ear growth rate (Table 1; Setter et al., 2011). This relationship suggests a role of ABA in the tendency of some genotypes to limit partitioning to ear and silk growth during water deficit, which increases the anthesis-silking interval (ASI), and is associated with decreased grain yield in drought environments (Ribaut et al., 2009). Analogous relationships between water deficit and arresting of reproductive organ growth are found in rice, sorghum, and other crops (Matthews et al., 1990; Liu et al., 2004, 2006; Leport et al., 2006).
Table 1.
Genotypic correlations between ABA levels and growth rates in ears and silks in a recombinant inbred line (RIL) population (Cimmyt P1 X P2) in two trials (2001 and 2002) under severe water deficit at flowering at the Tlaltizapan, Mexico field station.
| Silk ABA 0 DAA in 2001 | Silk ABA 7 DAA in 2001 | Silk ABA 0 DAA in 2002 | Ear ABA 0 DAA in 2002 | |
|---|---|---|---|---|
| Ear growth rate 0 to 7 day after anthesis | −0.90 | −0.62 | −0.82 | −0.57 |
| Silk growth rate 0 to 7 day after anthesis | −0.51 | −0.56 | −0.42 | −0.87 |
Tissues were harvested at the indicated days after anthesis (DAA). (Source: Setter and Ribault, unpublished data).
For genes that are regulated by ABA, the role of these gene products in crop performance is yet to be well established. For example, there is abundant evidence that, among the genes regulated by ABA signaling, those encoding members of the late embryogenesis abundant (LEA) family are among the most rapidly and substantially upregulated by ABA (Quatrano et al., 1997). LEA proteins were first identified as abundant proteins that accumulate during the late stages of seed formation when desiccation is about to begin. They were among the first ABA-regulated proteins for which promoter ABA response elements (ABRE) and associated elements in upstream DNA sequence were determined. Nevertheless, the mechanism(s) and value of LEA proteins and specialized compatible solutes in drought tolerance of tissues have not been established except for tissues undergoing extreme desiccation (Wise, 2003; Wise and Tunnacliffe, 2004; Goyal et al., 2005; Tunnacliffe and Wise, 2007; Bies-Etheve et al., 2008). However, even when a crop is subjected to a severe drought stress that diminishes yield by 70% or more, its organs usually do not reach water potentials and levels of desiccation in the range experienced by drying seeds where these stabilizing agents are effective. This argues against the idea that more ABA accumulation would be favorable as a stimulus for upregulating LEA and other agents for macromolecular stabilization. Moreover, as discussed above, studies have indicated that there is a negative correlation between ABA accumulation in floral tissues and kernel set. Therefore, in contrast to the leaves, in the case of developing flowers and young sinks, genotypes with high levels of ABA are less likely to perform well in drought environments.
Timing of tissue sampling for ABA
A further consideration in the use of constituent quantification for phenotyping is the dependence on timing of tissue sampling. Again referring to the ABA case study, leaf ABA levels depend on environmental and tissue development conditions. For phenotyping, a further consideration is that the levels of ABA measured in a tissue reflect the steady state of dynamically changing rates of ABA synthesis and catabolism (Ren et al., 2007). Quantification of free ABA provides an instantaneous measure of the system. Such an estimate is inherently dependent on the environment, with a likelihood of strong influence from genotype-by-environment interaction (GEI). Careful control of time of day and stage of soil water dry-down may help to control these influences. Alternatively, to obtain phenotypic values that have a relatively strong genetic component (and high heritability), it can be beneficial to average out the instantaneous environmental effects. A more time-averaged or integrative measure of ABA status might be obtained with a composite estimate of the sum of ABA plus its catabolites. The primary routes of ABA catabolism are 8′-hydroxylation to form phaseic acid and downstream products, and glucosyl esterification of the carboxyl group to form ABA-glucose ester (ABA-GE; Nambara and Marion-Poll, 2005; Priest et al., 2006; Yang and Zeevaart, 2006). The products of hydroxylation (phaseic acid and dihydrophaseic acid) also form glucose esters and other conjugates. To the extent that these catabolites are biologically inactive and accumulate into vacuoles or other compartments, their collective level could represent a valuable estimate of long-term flux through the ABA synthesis pathway. Following this logic, Setter et al. (2011) phenotyped ABA, ABA-GE, and phaseic acid in a diverse panel of maize genotypes that was used for association genetic analysis. While the phaseic acid data revealed a new marker-trait association unrelated to ABA, in this situation the addition of these ABA catabolites to the analysis did not strengthen any of the trait-marker associations that were obtained from ABA data alone. Development of cost-effective methods for a broader suite of catabolites has the potential to further improve this strategy.
Methods for abscisic acid quantification
The prospects for an expanded range of substances that can be phenotyped have been steadily improving. In the case of ABA, methods for determining the levels of and the hormone and its metabolites can be classified into two general categories: (i) physical-chemical methods; and (ii) immunochemical methods. Advantages of physical-chemical methods include the fact that they are based on fundamental properties of the compounds and, in the case of the more advanced methods that employ mass spectrometry (MS), their chemical specificity. Advantages of immunochemical methods include their low cost and ease of scale-up for high-throughput projects. The levels of ABA are usually much too low (typically a few nmol g−1 dry weight) relative to other substances in an extract for methods based solely on high performance liquid chromatography (HPLC) with ultraviolet (or photodiode array) detection to have sufficient selectivity. Such methods are in common use for carotenoids and other substances which are chemically related to ABA but are much more abundant (Harjes et al., 2008). However, they do not in themselves provide sufficient separation and selectivity for ABA and other plant hormone work. Good selectivity can be achieved with gas chromatography-mass spectrometry and, when used with selected ion monitoring (GC-MS/SIM) and robotics, throughput can be satisfactory for large-scale projects. However, some metabolites, notably ABA-GE, are not stable at GC temperatures, and the method is not well suited to profiling a wide range of hormone metabolites in a single pass. A method for plant hormone profiling has been developed that is capable of analyzing in a single run ABA and its metabolites, as well as hormones in the cytokinin, gibberellin, and auxin families (Chiwocha et al., 2003; Ross et al., 2004). This method utilizes reverse-phase HPLC separation, coupled to electrospray ionization and MS with multiple reaction monitoring (MRM). Although chromatography only partially separates compounds from one another, a high level of specificity is achieved by MRM, wherein each ionized compound in a plant sample gives a distinct precursor-to-product ion transition that is diagnostic of that particular compound. An important attribute of the system is the development and use of deuterated internal standards to correct for workup variability. The system is capable of quantifying ABA levels in the 10−12 g range, which permits a tissue sample of about 50 mg dry weight to be analyzed for the whole profile of hormones and their conjugates. Cost is a limitation for phenotyping projects that involve large numbers of samples. The system utilizes elaborate instrumentation and requires about 40 min per sample, leading to a rather high cost per sample (Zaharia and Abrams, 2011).
A second category of methods for ABA analysis involves immunochemical procedures with labeling from either a radioisotope (radioimmunoassay, RIA) or an enzyme (enzyme-linked immunosorbent assay, ELISA). These methods take advantage of the high level of binding specificity of antibodies to discriminate between substances in a complex mixture. In RIA, a radioactively labeled form of the analyte (e.g., ABA) is mixed with the sample to provide quantification, while in ELISA an enzyme reporter system is used to generate a measurable signal indicating presence of the analyte. Both immunochemical systems have been widely used for ABA quantification (Mertens et al., 1983; Walker-Simmons, 1987; Quarrie et al., 1988; Vernieri et al., 1989; Perata et al., 1990; Philosoph-Hadas et al., 1993; Banowetz et al., 1994; Xie et al., 1996; Kalantari et al., 2001; Hradecka et al., 2007).
An advantage of immunochemical analyses over physical-chemical systems is the low detection limit. The mid-point of the assay range for ABA ELISA is about 0.2 × 10−12 mol (Setter and Parra, 2010). This means that tissue samples as small as 1 mg dry weight are sufficient, permitting field sampling of small, but representative, portions of leaves or floral parts whose removal does not significantly affect plant growth in the field plot for yield and other phenotyping data. In practice, samples are usually composites of several plants per plot and of sufficient size for them to be used for analysis of multiple constituents in addition to ABA. In addition to analyzing ABA, it is also possible to use ABA immunoassay to analyze ABA catabolites in plant samples. ABA-GE can be assayed by chromatographically separating ABA from ABA-GE, then using alkaline hydrolysis to cleave the ester linkage between ABA and glucose, and then imunoassaying ABA as usual. Phaseic acid (PA) and its glucose ester can be analyzed with immunoassay as well, using a monoclonal antibody directed against the conjugated form of PA (Gergs et al., 1993; Setter et al., 2011). To date, there are no reports of the development of antibodies directed against the other major ABA catabolite, dihydrophaseic acid (DPA). If an anti-DPA antibody were to be produced, investigators would have a set of antibodies that could be used to immunoassay a full profile of ABA and its most abundant catabolites.
