Abstract
Gene expression can be regulated by chromatin modifiers, transcription factors and proteins that modulate DNA architecture. Among the latter, AT-hook transcription factors have emerged as multifaceted regulators that can activate or repress broad A/T-rich gene networks. Thus, alterations of AT-hook genes could affect the transcription of multiple genes causing global cell dysfunction. Here we report that targeted deletions of mouse AKNA, a hypothetical AT-hook-like transcription factor, sensitize mice to pathogen-induced inflammation and cause sudden neonatal death. Compared with wild-type littermates, AKNA KO mice appeared weak, failed to thrive and most died by postnatal day 10. Systemic inflammation, predominantly in the lungs, was accompanied by enhanced leukocyte infiltration and alveolar destruction. Cytologic, immunohistochemical and molecular analyses revealed CD11b+Gr1+ neutrophils as major tissue infiltrators, neutrophilic granule protein, cathelin-related antimicrobial peptide and S100A8/9 as neutrophil-specific chemoattracting factors, interleukin-1β and interferon-γ as proinflammatory mediators, and matrix metalloprotease 9 as a plausible proteolytic trigger of alveolar damage. AKNA KO bone marrow transplants in wild-type recipients reproduced the severe pathogen-induced reactions and confirmed the involvement of neutrophils in acute inflammation. Moreover, promoter/reporter experiments showed that AKNA could act as a gene repressor. Our results support the concept of coordinated pathway-specific gene regulation functions modulating the intensity of inflammatory responses, reveal neutrophils as prominent mediators of acute inflammation and suggest mechanisms underlying the triggering of acute and potentially fatal immune reactions.
Keywords: in vivo, mouse genetics, gene network regulation, inflammation, pathogens, innate reactions
Introduction
Cell responses to extracellular cues depend on coordinated and timely regulation of pathway-specific gene networks, whose expression is controlled by master regulators 1, 2, 3. Examples of such regulators include AT-hook proteins, a family of transcription factors that target genes with A/T-rich promoters 4, 5, 6 and orchestrate their activation or repression 7, 8, 9, 10, 11. Notably, AT-hook proteins have been mainly recognized within the high mobility group (HMG) family 12, 13. However, non-HMG AT-hook proteins continue to be identified and characterized 14, 15. Among these, proteins with AT-hook-like motifs (ALMs) have been found to be capable of binding A/T-rich gene targets and regulating their transcription 16, 17, 18. In keeping with this notion, the human AKNA gene is a non-HMG transcription factor with N- and C-terminus AT-hook motifs 9, 19, whereas its murine ortholog depicts putative N-terminus AT-hook and C-terminus ALM sequences. Although the structural identity of murine AKNA's ALM remains hypothetical and its physiopathological resemblance to the human protein needs to be established, inferred evidence 9, 19 suggests that mouse AKNA may be able to bind pathway-specific genes and regulate their expression. Thus, it is possible that AKNA expression plays a role in mechanisms that, if altered, could result in systemic and potentially fatal disorders. In keeping with this hypothesis, human AKNA is encoded by a single gene located within the FRA9E region of chromosome 9q32 19, a common fragile site (CFS) linked to loss-of-function mutations that often lead to inflammatory and neoplastic diseases 20, 21, 22. The recent finding that single nucleotide polymorphisms (SNP) within the human AKNA AT-hook domain increase the risk of cervical cancer 23 lends support to this concept. Furthermore, mouse Fra4C2 on chromosome 4 shares a CFS of synteny with the human FRA9E region 24, which underscores the evolutionary resemblance of conserved chromosome instability and suggests potential biological parallels between them. Based on this reasoning, we engineered two independent gene-targeting mouse models to assess in vivo the physiological significance of AKNA gene expression. We found that the phenotypes resulting from the deletion of the putative C-terminus ALM sequence (AKNA KO) or disruption of AKNA's exon 3 (AKNA KO2) were by and large similar: (1) mice died prematurely at neonatal age; (2) probable causes of sudden death included acute inflammatory reactions and alveolar destruction; (3) triggering of the observed inflammation appeared to be pathogen-induced; (4) systemic neutrophil mobilization and alveolar infiltration were routinely observed; and (5) concerted activation of neutrophil-specific chemokine, cytokine and proteolytic enzyme expression seemed to be the norm. The central goal of the present study was to provide experimental support to the hypothesis that AKNA expression plays an important role in the mechanisms that regulate the magnitude of inflammatory responses to pathogens.