Osmotic Solute Accumulation and Osmotic Adjustment
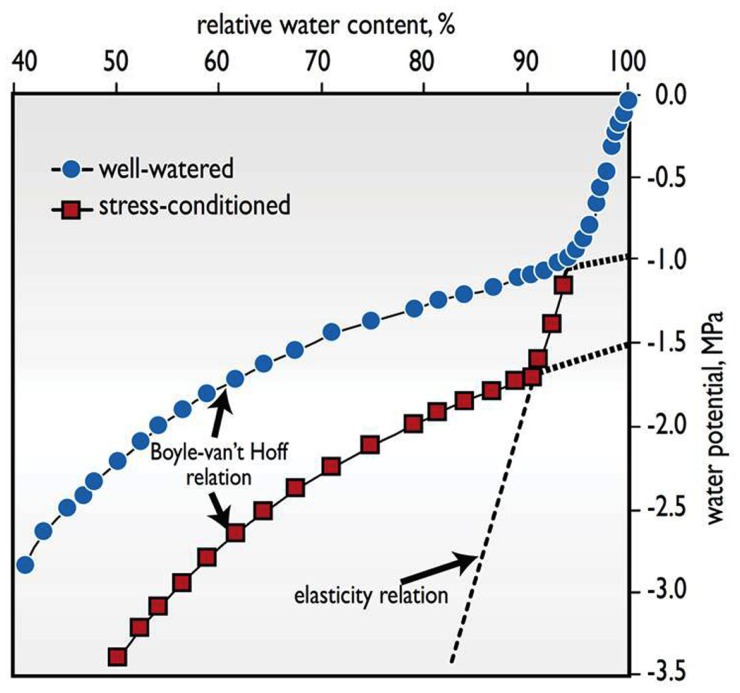
Osmotic adjustment refers to the accumulation of osmotically active solutes in response to the imposition of stress. It is a potential contributor to drought tolerance, and a sizable body of literature about it has developed. The potential benefit OA provides to a plant is that it helps cells retain water in the face of decreasing water potential that would otherwise result in cell shrinkage, distortion, and plasmolysis (Figure 1).
Figure 1.
The advantages of accumulating osmotically active solutes is illustrated with the response of total water potential (Ψw) to a lowering of relative water content (RWC) in wheat leaves, comparing a plant that was well-watered with another that was stress-conditioned by several cycles of water deficit and rewatering over a number of weeks, causing it to accumulate additional osmotically active solutes (Adapted from Melkonian et al., 1982). Two phases are identified. In tissue equilibrated with free water (Ψw ≈ 0 MPa), a large positive turgor pressure balances the osmotic component of water potential. As water is lost, turgor decreases steeply, reflecting the rather stiff cell walls in mature organs, in accordance with the elasticity relation. At RWCs below the point of zero turgor (the inflection point), the relationship is dictated by the less steep Boyle-van’t Hoff relation between osmotic solute activity and water potential, Ψw ≈ −RTC, where R is the ideal gas constant, T is the absolute temperature, and C is the concentration in moles of osmotically active solutes per kg of water. Tissues that accumulate more solutes via the process of osmotic adjustment, such as the stress-conditioned leaf shown here, shift the Boyle-van’t Hoff curve to lower water potentials. As a consequence, the tissue can maintain positive turgor over a wider Ψw range and incur less shrinkage at still lower levels of Ψw below the turgor loss point. Such shrinkage and low Ψw can damage the integrity of cell membranes and other cell constituents, and lead to cell death as in stress-induced leaf necrosis.
Although currently available tools to assess OA have enabled progress to be made in developing our understanding of the physiological role of OA and in identifying genotypic differences, studies that have involved large-scale phenotyping of populations have had mixed results. As discussed below, this may be due, in part, to a lack of easy-to-use sampling and analytical methods. Nevertheless, several studies have reported identification of QTLs for OA and related traits in crops (Table 2).
Table 2.
Studies in which QTLs were identified for osmotic adjustment (OA) and related traits.
| Crop | QTLs identified |
Citation | ||
|---|---|---|---|---|
| OA | Solute potential | Others | ||
| Barley | x | x | Δ13C, soluble carbohydrate | Diab et al. (2004) |
| Barley | x | x | RWC | Teulat et al. (1998) |
| Barley | x | x | Carbohydrate, RWC | Teulat et al. (2001) |
| Cotton | x | Δ13C, yield, canopy temperature, chlorophyll | Saranga et al. (2004) | |
| Cotton | x | Saranga et al. (2001) | ||
| Rice | x | x | Lethal osmotic potential | Lilley et al. (1996) |
| Rice | x | Robin et al. (2003) | ||
| Rice | x | Root traits | Zhang et al. (2001) | |
| Sunflower | x | x | Turgor, RWC | Kiani et al. (2007) |
RWC, relative water content.
A major difficulty of current methods of measuring OA is that it is necessary to harvest fresh tissue and immediately perform several operations on it while keeping it alive, keeping cell membranes intact, and not disturbing metabolism. For field studies, this generally requires care to prevent overheating of specimens during handling, and the availability on-site of apparatus such as a balance to weigh fresh tissue and an osmometer to measure solute potential. Also, since samples for all plots should be taken at a defined number of days after water deficit, the need to sample and analyze fresh tissue means the investigator must perform the analyses on-the-spot and cannot put the samples in storage for analysis at a later, more convenient time.
Several studies have assessed the adequacy of various methods for quantifying OA. In a comprehensive comparison of four methods for evaluating OA, Babu et al. (1999) considered two of them feasible for phenotyping work. In the first of these methods, the tissue relative water content (RWC) and osmotic potential are measured, then the osmotic potential is extrapolated to an RWC of 100% using an assumed RWC-solute potential relationship. In the second, the plants (or portions thereof) are fully rehydrated overnight. Then the osmotic potential is measured on samples. It is assumed that the rehydration does not alter solute concentrations significantly, even though it takes several hours and sometimes involves floating specimens on water, allowing the possibility of solute efflux. Measurement of osmotic potential involves several steps that must be carried out immediately and with care to avoid evaporative losses. Sap is obtained either by pressing it out of fresh tissue or releasing it from freeze-thawed tissue, then transferring it to a dew point or freezing point osmometer. Measurement of RWC is also a substantial undertaking under field conditions. Usually, tissue specimens are placed into preweighed vials and the vials are weighed soon after collection to obtain the initial fresh weight. Samples are then incubated in water for several hours to rehydrate fully, are blotted to remove unabsorbed surface water, weighed again, dried in an oven, and weighed a final time. With so many manipulations and measurements, the final calculated value for RWC inevitably incurs some loss of precision.
Another potential weakness of OA phenotyping is that it is a composite property that has many contributing components. A wide range of osmotically active solutes can contribute to osmotic potential and to OA. This can confound identification of OA QTLs and the genes responsible for OA. An alternative approach that is becoming ever more realistic with the advent of high-throughput analytical methods for large numbers of metabolites (“metabolomics”) is to analyze the main constituents that contribute to OA. There have only been a few studies where the substances contributing to the osmotic component of water potential have been determined (Table 3). The main categories of osmotic solute are sugars, mineral ions such as potassium, and nitrate, organic acids, and amino acids. In most cases, potassium along with its anion partners is the main class of osmotically active solute. In certain cases, such as the leaves of sorghum and maize, sucrose and other sugars are important contributors to OA, while the contributions of proline and other amino acids are relatively small on a bulk tissue basis. Hence, it is feasible to quantify each of the major contributors to OA in phenotyping projects.
Table 3.
Contributions to total solute potential by sugars, potassium (K) salts, and proline in mature leaves of plants subjected to water deficit.
| Family | Species | Common name | Osmotic adjustment (MPa) | Contribution to solute potential (MPa) |
Citation | ||
|---|---|---|---|---|---|---|---|
| Sugars | K-salts | Proline | |||||
| Asteracea | Helianthus annuus | Sunflower | 0.17 | −0.04 | −0.84 | nd | Jones et al. (1980) |
| Asteracea | Heteropogon contortus | Spear grass | 0.39 | −0.09 | −0.30 | −0.02 | Ford and Wilson (1981) |
| Brassicacea | Brassica napus, B. junea | Canola, juncea | 0.39 | −0.24 | −0.64 | −0.03 | Ma et al. (2004), Ma and Turner (2006) |
| Euphorbeacea | Manihot esculenta | cassava | 0.27 | −0.14 | −0.85 | 0.00 | Alves and Setter (2004) |
| Leguminacea | Cicer arietinum | Chickpea | 0.76 | 0.02 | nd | nd | Basu et al. (2007) |
| Leguminacea | Macroptilium atropurpuueum | Siratro | 0.34 | −0.09 | −0.41 | 0.00 | Ford and Wilson (1981) |
| Leguminacea | Trifolium alexandrinum | Berseem clover | 0.41 | −0.23 | −0.88 | −0.12 | Iannucci et al. (2002) |
| Leguminacea | Trifolium incarnatum | Crimson clover | 0.24 | −0.20 | −0.71 | −0.09 | Iannucci et al. (2002) |
| Leguminacea | Trifolium resupinatum | Persion clover | 0.20 | −0.20 | −0.70 | −0.08 | Iannucci et al. (2002) |
| Leguminacea | Trifolium squarrosum | Squarrosum clover | 0.52 | −0.30 | −1.11 | −0.11 | Iannucci et al. (2002) |
| Malvacea | Gossypium hirsutum | Cotton | 0.30 | −0.02 | −0.41 | nd | Cutler and Rains (1978) |
| Poacea | Cenchuus cillaris | Buffell grass | 0.71 | −0.05 | −0.68 | −0.02 | Ford and Wilson (1981) |
| Poacea | Panicum maximum | Green panic | 0.55 | −0.06 | −0.38 | −0.04 | Ford and Wilson (1981) |
| Poacea | Pennisetum glaucum | Pearl millet | 0.40 | −0.11 | −0.88 | −0.06 | Kusaka et al. (2005) |
| Poacea | Sorghum bicolor | Sorghum | 0.49 | −0.25 | −0.66 | nd | Jones et al. (1980) |
| Poacea | Triticum durum | Durum wheat | 0.02 | −0.16 | −0.86 | −0.06 | Bajji et al. (2001) |
| Poacea | Triticum durum | Durum wheat | 0.39 | −0.22 | −0.24 | −0.05 | Rascio et al. (1994) |
| Poacea | Triticum durum | Durum wheat | 0.08 | −0.16 | −0.74 | −0.18 | Kameli and Lösel (1995) |
| Rhamnacea | Ziziphus mauritiana | Indian Jujube | none | −0.13 | −0.23 | −0.02 | Arndt et al. (2000) |
| Rosacea | Prunus persica | Peach | 0.15 | −0.05 | −0.43 | 0.00 | Arndt et al. (2000) |
| Vitaceae | Vitus vinifera | Grape | 0.41 | −0.84 | −0.61 | nd | Patakas et al. (2002) |
Values are referenced to RWC=100%. Potassium is assumed to have an equivalent concentration of monovalent anion partner. (nd, not determined).
Measurement of osmotically active solutes
Determination of the component compounds which contribute to OA can be straightforward and done on a large scale. For example, in studies of maize, a population of over 200 recombinant inbred lines (RILs) was sampled at three stages of stress in 2 years with replication such that over 10,000 samples were analyzed (T. Setter and J.-M. Ribaut, unpublished data). Sampling involved cutting disks from mature leaves of several plants representative of each plot and immersing them in tubes containing ice-chilled 80% methanol. In this solvent, metabolic changes are halted by the cold and by the enzyme denaturing action of methanol. Samples for determination of osmolyte can be stored safely in the solvent for weeks, permitting the research team to schedule further work on them at a convenient time. In mature leaves with living cells (Ψw above the lethal point), the cell walls are elastic but sufficiently rigid to prevent significant shrinkage at the prevailing RWCs, so that passive concentration of solutes on a leaf area basis is not a significant contributor to the measured content of solutes.
Sugars
Several methods are available for analyzing organic osmolytes. If there is likelihood that a particular compound or related group of compounds are of primary interest, it is possible to use targeted assays. The analysis of sugars can be done using coupled enzyme reagents that use glucose oxidase/peroxidase, or hexokinase/glucose-6-phosphate-dehydrogenase, and invertase, to develop a chromogen whose color is determined in a 96-well colorimeter (Cairns, 1987; Setter et al., 2001; Alves and Setter, 2004). These steps are easy to scale up to the desired number of samples by using 12- or 96-channel pipetting and automated data transfer from colorimeter to computer. Three QTLs for leaf sugar accumulation were identified using this approach (Table 4). In support of the hypothesis that sugar accumulation contributed to leaf stability during water deficit, it was observed that the accumulation of leaf sugar had significant genetic correlation with the maintenance of leaf chlorophyll at 3 weeks into drought, as expected for a protective osmolyte effect (Table 4, lower panel).
Table 4.
Upper panel: QTLs identified for leaf sugar accumulation in a maize RIL population (Cimmyt P1 X P2) subjected to severe water deficit during flowering. Lower panel: genetic correlations between leaf sucrose and leaf chlorophyll.
| Trait | Year of trial | Chromosome bin of QTL | |
|---|---|---|---|
| Leaf suc 3W | 2001 | 8.04 | |
| Leaf suc 4W | 2001 | 8.04 | |
| Leaf glc 2W | 2001 | 8.04 | |
| Leaf suc 2W | 2001 | 8.04 | |
| Leaf glc 3W | 2002 | 8.04 | |
| Leaf glc 4W | 2002 | 8.04 | |
| Leaf suc 3W | 2002 | 8.04 | |
| Leaf total sugar 3W | 2002 | 8.04 | |
| Leaf total sugar 2W | 2001 | 9.04 | |
| Leaf total sugar 4W | 2001 | 9.04 | |
| Leaf suc 4W | 2001 | 9.04 | |
| Leaf suc 2W | 2002 | 9.04 | |
| Leaf total sugar 2W | 2002 | 9.04 | |
| Leaf total sugar 3W | 2002 | 9.04 | |
| Leaf suc 3W | 2002 | 9.04 | |
| Leaf suc 4W | 2001 | 10.03 | |
| Leaf total sugar 4W | 2001 | 10.03 | |
| Leaf glc 2W | 2002 | 10.03 | |
| Leaf suc 2W | 2002 | 10.03 | |
| Leaf suc 4W | 2002 | 10.03 | |
| Leaf total sugar 2W | 2002 | 10.03 | |
| Leaf total sugar 4W | 2002 | 10.03 | |
| Genetic correlations | Leaf suc 3W | ||
| Leaf chlorophyll 2W | 0.80 | ||
| Leaf chlorophyll 3W | 0.53 | ||
| Leaf chlorophyll 4W | 0.98 |
Trials were in 2001 and 2002 at Tlaltizapan field station, Mexico. Leaf disks were sampled at 2, 3, and 4 weeks after withholding irrigation (2W, 3W, 4W) and analyzed with coupled enzyme procedures for sucrose (suc), glucose (glc), and total sugar, expressed per unit leaf area. (Source: Setter and Ribaut, unpublished data).
Metabolomic approaches
An alternative approach to targeted assays is use methods that are capable of assaying a wide range of compounds in a single step. Such metabolomic approaches are capable of quantifying hundreds of metabolites in a single chromatographic operation, though they require a substantial budget and justification for using a broad exploration of constituent composition. Recent reviews describe a wide variety of powerful analytical systems that have been developed for this purpose (Stitt and Fernie, 2003; Sumner et al., 2003; Schauer and Fernie, 2006; Dixon et al., 2007; Shulaev et al., 2008; Saito and Matsuda, 2010; Hall, 2011). For the analysis of organic osmotic solutes, GC-MS with either quadrupole or time-of-flight (TOF) mode is capable of quantifying several hundred compounds of diverse classes including sugars, sugar alcohols, organic acids, amino acids, and fatty acids. However, samples require derivatization to make them volatile for GC analysis. HPLC-MS is a favored alternative not requiring derivatization and more suitable for unstable compounds (Tohge et al., 2011). For phenotyping a narrower range of compounds, nuclear magnetic resonance (NMR) has the advantage of not requiring separation and providing high-throughput.