Results
Growth retardation, alveolar airspace enlargement and postnatal lethality
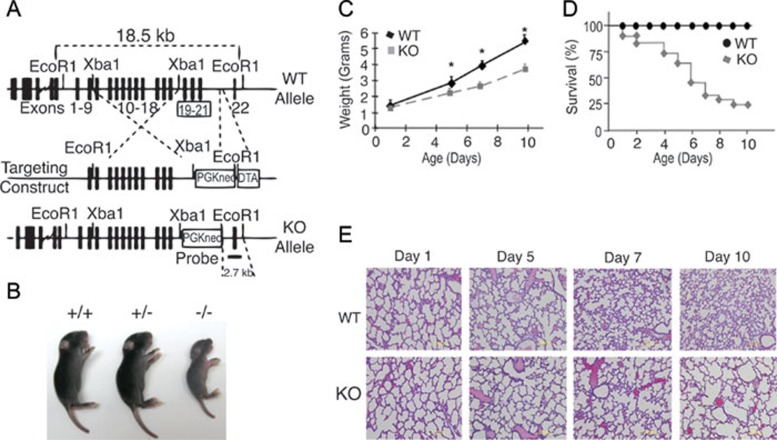
Knockout (KO) mice with targeted deletions of exons 19 through 21, which include a theoretical C-terminus ALM sequence were generated (Figure 1A and Supplementary information, Figure S1A and S1B) and bred for 10 generations into the C57BL/6 background to obtain genetically homogeneous (99%) mouse strains. By postnatal days 4-5 we observed that, irrespective of sex, KO mice were smaller than wild-type (WT) and heterozygous littermates (Figure 1B). As shown by quantitative weight and survival curves (Figure 1C and 1D), KO mice failed to thrive and most of them died within 10 days after birth. By contrast, heterozygous animals were indistinguishable from WT littermates in all parameters tested throughout this study. Thus, unless otherwise specified, subsequent analyses only compare WT with KO mice.
Figure 1.
Growth retardation and postnatal lethality in AKNA KO mice. (A) Targeting strategy to delete AKNA exons 19-21 (framed). The probe used to concomitantly detect the 18.5 kb WT and 2.7 kb KO alleles is indicated. (B) Representative photographs of neonatal WT (+/+), heterozygous (+/−) and KO (−/−) mice. (C) Weight curves of WT and AKNA KO mice at postnatal days 1, 5, 7 and 10. Bars indicate SEM. Total number of mice evaluated (n = 20, 10 WT and 10 KO mice). The symbol * denotes high statistical significance (P < 0.05). (D) Kaplan-Meier curves of survival rates for 41 WT and 41 KO mice (n = 82, with a log-rank P < 0.01). (E) HE-stained lung sections comparing WT (upper panel) and KO (lower panel) mice at postnatal days 1–10. Quantitative chord length (alveolar airspace) values at each postnatal stage are provided in Supplementary information, Figure S2.