Apart from sugars and sugar alcohols, the main contributors to the organic osmolyte pool are organic acids (Hummel et al., 2010), amino acids, and, in some species, quaternary ammonium compounds (QACs). High-throughput analysis of organic acids can also be performed using coupled enzyme procedures, as described above for sugars. For example, malate and citrate dehydrogenase and associated colorimetric reagents can be used. However, these assays operate in parallel mode. Thus, as more analytes are added to a project, assay of a full profile of them on a large set of samples involves splitting samples into aliquots and running numerous separate assays, which becomes cumbersome and costly. With appropriate choice of analytics, metabolomics methods are able to include these additional analytes.
There are a few published examples of metabolomics used in plants. Among those studies involving stress responses, Gagneul et al. (2007) studied the effect of salt treatment in the halophyte Limonium latifolium using a combination of GC-MS for organic substances, HPLC-fluorescence detection for amino acids, proton NMR for QACs, flame photometry for sodium and potassium, and salicylate-colorimetry for nitrate. They concluded that, contrary to expectations, organic solute accumulation is predominantly constitutive and only slightly modulated by salinity. The major contributors to osmolarity were inorganic solutes and, although present, the compatible solutes proline, QACs, and inositols were rather minor. Sanchez et al. (2008), studying Lotus japonicus, also used GC-MS to assess salinity effects on organic solutes, whereas they used ICP to analyze inorganic ions. They found that L. japonicus had broad shifts in metabolism in response to salinity, with decreases in potassium and organic acids, and increases in many amino acids, sugars, and polyols. Schauer et al. (2006) reported a QTL study of tomato fruit morphology and metabolites. By using GC-MS to quantify 74 metabolites, they identified 889 QTLs for metabolite levels, offering the possibility that this approach could be used to modify fruit composition and quality.
A further advantage of the metabolomics approach is that clusters of correlated metabolites may be identified that are either part of a common metabolic pathway or whose accumulation is coordinately regulated. For instance, in comparisons of barley cultivars differing in salt tolerance, Widodo et al. (2009) found that the tolerant cultivar responded to salinity by accumulating a broad range sugars, polyols, and organic acids and maintaining relatively low levels of amino acids. Using MS systems to quantify over 500 metabolites in Arabidopsis leaves subjected to water deficit, Urano et al. (2009) found that amino acid levels responded coordinately and involved ABA signaling, whereas raffinose oligosaccharide accumulation responded differently. Co-regulated clusters may provide a more reliable and robust phenotypic trait than any individual compound, thus providing more utility for phenotyping.
Potassium
The single most abundant osmotic solute in water stressed plant tissue is usually potassium. Flame photometry, atomic absorption spectrometry and inductively coupled plasma-atomic emission spectrometry (known as ICP) are usually the methods of choice (Munns et al., 2010).
For profiling the levels of potassium and other mineral ions, ICP spectroscopy is the most commonly used method. Although usually performed in relatively small numbers of samples, it can be automated to increase the rate of throughput, as has been done on large “ionomics” projects (Lahner et al., 2003; Salt et al., 2008).
A further consideration in the analysis of metabolites is that metabolite concentrations tend to be variable with respect to time of sampling, age of an organ, and environmental conditions. To characterize a genotype, investigators might be able to use full-scale metabolomics to identify a smaller set of key diagnostic compounds such that it is cost-effective to determine levels of a small number of key compounds in several organs and at several time-points rather than to determine the whole metabolic profile for just one sample. For phenotyping projects that involve hundreds or thousands of samples, it may be valuable to use more targeted, less expensive methods for all samples, and reserve full-scale metabolomics for a subset.
Antioxidants
In leaves, water deficit can result in excess electron flow to the production of reactive oxygen species (ROS) which in turn damages leaf membranes and proteins (Demmig-Adams and Adams, 2002). While high-throughput luminol chemiluminescence methods are available to assay the composite ROS levels in tissues subjected to stress, the functions of ROS are multifaceted and include signaling (Mittler et al., 2011), thereby complicating their interpretation. It is plausible that metabolomics may provide valuable phenotypic information on the spectrum of antioxidants and photoprotectants in a genotype. For example, studies indicate that the xanthophyll carotenoids perform a critical photoprotectant role (Demmig-Adams and Adams, 2002), and methods for metabolite profiling of carotenoids are now available (Fraser et al., 2007).
Compatible Solutes
Compatible solutes are small molecular weight osmolytes which are highly soluble in the cell solution and do not interfere with cellular metabolism, even at high concentrations. They differ in this regard from inorganic solutes, which can disrupt protein and membrane structure at high concentrations. Examples of compatible solutes include proline, QACs (glycine betaine), and sugar alcohols (mannitol, pinitol). Given their putative properties, they have been of interest with respect to genetic improvement of crops for a long time. The possibility of manipulating the levels of compatible solutes through transgenic metabolic engineering has been considered to be within grasp because the genes encoding their synthesis and catabolism have been cloned. While such overexpression transgenics have had limited success in improving drought tolerance (Abebe et al., 2003; Su et al., 2006; Chen and Murata, 2008; Szabados and Savoure, 2010; Sanchez et al., 2012), it might still be of interest to phenotype populations to identify alleles that contribute to compatible solute synthesis. In addition to the metabolite and protein assays described above, another strategy might be to analyze the activities of key enzymes in compatible solute synthesis or catabolism. Recently, methods have been developed to vastly increase the throughput of such assays, so that it is now feasible to use this approach in phenotyping. For example, Gibon et al. (2004) reported the development of a robot-based system for assay of the activity of 23 enzymes that are involved in central carbon and nitrogen metabolism.
Late Embryogenesis Abundant Proteins
Studies of plant organs that undergo extreme desiccation, such as dry seeds and resurrection plants, have led to the discovery of a broad class of proteins called “LEA proteins.” LEAs are highly expressed during desiccation and are thought to play a role in desiccation tolerance, although the mechanisms for this are not yet known. LEAs are unstructured, unfolded proteins that are highly hydrophilic and remain water soluble even when heated to 80°C. This is due to their high content of glycine, glutamic acid, glutamine and lysine the near absence of cysteine, and their unique peptide profiles (Wise and Tunnacliffe, 2004; Tunnacliffe and Wise, 2007). But there is evidence that during stress, LEAs can develop a secondary structure and interact with proteins and membranes, and this may explain their function in drying tissue. Under such conditions, they might serve a chaperone function to form three-dimensional structures that could provide stability to cellular protein and membrane systems, to prevent enzymes and other structures from aggregating and denaturing (Goyal et al., 2005). In the highly dehydrated state, they might also serve as a molecular shield to prevent excessively close proximity of proteins, or they might serve a water-binding role that helps create a more favorable environment for protein stability during desiccation. Studies of transgenic plants in which LEA polypeptides have been overexpressed have been reported to contribute to tolerance of water loss (Sivamani et al., 2000; Cheng et al., 2002; Xiao et al., 2007). Much of this work involves severe water loss, such as in desiccation. Hence it is possible that their roles only come into play when a tissue experiences severe dehydration, such as during advanced stages of a drought episode, where soil water is depleted to a severely low water potential and leaves desiccate to an even lower water potential. In these cases, LEA proteins may act together with compatible solutes to prevent tissue death, and permit subsequent rehydration to a viable state, if and when deeper rooting or precipitation occurs. Hence, with respect to crop growth during the vegetative stage, the situations in which they might be most important are where more tolerant genotypes use LEA polypeptides to help avoid lethal desiccation and ensuing cell death, such as in conditions where susceptible genotypes suffer from leaf firing and tassel blasting.
Late embryogenesis abundant levels have been measured using immunochemical approaches. By taking advantage of the fact that members of each LEA family of polypeptides have a few conserved domains, antibodies developed against a conserved domain are useful in detecting several members of the family (Close and Chandler, 1990). Assays using such antibodies have been developed for protein gel blots and ELISA (Jayaprakash et al., 1998; Volaire, 2003; Yang et al., 2007; Pinheiro et al., 2008). Recently, proteomic methods wherein polypeptides are separated by two-dimensional polyacrylamide gel electrophoresis followed by identification by liquid chromatography-mass spectrometry (LC-MS) have been used to analyze the levels of LEA polypeptides in plant tissues (March et al., 2007). As with other “omics” methods, proteomics provides the capability to survey the levels of the whole family of LEA polypeptides, which are numerous; for example, there are 50 members in Arabidopsis LEA family (Bies-Etheve et al., 2008).