Post-mortem examination showed that although organs from their WT littermates appeared normal, KO mice had notable signs of systemic inflammation, which was particularly prominent in the lungs. Remarkably, histopathologic analysis at postnatal days 1-5 showed no significant differences in alveolar formation 25 between WT and KO mice (Figure 1E). However, lung sections at postnatal day 7 revealed significant enlargement of the alveolar airspace in KO mice and this became more pronounced by day 10 (Figure 1E), as quantitatively confirmed by chord length measurements (Supplementary information, Figure S2). Despite the abrupt onset and severity of the pulmonary syndrome observed in KO mice, there was no evidence of inherent defects in alveolar development. For instance, alveolar deposit of Elastin fibers was comparable in both WT and KO mice (Supplementary information, Figure S3A). Also, lung mRNA levels of critical genes such as vascular endothelial growth factor (VEGF) 26 and transforming growth factor-β (TGF-β) 27 in KO mice were virtually identical to those of WT littermates (Supplementary information, Figure S3B). Furthermore, the number of apoptotic bodies, an indicator of tissue remodeling and repair 28, was somewhat similar between WT and KO mice (Supplementary information, Figure S3C). These data are consistent with the normal alveolar formation observed in AKNA KO mice from birth to postnatal day 5 (Figure 1E) and suggest the involvement of extrinsic cell and molecular mediators of acute alveolar damage.
Involvement of neutrophils in AKNA KO physiopathology
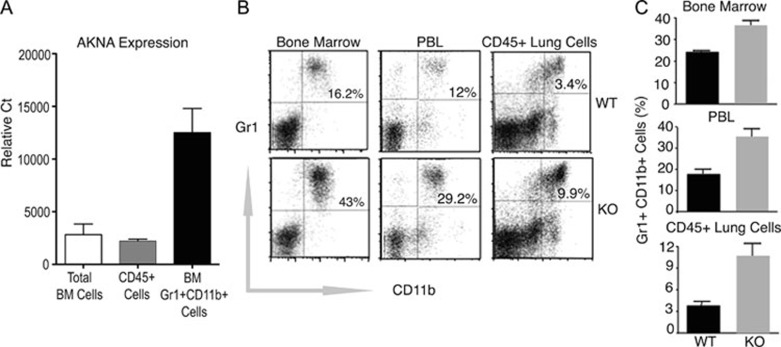
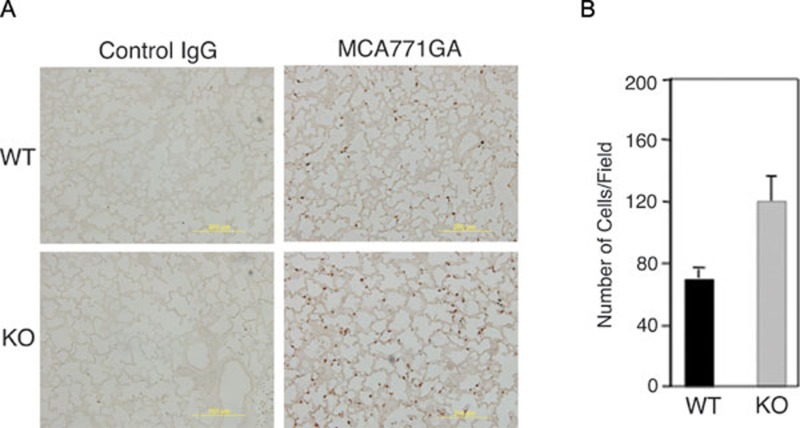
Because AKNA is highly expressed by CD11b+Gr1+ neutrophils (Figure 2A) and these cells are known mediators of alveolar inflammation 29, 30, 31, we reasoned that the lung abnormalities observed in KO mice could be the result of altered neutrophil output, circulation and/or alveolar recruitment. In support of this hypothesis, Figure 2B and 2C show that the number of bone marrow, peripheral blood and lung CD11b+Gr1+ cells in KO mice was 2.5 to 3-fold higher than those of WT littermates. The fact that no other leukocyte populations appeared to be affected (Supplementary information, Figure S4A and S4B) underscores the plausible contribution of neutrophils to phenotypes observed for AKNA deficiency. To test whether the observed peripheral neutrophil mobilization indeed leads to enhanced alveolar infiltration, lung tissue sections from WT and KO mice were probed with the neutrophil-specific antibody MCA771GA 32. Immunohistochemical results in Figure 3A and 3B show that the number of MCA771GA+ cells present in the alveolar microenvironment was quantitatively higher in KO mice, further supporting the involvement of neutrophils in the lung pathology observed in AKNA KO mice.