Reproductive and Storage Organ Development
Many important traits that contribute to superior crop performance in stress environments involve better developmental regulatory systems whereby deep root growth is stimulated and lateral rooting decreased, or development of sink organs that will contribute to yield is sustained rather than aborted. These traits can be especially challenging to phenotype using analytic methods due to their complex regulatory networks and the large number of component factors. While quantification of easily scored morphological traits such as root length and plant height are straightforward, there may be value in quantifying processes that underpin these growth processes and the impact of stress on them. Methods to phenotype processes that contribute to seed-set, fruit-set and tuber-set are of interest for many crops, as are similar developmental attributes that are often diminished in stress, and genetic selection for these traits has been successful in improving crop yield in drought stress Ribaut et al. (2009). While morphological measures of these traits can be effective (e.g., ASI in maize, spikelet fertility in rice, and components of yield in most crops), it is possibly of interest to have diagnostic tests that would involve sampling tissue at the moment it is engaged in developmental decision-making, so that genotypes could be evaluated more directly for important stress tolerance behavior. For example, in maize and rice, a common response to water deficit at flowering is decreased cell wall invertase (INCW) activity (Ji et al., 2005; Boyer and McLaughlin, 2007). Given that expression of INCW represents one of many genes that are part of the growth-arrest syndrome, measurement of INCW levels can be considered a diagnostic test for this growth-arrest phenotype. Many other diagnostic test candidates can be considered, including other growth-specific proteins such as cyclins (for cell division), expansins (for cell expansion growth), and starch synthase (for starch accumulation).
In addition to direct participants in growth, the level of expression of key signaling or transcription factor proteins could also be considered valuable in this phenotyping strategy. Candidates for diagnostic tests might be identified from transcript profiling studies that are being carried out on reproductive tissue exposed to drought. For example, studies by Yu and Setter (2003) in maize and Agarwal et al. (2007) in rice have profiled transcription in pedicels, endosperms, and panicles during their early stage of development, and have found sets of transcripts that are consistently up- or downregulated in response to stress and studies by Fujita et al. (2010) have identified transcripts expressed at specific stages of reproductive organ development. In a review of over 100 transcriptomic studies of plant response to drought and salinity, Deyholos (2010) concluded that with future improvements, transcriptomics could be valuable as a screening tool for candidate gene discovery. This approach is consistent with the strategy of identifying a small number of key transcripts (e.g., Rabbani et al., 2003; Boyer and McLaughlin, 2007) which could be used to construct a diagnostic test of stress tolerance attributes for use in phenotyping large populations. Tools for handling hundreds of RNA samples are available. For example, real-time polymerase chain reaction (RT-PCR) can be run on 384 plates fairly economically and reliably.
Other methods are under development to extend transcriptomic assays from being highly focused on just a few samples (profiling tens of thousands of genes) toward a capability of efficiently handling thousands of samples. An advantage of such an approach is that it might reveal genetic variability for processes underlying the decision-making involved in reproductive set versus abortion. This approach could be particularly effective for developmental traits such as flower and seed-set, which involve complex signaling and gene expression networks rather than clear-cut metabolic pathways. Methods for morphological measurement of growth or components of yield at final harvest as currently used might miss some of this information. As the cost of transcript profiling by RNA sequencing (RNAseq) and other genomics methods continue to decrease, the prospects of using these methods for phenotyping will be enhanced. This may enable clustering of RNAseq (or proteomic) data points to identify higher order patterns in profiles of gene expression that correlate with superior stress tolerance. Such systems approaches might capture trait information on a complex process such as sustained floral development that could not be explained by quantifying a small set of transcripts.
Conclusion
Investigators now have a wide range of analytical tools to use in measuring metabolites, proteins and transcripts in plant tissues. The tools range from inexpensive to costly, from single-purpose to broad based profiling in “omic” mode. The most appropriate choice for phenotyping will depend on a project’s goals, the relative merit of each analytical approach, the cost, and any trade-offs between phenotyping a large number of entries at low cost per analysis versus a smaller number in great detail.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abebe T., Guenzi A. C., Martin B., Cushman J. C. (2003). Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol. 131, 1748–1755 10.1104/pp.102.003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P., Arora R., Ray S., Singh A. K., Singh V. P., Takatsuji H., Kapoor S., Tyagi A. K. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485 10.1007/s11103-007-9199-y [DOI] [PubMed] [Google Scholar]
- Alves A. A. C., Setter T. L. (2004). Abscisic acid accumulation and osmotic adjustment in cassava under water deficit. Environ. Exp. Bot. 51, 259–271 10.1016/j.envexpbot.2003.11.005 [DOI] [Google Scholar]
- Arndt S. K., Wanek W., Clifford S. C., Popp M. (2000). Contrasting adaptations to drought stress in field-grown Ziziphus mauritiana and Prunus persica trees: water relations, osmotic adjustment and carbon isotope composition. Aust. J. Plant Physiol. 27, 985–996 [Google Scholar]
- Babu R., Pathan M., Blum A., Nguyen H. (1999). Comparison of measurement methods of osmotic adjustment in rice cultivars. Crop Sci. 39, 150–158 10.2135/cropsci1999.0011183X003900010024x [DOI] [Google Scholar]
- Bajji M., Lutts S., Kinet J.-M. (2001). Water deficit effects on solute contribution to osmotic adjustment as a function of leaf ageing in three durum wheat (Triticum durum Desf.) cultivars performing differently in arid conditions. Plant Sci. 160, 669–681 10.1016/S0168-9452(00)00443-X [DOI] [PubMed] [Google Scholar]
- Banowetz G. M., Hess J. R., Carman J. G. (1994). A monoclonal antibody against the plant growth regulator, abscisic acid. Hybridoma 13, 537–541 [DOI] [PubMed] [Google Scholar]
- Basu P.-S., Berger J. D., Turner N. C., Chaturvedi S. K., Ali M., Siddique K. H. M. (2007). Osmotic adjustment of chickpea (Cicer arietinum) is not associated with changes in carbohydrate composition or leaf gas exchange under drought. Ann. Appl. Biol. 150, 217–225 10.1111/j.1744-7348.2007.00119.x [DOI] [Google Scholar]
- Bies-Etheve N., Gaubier-Comella P., Debures A., Lasserre E., Jobet E., Raynal M., Cooke R., Delseny M. (2008). Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 107–124 10.1007/s11103-008-9304-x [DOI] [PubMed] [Google Scholar]
- Boyer J. S., McLaughlin J. E. (2007). Functional reversion to identify controlling genes in multigenic responses: analysis of floral abortion. J. Exp. Bot. 58, 267–277 10.1093/jxb/erl177 [DOI] [PubMed] [Google Scholar]
- Cairns A. J. (1987). Colorimetric microtiter plate assay of glucose and fructose by enzyme-linked formazan production: applicability to the measurement of fructosyl transferase activity in higher plants. Anal. Biochem. 167, 270–278 10.1016/0003-2697(87)90163-1 [DOI] [PubMed] [Google Scholar]
- Chen T. H. H., Murata N. (2008). Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci. 13, 499–505 10.1016/j.tplants.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Cheng Z., Targolli J., Huang X., Wu R. (2002). Wheat LEA genes, PMA80 and PMA1959, enhance dehydration tolerance of transgenic rice (Oryza sativa L.). Mol. Breed. 10, 71–82 10.1023/A:1020329401191 [DOI] [Google Scholar]
- Chiwocha S. D. S., Abrams S. R., Ambrose S. J., Cutler A. J., Loewen M., Ross A. R. S., Kermode A. R. (2003). A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. Plant J. 35, 405–417 10.1046/j.1365-313X.2003.01800.x [DOI] [PubMed] [Google Scholar]
- Close T. J., Chandler P. M. (1990). Cereal dehydrins serology gene mapping and potential functional roles. Aust. J. Plant Physiol. 17, 333–344 10.1071/PP9900333 [DOI] [Google Scholar]
- Condon A. G., Reynolds M. P., Rebetzke G. J., van Ginkel M., Richards R. A., Farquhar G. D. (2007). “Using stomatal aperture-related traits to select for high yield potential in bread wheat,” in Developments in Plant Breeding, Vol. 12, eds Buck H. T., Nisi J. E., Salomón N. (Dordrecht: Springer; ), 617–624 [Google Scholar]
- Condon A. G., Richards R. A., Rebetzke G. J., Farquar G. D. (2004). Breeding for high water-use efficiency. J. Exp. Bot. 55, 2447–2460 10.1093/jxb/erh277 [DOI] [PubMed] [Google Scholar]
- Cutler J. M., Rains D. W. (1978). Effects of water stress and hardening on the internal water relations and osmotic constituents of cotton leaves. Physiol. Plant. 42, 261–268 10.1111/j.1399-3054.1978.tb02558.x [DOI] [Google Scholar]
- Demmig-Adams B., Adams W. W., III. (2002). Antioxidants in photosynthesis and human nutrition. Science 298, 2149–2153 10.1126/science.1078002 [DOI] [PubMed] [Google Scholar]
- Deyholos M. K. (2010). Making the most of drought and salinity transcriptomics. Plant Cell Environ. 33, 648–654 10.1111/j.1365-3040.2009.02092.x [DOI] [PubMed] [Google Scholar]
- Diab A. A., Teulat-Merah B., This D., Ozturk N. Z., Benscher D., Sorrells M. E. (2004). Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor. Appl. Genet. 109, 1417–1425 10.1007/s00122-004-1755-0 [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Bouton J. H., Narasimhamoorthy B., Saha M., Wang Z.-Y., Maya G. D. (2007). Beyond structural genomics for plant science. Adv. Agronomy 95, 77–161 10.1016/S0065-2113(07)95002-6 [DOI] [Google Scholar]
- Ford C. W., Wilson J. R. (1981). Changes in levels of solutes during osmotic adjustment to water stress in leaves of 4 tropical pasture species. Aust. J. Plant Physiol. 8, 77–92 10.1071/PP9810077 [DOI] [Google Scholar]
- Fraser P. D., Enfissi E. M. A., Goodfellow M., Eguchi T., Bramley P. M. (2007). Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant J. 49, 552–564 10.1111/j.1365-313X.2006.02949.x [DOI] [PubMed] [Google Scholar]
- Fujita M., Horiuchi Y., Ueda Y., Mizuta Y., Kubo T., Yano K., Yamaki S., Tsuda K., Nagata T., Niihama M., Kato H., Kikuchi S., Hamada K., Mochizuki T., Ishimizu T., Iwai H., Tsutsumi N., Kurata N. (2010). Rice expression atlas in reproductive development. Plant Cell Physiol. 51, 2060–2081 10.1093/pcp/pcq165 [DOI] [PubMed] [Google Scholar]
- Gagneul D., Aienouche A., Duhaze C., Lugan R., Larher F. R., Bouchereau A. (2007). A reassessment of the function of the so-called compatible solutes in the halophytic Plumbaginaceae Limonium latifolium. Plant Physiol. 144, 1598–1611 10.1104/pp.107.099820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergs U., Hagemann K., Zeevaart J. A. D., Weiler E.-W. (1993). The determination of phaseic acid by monoclonal antibody-based enzyme immunoassay. Bot. Acta 106, 404–410 [Google Scholar]
- Gibon Y., Blaesing O. E., Hannemann J., Carillo P., Hohne M., Hendriks J. H. M., Palacios N., Cross J., Selbig J., Stitt M. (2004). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16, 3304–3325 10.1105/tpc.104.025973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani S., Sanguineti M. C., Tuberosa R., Bellotti M., Salvi S., Landi P. (2005). Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J. Exp. Bot. 56, 3061–3070 10.1093/jxb/eri303 [DOI] [PubMed] [Google Scholar]
- Goyal K., Walton L.-J., Tunnacliffe A. (2005). LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157 10.1042/BJ20041931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. D. (2011). “Plant metabolomics in a nutshell: potential and future challenges,” in Annual Plant Reviews, Vol. 43, ed. Hall R. D. (Ames: Wiley-Blackwell; ), 1–24 [Google Scholar]
- Harjes C. E., Rocheford T. R., Bai L., Brutnell T. P., Kandianis C. B., Sowinski S. G., Stapleton A. E., Vallabhaneni R., Williams M., Wurtzel E. T., Yan J., Buckler E. S. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333 10.1126/science.1150255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hradecka V., Novak O., Havlicek L., Strnad M. (2007). Immunoaffinity chromatography of abscisic acid combined with electrospray liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 847, 162–173 10.1016/j.jchromb.2006.09.034 [DOI] [PubMed] [Google Scholar]
- Hummel I., Pantin F., Sulpice R., Piques M., Rolland G., Dauzat M., Christophe A., Pervent M., Bouteille M., Stitt M., Gibon Y., Muller B. (2010). Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol. 54, 357–372 10.1104/pp.110.157008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannucci A., Russo M., Arena L., Di-Fonzo N., Martiniello P. (2002). Water deficit effects on osmotic adjustment and solute accumulation in leaves of annual clovers. Eur. J. Agron. 16, 111–122 10.1016/S1161-0301(01)00121-6 [DOI] [Google Scholar]
- Jayaprakash T. L., Ramamohan G., Krishnaprasad B. T., Ganeshkumar, Prasad T. G., Mathew M. K., Udayakumar M. (1998). Genotypic variability in differential expression of lea2 and lea3 genes and proteins in response to salinity stress in fingermillet (Eleusine coracana Gaertn) and rice (Oryza sativa L.) seedlings. Ann. Bot. 82, 513–522 10.1006/anbo.1998.0712 [DOI] [Google Scholar]
- Ji X., Dong B., Shiran B., Talbot M. J., Edlington J. E., Hughes T., White R. G., Gubler F., Dolferus R. (2011). Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol. 156, 647–662 10.1104/pp.111.176164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X.-M., Raveendran M., Oane R., Ismail A., Lafitte R., Bruskiewich R., Cheng S.-H., Bennett J. (2005). Tissue-specific expression and drought responsiveness of cell-wall invertase genes of rice at flowering. Plant Mol. Biol. 59, 945–964 10.1007/s11103-005-2415-8 [DOI] [PubMed] [Google Scholar]
- Jones M. M., Osmond C. B., Turner N. C. (1980). Accumulation of solutes in leaves of sorghum Sorghum-bicolor and sunflower Helianthus-annuus cultivar Hysun-10 in response to water deficits. Aust. J. Plant Physiol. 7, 193–206 10.1071/PP9800181 [DOI] [Google Scholar]
- Kalantari S., Turcotte C., Fliss I., Makhlouf J., Arul J. (2001). A new indirect ELISA method to measure abscisic acid. HortScience 36, 546 [Google Scholar]
- Kameli A., Lösel D. M. (1995). Contribution of carbohydrates and other solutes to osmotic adjustment in wheat leaves under water stress. J. Plant Physiol. 145, 363–366 10.1016/S0176-1617(11)81903-6 [DOI] [Google Scholar]
- Kiani S. P., Talia P., Maury P., Grieu P., Heinz R., Perrault A., Nishinakamasu V., Hopp E., Gentzbittel L., Paniego N., Sarrafi A. (2007). Genetic analysis of plant water status and osmotic adjustment in recombinant inbred lines of sunflower under two water treatments. Plant Sci. 172, 773–787 10.1016/j.plantsci.2006.12.007 [DOI] [Google Scholar]
- Kusaka M., Ohta M., Fujimura T. (2005). Contribution of inorganic components to osmotic adjustment and leaf folding for drought tolerance in pearl millet. Physiol. Plant 125, 474–489 10.1111/j.1399-3054.2005.00578.x [DOI] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E. L., Abid K. B., Rogers E. E., Guerinot M. L., Harper J. F., Ward J. M., McIntyre L., Schroeder J. I., Salt D. E. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21, 1215–1221 10.1038/nbt865 [DOI] [PubMed] [Google Scholar]