Figure 2.
Involvement of neutrophils in AKNA KO pathology. (A) The qRT-PCR results showing WT AKNA expression by murine bone marrow cells, CD45+ peripheral blood leukocytes (PBL) and pure bone marrow CD11b+Gr1+ neutrophils. Results are shown for three independent samples (n = 3) of each cell population ± SEM. P values < 0.05. (B) Comparative flow cytometry of CD11b+Gr1+ neutrophils levels between WT and AKNA KO mice in bone marrow, PBL and CD45+-gated lung cell suspensions. Values are presented (upper right quadrants) as the percentage of Gr1+ (y-axis) and CD11b+ (x-axis) cells. (C) Quantitative assessment of CD11b+Gr1+ neutrophils present in bone marrow, PBL and lung infiltrates of WT and KO mice. Bars indicate ± SEM (n = 6, P < 0.05).
Figure 3.
Presence of neutrophils in the alveolar compartment. (A) Immunohistochemistry (IHC) of paraffin-embedded lung sections from WT (upper panels) and KO (lower panels) mice at postnatal day 7, comparing levels of neutrophil alveolar infiltration by the specific reactivity of monoclonal antibody MCA771GA (right panels). Staining with isotype-matched control IgG (left panels) was also performed to validate MCA771GA antibody specificity. (B) Quantitative measurements of MCA771GA+ alveolar infiltrates performed by double-blind counts of three independent readings of five distinct IHC fields. Bars indicate ± SEM (n = 15, P < 0.05).
Relevance of AKNA to gene transcription
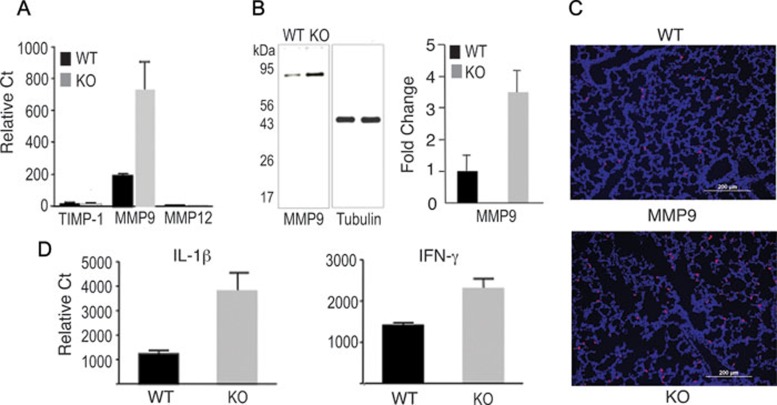
Bearing in mind that mouse AKNA possesses a theoretical C-terminus ALM, we reasoned that the deletion of this motif could affect a broad range of gene targets connected to neutrophil functions. To investigate this possibility, we compared mRNA levels of WT and KO mice using cDNA arrays and found a significant enhancement of protease, cytokine and chemotactic factor gene expression in the KO mice. These findings were confirmed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), which revealed that matrix metalloprotease 9 (MMP9) expression was at least 3-fold higher in KO than in WT mice (Figure 4A). Immunoblot experiments also showed that MMP9 protein levels were correspondingly higher in KO than in WT mice (Figure 4B). In addition, results in Figure 4A show that the high MMP9 expression in KO mice was in striking contrast with the relative absence of macrophage MMP12/elastase and tissue inhibitor of MMP (TIMP-1) 33, 34. Given that MMP9 is a neutrophil collagenase 35, which is known to be involved in inflammatory reactions that lead to alveolar damage 36, these results could be expected. Therefore, we sought to determine whether the significant MMP9 upregulation observed in AKNA KO mice could be histologically corroborated within the alveolar compartment. To that end, we performed immunofluorescent microscopic analyses on identical serial lung sections where MCA771GA+ neutrophil infiltration was detected (Figure 3A and 3B) and confirmed the higher MMP9 expression in KO mice (Figure 4C). Although dual detection of MCA771GA+ neutrophils and MMP9 expression cannot be achieved with currently available reagents, the combined data from Figures 3 and 4 provide a histological correlation between neutrophil infiltration and proteolytic enzyme production within the damaged alveolar microenvironment.