- Landi P., Sanguineti M. C., Liu C., Li Y., Wang T. Y., Giuliani S., Bellotti M., Salvi S., Tuberosa R. (2007). Root-ABA1 QTL affects root lodging, grain yield, and other agronomic traits in maize grown under well-watered and water-stressed conditions. J. Exp. Bot. 58, 319–326 10.1093/jxb/erl161 [DOI] [PubMed] [Google Scholar]
- Lebreton C., Lazic-Jancic V., Steed A., Pekic S., Quarrie S. A. (1995). Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J. Exp. Bot. 46, 853–865 10.1093/jxb/46.7.853 [DOI] [Google Scholar]
- Leport L., Turner N. C., Davies S. L., Siddique K. H. M. (2006). Variation in pod production and abortion among chickpea cultivars under terminal drought. Eur. J. Agron. 24, 236–246 10.1016/j.eja.2005.08.005 [DOI] [Google Scholar]
- Lilley J. M., Ludlow M. M., McCouch S. R., O’Toole J. C. (1996). Locating QTL for osmotic adjustment and dehydration tolerance in rice. J. Exp. Bot. 47, 1427–1436 10.1093/jxb/47.9.1427 [DOI] [Google Scholar]
- Liu F., Jensen C. R., Andersen M. N. (2004). Pod set related to photosynthetic rate and endogenous aba in soybeans subjected to different water regimes and exogenous ABA and BA at early reproductive stages. Ann. Bot. 94, 405–411 10.1093/aob/mch157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-X., Liao D.-Q., Oane R., Estenor L., Yang X. E., Li Z.-C., Bennett J. (2006). Genetic variation in the sensitivity of anther dehiscence to drought stress in rice. Field Crops Res. 97, 87–100 10.1016/j.fcr.2005.08.019 [DOI] [Google Scholar]
- Ma Q., Turner D. W. (2006). Osmotic adjustment segregates with and is positively related to seed yield in F3 lines of crosses between Brassica napus and B. juncea subjected to water deficit. Aust. J. Exp. Agric. 46, 1621–1627 10.1071/EA05247 [DOI] [Google Scholar]
- Ma Q., Turner D. W., Levy D., Cowling W. A. (2004). Solute accumulation and osmotic adjustment in leaves of Brassica oilseeds in response to soil water deficit. Aust. J. Agric. Res. 55, 939–945 10.1071/AR03183 [DOI] [Google Scholar]
- March T. J., Able J. A., Schultz C. J., Able A. J. (2007). A novel late embryogenesis abundant protein and peroxidase associated with black point in barley grains. Proteomics 7, 3800–3808 10.1002/pmic.200700456 [DOI] [PubMed] [Google Scholar]
- Matthews R. B., Reddy D. M., Rani A. U., Azam-Ali S. N., Peacock J. M. (1990). Response of four sorghum lines to mid-season drought. I. Growth, water use and yield. Field Crops Res. 25, 279–296 10.1016/0378-4290(90)90011-Y [DOI] [Google Scholar]
- Melkonian J. J., Wolfe J., Steponkus P. L. (1982). Determination of the volumetric modulus of elasticity of wheat Triticum-aestivum cultivar yorkstar leaves by pressure volume relations and the effect of drought conditioning. Crop Sci. 22, 116–123 10.2135/cropsci1982.0011183X002200010027x [DOI] [Google Scholar]
- Mertens R., Deus-Neumann B., Weiler E. W. (1983). Mono clonal antibodies for the detection and quantitation of the endogenous plant growth regulator abscisic-acid. FEBS Lett. 160, 269–272 10.1016/0014-5793(83)80980-6 [DOI] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Mugo S. N., Smith M. E., Banziger M., Setter T. L., Edmeades G. O., Elings A. (1998). Performance of early maturing katumani and kito maize composites under drought at the seedling and flowering stages. Afr. Crop Sci. J. 6, 329–344 [Google Scholar]
- Munns R., Wallace P. A., Teakle N. L., Colmer T. D. (2010). “Measuring soluble ion concentrations (Na+, K+, Cl-) in salt-treated plants,” in Plant Stress Tolerance, Methods in Molecular Biology, ed. Sunkar R. (New York: Springer Science+Business Media; ), 371–382 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 10.1146/annurev.arplant.56.032604.144046 [DOI] [PubMed] [Google Scholar]
- Ober E., Sharp R. (2007). “Regulation of root growth responses to water deficit,” in Advances in Molecular Breeding Towards Salinity and Drought Tolerance, eds Jenks M. A., Hasegawa P. M., Jain S. M. (Dordrecht: Springer; ), 33–53 [Google Scholar]
- Ober E. S., Setter T. L., Madison J. T., Thompson J. F., Shapiro P. S. (1991). Influence of water deficit on maize endosperm development: enzyme activities and rna transcripts of starch and zein synthesis, abscisic acid, and cell division. Plant Physiol. 97, 154–164 10.1104/pp.97.1.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patakas A., Nikolaou N., Zioziou E., Radoglou K., Noitsakis B. (2002). The role of organic solute and ion accumulation in osmotic adjustment in drought-stressed grapevines. Plant Sci. 163, 361–367 10.1016/S0168-9452(02)00140-1 [DOI] [Google Scholar]
- Perata P., Vernieri P., Armellini D., Bugnoli M., Presentini R., Picciarelli P., Alpi A., Tognoni F. (1990). A monoclonal antibody for the detection of conjugated forms of abscisic acid in plant tissues. J. Plant Growth Regul. 9, 1–6 10.1007/BF02041934 [DOI] [Google Scholar]
- Philosoph-Hadas S., Hadas E., Aharoni N. (1993). Characterization and use in ELISA of a new monoclonal antibody for quantitation of abscisic acid in senescing rice leaves. Plant Growth Regul. 12, 71–78 10.1007/BF00144585 [DOI] [Google Scholar]
- Pinheiro C., de Carvalho M. H. C., Bartels D., Ricardo A. P., Chaves M. M. (2008). Dehydrins in Lupinus albus: pattern of protein accumulation in response to drought. Funct. Plant Biol. 35, 85–91 10.1071/FP07179 [DOI] [PubMed] [Google Scholar]
- Priest D. M., Ambrose S. J., Vaistij F. E., Elias L., Higgins G. S., Ross A. R. S., Abrams S. R., Bowles D. J. (2006). Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 46, 492–502 10.1111/j.1365-313X.2006.02701.x [DOI] [PubMed] [Google Scholar]
- Quarrie S. A., Whitford P. N., Appleford N. E. J., Wang T. L., Cook S. K., Henson I. E., Loveys B. R. (1988). A monoclonal antibody to s abscisic acid its characterization and use in a radioimmunoassay for measuring abscisic acid in crude extracts of cereal and lupine leaves. Planta 173, 330–339 10.1007/BF00401020 [DOI] [PubMed] [Google Scholar]
- Quatrano R. S., Bartels D., Ho T.-H. D., Pages M. (1997). New insights into ABA-mediated process. Plant Cell 9, 470–475 10.2307/3870499 [DOI] [Google Scholar]
- Rabbani M.-A., Maruyama K., Abe H., Khan M.-A., Katsura K., Ito Y., Yoshiwara K., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133, 1755–1767 10.1104/pp.103.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascio A., Platani C., Scalfati G., Tonti A., Fonzo N. (1994). The accumulation of solutes and water binding strength in durum wheat. Physiol. Plant. 90, 715–721 10.1034/j.1399-3054.1994.900413.x [DOI] [Google Scholar]
- Ren H., Gao Z., Chen L., Wei K., Liu J., Fan Y., Davies W. J., Jia W., Zhang J. (2007). Dynamic analysis of ABA accumulation in relation to the rate of ABA catabolism in maize tissues under water deficit. J. Exp. Bot. 58, 211–219 10.1093/jxb/erl117 [DOI] [PubMed] [Google Scholar]
- Ribaut J.-M., Betran J., Monneveux P., Setter T. (2009). “Drought tolerance in maize,” in Handbook of Maize: Its Biology, eds Bennetzen J. L., Hake S. C. (New York: Springer; ), 311–344 [Google Scholar]
- Robin S., Pathan M. S., Courtois B., Lafitte R., Carandang S., Lanceras S., Amante M., Nguyen H. T., Li Z. (2003). Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theor. Appl. Genet. 107, 1288–1296 10.1007/s00122-003-1360-7 [DOI] [PubMed] [Google Scholar]
- Ross A. R. S., Ambrose S. J., Cutler A. J., Feurtado J. A., Kermode A. R., Nelson K., Zhou R., Abrams S. R. (2004). Determination of endogenous and supplied deuterated abscisic acid in plaint tissues by high-performance liquid chromatography-electro-spray ionization tandem mass spectrometry with multiple reaction monitoring. Anal. Biochem. 329, 324–333 10.1016/j.ab.2004.02.026 [DOI] [PubMed] [Google Scholar]
- Saito K., Matsuda F. (2010). Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489 10.1146/annurev.arplant.043008.092035 [DOI] [PubMed] [Google Scholar]
- Salt D. E., Baxter I., Lahner B. (2008). Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 59, 709–733 10.1146/annurev.arplant.59.032607.092942 [DOI] [PubMed] [Google Scholar]