Figure 4.
Metalloprotease and cytokine gene expression. (A) The qRT-PCR experiments measuring TIMP-1, MMP9 and MMP12 expression in the lungs of WT and AKNA KO mice. Bars indicate ± SEM; n = 8 in all RT-PCR analyses; P < 0.05. (B) Left panel: immunoblot results of MMP9 and control tubulin expression levels in WT and KO mice. Right panel: quantitative scans of MMP9 protein bands obtained from four independent WT and KO immunoblots (n = 8) ± SEM; P < 0.05. (C) Representative immunofluorescent histopathologic analysis comparing the production of MMP9 within the alveolar microenvironment between WT and KO mice. (D) Results of qRT-PCR experiments comparing the expression of IL-1β (n = 8, P < 0.01) and IFN-γ (n = 6, P < 0.01) between WT and KO mice. Bars indicate ± SEM.
Comparative qRT-PCR experiments using WT and AKNA-deficient mice corroborated a selective upregulation of interleukin-1β (IL-1β) and interferon-γ (IFN-γ) expression in the lungs of KO mice similar to what we found for MMP9 (Figure 4D). These findings are significant because both cytokines exert high proinflammatory functions, and IL-1β is known to facilitate neutrophil recruitment to the lungs, is activated by MMP9, and is a major contributor to alveolar damage 37. Such selective activation of IL-1β and IFN-γ expression, resulting from AKNA deficiency, is further substantiated by the relatively unaltered expression of other cytokines such as tumor necrosis factor-α (TNF-α) and IL-13 (Supplementary information, Figure 4C).
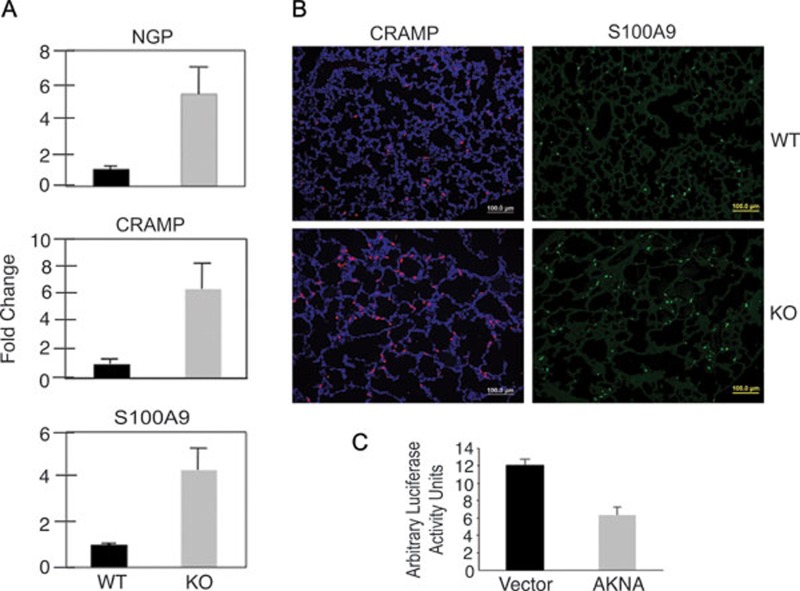
In support of the active mobilization and recruitment of neutrophils into the alveolar microenvironment observed in AKNA-deficient mice, qRT-PCR results confirmed higher neutrophilic granule protein (NGP), cathelin-related antimicrobial peptide (CRAMP) and S100A9 gene expression in KO than in WT mice (Figure 5A). Lack of suitable reagents precluded in situ comparisons of NGP expression in the alveolar compartment between WT and KO mice. However, detection of CRAMP and S100A9 by immunofluorescent microscopy on lung tissue sections corroborated higher expression in KO than in WT alveoli (Figure 5B). These findings are important because NGP, CRAMP and S100A9 share biological properties that are important for neutrophil activation and alveolar infiltration. For instance, NGP is a key constituent of neutrophil secondary granules and has been implicated in neutrophil-mediated lung inflammation 38; CRAMP is an angiogenic protein that exerts direct effects on neutrophil migration and is activated by MMP-dependent cleavage 39, 40; S100A9 is a member of the calcium-binding protein family involved in neutrophil recruitment to the inflammatory lung microenvironment and whose activity also depends on MMP9 proteolytic processing 41.