- Sanchez D. H., Lippold F., Redestig H., Hannah M. A., Erban A., Kraemer U., Kopka J., Udvardi M. K. (2008). Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 53, 973–987 10.1111/j.1365-313X.2007.03381.x [DOI] [PubMed] [Google Scholar]
- Sanchez D. H., Schwabe F., Erban A., Udvardi M. K., Kopka J. (2012). Comparative metabolomics of drought acclimation in model and forage legumes. Plant Cell Environ. 35, 136–149 10.1111/j.1365-3040.2011.02423.x [DOI] [PubMed] [Google Scholar]
- Saranga Y., Jiang C. X., Wright R. J., Yakir D., Paterson A. H. (2004). Genetic dissection of cotton physiological responses to arid conditions and their inter-relationships with productivity. Plant Cell Environ. 27, 263–277 10.1111/j.1365-3040.2003.01134.x [DOI] [Google Scholar]
- Saranga Y., Menz M., Jiang C.-X., Wright R. J., Yakir D., Paterson A. H. (2001). Genomic dissection of genotype X environment interactions conferring adaptation of cotton to arid conditions. Genome Res. 11, 1988–1995 10.1101/gr.157201 [DOI] [PubMed] [Google Scholar]
- Schauer N., Fernie A. R. (2006). Plant metabolomics: towards biological function and mechanism. Trends Plant Sci. 11, 508–516 10.1016/j.tplants.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., Pleban T., Perez-Melis A., Bruedigam C., Kopka J., Willmitzer L., Zamir D., Fernie A. R. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454 10.1038/nbt1192 [DOI] [PubMed] [Google Scholar]
- Setter T. L., Flannigan B. A., Melkonian J. (2001). Loss of kernel set due to water deficit and shade in maize: carbohydrate supplies, abscisic acid, and cytokinins. Crop Sci. 41, 1530–1540 10.2135/cropsci2001.4151530x [DOI] [Google Scholar]
- Setter T. L., Parra R. (2010). Relationship of carbohydrate and abscisic acid levels to kernel set in maize under post pollination water deficit. Crop Sci. 50, 980–988 10.2135/cropsci2009.07.0391 [DOI] [Google Scholar]
- Setter T. L., Yan J., Warburton M., Ribaut J.-M., Xu Y., Sawkins M., Buckler E. S., Zhang Z., Gore M. A. (2011). Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. J. Exp. Bot. 62, 701–716 10.1093/jxb/erq308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R. E., Wu Y., Voetberg G. S., Saab I. N., Lenoble M. E. (1994). Confirmation that abscisic acid accumulation is required for maize primary root elongation at low water potentials. J. Exp. Bot. 45, 1743–1751 [Google Scholar]
- Shulaev V., Cortes D., Miller G., Mittler R. (2008). Metabolomics for plant stress response. Physiol. Plant 132, 199–208 10.1111/j.1399-3054.2007.01025.x [DOI] [PubMed] [Google Scholar]
- Sivamani E., Bahieldin A., Wraith J. M., Al-Niemi T., Dyer W. E., Ho T.-H. D., Qu R. (2000). Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155, 1–9 10.1016/S0168-9452(99)00247-2 [DOI] [PubMed] [Google Scholar]
- Stitt M., Fernie A. R. (2003). From measurements of metabolites to metabolomics: an ‘on the fly’ perspective illustrated by recent studies of carbon-nitrogen interactions. Curr. Opin. Biotechnol. 14, 136–144 10.1016/S0958-1669(03)00023-5 [DOI] [PubMed] [Google Scholar]
- Su J., Hirji R., Zhang L., He C., Selvaraj G., Wu R. (2006). Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. J. Exp. Bot. 57, 1129–1135 10.1093/jxb/erj133 [DOI] [PubMed] [Google Scholar]
- Sumner L. W., Mendes P., Dixon R. A. (2003). Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62, 817–836 10.1016/S0031-9422(02)00708-2 [DOI] [PubMed] [Google Scholar]
- Szabados L., Savoure A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- Teulat B., Borries C., This D. (2001). New QTLs identified for plant water status, water-soluble carbohydrate and osmotic adjustment in a barley population grown in a growth-chamber under two water regimes. Theor. Appl. Genet. 103, 161–170 10.1007/s001220100619 [DOI] [Google Scholar]
- Teulat B., This D., Khairallah M., Borries C., Ragot C., Sourdille P., Leroy P., Monneveux P., Charrier A. (1998). Several QTLs involved in osmotic-adjustment trait variation in barley (Hordeum vulgare L.). Theor. Appl. Genet. 96, 88–698 10.1007/s001220050790 [DOI] [Google Scholar]
- Tohge T., Mettler T., Arrivault S., Carroll A. J., Stitt M., Fernie A. (2011). From models to crop species: caveats and solutions for translational metabolomics. Front. Plant Sci. 2:61. 10.3389/fpls.2011.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A., Wise M. J. (2007). The continuing conundrum of the LEA proteins. Naturwissenschaften 94, 791–812 10.1007/s00114-007-0254-y [DOI] [PubMed] [Google Scholar]
- Urano K., Maruyama K., Ogata Y., Morishita Y., Takeda M., Sakurai N., Suzuki H., Saito K., Shibata D., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57, 1065–1078 10.1111/j.1365-313X.2008.03748.x [DOI] [PubMed] [Google Scholar]
- Vernieri P., Perata P., Armellini D., Bugnoli M., Presentini R., Lorenzi R., Ceccarelli N., Alpi A., Tognoni F. (1989). Solid phase radioimmunoassay for the quantitation of abscisic acid in plant crude extracts using a new monoclonal antibody. J. Plant Physiol. 134, 441–446 10.1016/S0176-1617(89)80007-0 [DOI] [Google Scholar]
- Volaire F. (2003). Seedling survival under drought differs between an annual (Hordeum vulgare) and a perennial grass (Dactylis glomerata). New Phytol. 160, 501–510 10.1046/j.1469-8137.2003.00906.x [DOI] [PubMed] [Google Scholar]
- Walker-Simmons M. (1987). Aba levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–66 10.1104/pp.84.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ying J., Kuzma M., Chalifoux M., Sample A., McArthur C., Uchacz T., Sarvas C., Wan J., Dennis D. T., McCourt P., Huang Y. (2005). Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 43, 413–424 10.1111/j.1365-313X.2005.02463.x [DOI] [PubMed] [Google Scholar]
- Westgate M. E., Passioura J. B., Munns R. (1996). Water status and ABA content of floral organs in drought-stressed wheat. Aust. J. Plant Physiol. 23, 763–772 10.1071/PP9960763 [DOI] [Google Scholar]
- Widodo, Patterson J. H., Newbigin E., Tester M., Bacic A., Roessner U. (2009). Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, sahara and clipper, which differ in salinity tolerance. J. Exp. Bot. 60, 4089–4103 10.1093/jxb/erp243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M. (2003). LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4, 52. 10.1186/1471-2105-4-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M. J., Tunnacliffe A. (2004). POPP the question: what do LEA proteins do? Trends Plant Sci. 9, 13–17 10.1016/j.tplants.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Xiao B., Huang Y., Tang N., Xiong L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46 10.1007/s00122-007-0583-4 [DOI] [PubMed] [Google Scholar]
- Xie Z., Zheng Z.-F., Chen P.-Y., Zhang D. (1996). Preparation and application of monoclonal antibodies specific for abscisic acid methyl ester. Zhi Wu Sheng Li Xue Bao 22, 284–290 [Google Scholar]
- Yang S. H., Zeevaart J. A. D. (2006). Expression of ABA 8’-hydroxylases in relation to leaf water relations and seed development in bean. Plant J. 47, 675–686 10.1111/j.1365-313X.2006.02811.x [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang L.-S., Zhang X.-J., Shan L. (2007). Expression and purification of a dehydrin gene of wheat and preparation of its antibody. Prog. Biochem. Biophy. 34, 960–964 [Google Scholar]
- Yu L.-X., Setter T. L. (2003). Comparative transcriptional profiling of placenta and endosperm in developing maize kernels in response to water deficit. Plant Physiol. 131, 568–582 10.1104/pp.014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia L. I., Abrams S. (2011). Plant Hormone Profiling Available at: http://www.nrc-cnrc.gc.ca/eng/facilities/pbi/plant-hormone.html [accessed December 25, 2011].
- Zhang J., Zheng H. G., Aarti A., Pantuwan G., Nguyen T. T., Tripathy J. N., Sarial A. K., Robin S., Babu R. C., Nguyen B. D., Sarkarung S., Blum A., Nguyen H. T. (2001). Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor. Appl. Genet. 103, 19–29 10.1007/s001220100602 [DOI] [Google Scholar]