Figure 5.
Chemokine expression and gene regulation analyses. (A) The qRT-PCR results comparing the expression of neutrophil-specific chemokines NGP, CRAMP and S100A (n = 6, P < 0.05) between WT and AKNA KO mice. Bars represent ± SEM. (B) Immunofluorescent histologic analysis displaying the differential presence of chemokines CRAMP and S100A9 within the alveolar microenvironment of WT and KO mice. (C) Results of transient luciferase expression experiments in that the transcription of the reporter gene was placed under the control of the MMP9 promoter and evaluated in the absence (vector) or presence of AKNA-enforced overexpression (AKNA). The average of three independent experiments (n = 3, P < 0.01) for each condition is shown. Bars represent ± SEM.
Collectively, our findings suggest that AKNA plays a role in mechanisms that regulate the magnitude of acute inflammatory responses by coordinately repressing genes involved in neutrophil activation, mobilization and function. To test this hypothesis, promoter/reporter assays were performed to determine whether AKNA could modulate gene expression in a manner that could explain the transcriptional dysregulation observed in KO mice. Toward that end, a luciferase construct in which the expression of the reporter gene is driven by the MMP9 promoter 42 was transiently cotransfected with an AKNA expression plasmid (AKNA). Parallel cotransfections were comparatively conducted with the empty vector (Vector) to control for the specificity of the effect of AKNA on MMP9 promoter-driven reporter transcription. Figure 5C shows that, in contrast to vector transfectants, AKNA-expressing cells exhibited quantitatively lower expression of the luciferase reporter, which seemed evocative of a gene repression function. Further experimentation is necessary to precisely determine how mouse AKNA binds and regulates target gene promoters. However we cautiously postulate that the increased cytokine, chemotactic factor and proteolytic MMP9 expression resulting from AKNA deficiency may indeed reflect a loss of gene repression function.
Bone marrow transplants and neutrophil-specific alveolar damage
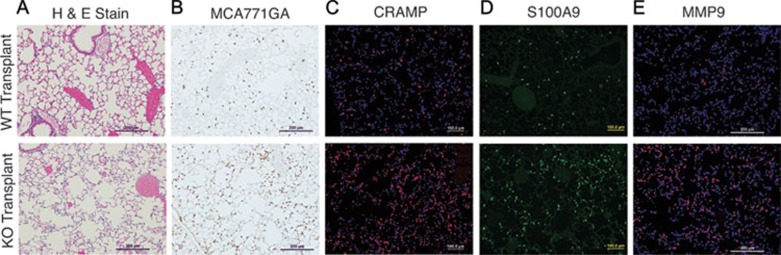
Although the preceding data support the role of neutrophils in the acute pulmonary inflammation observed in AKNA-deficient mice, we reasoned that adoptive transfer experiments were necessary to demonstrate the direct involvement of those cells. Toward that end, irradiated adult syngeneic mouse recipients were given bone marrow transplants from either WT or KO mouse donors. Three months after transplant, WT and KO mouse chimeras appeared healthy, with peripheral leukocytes confirmed to be more than 93% of donor origin (CD45.2) and without significant differences in the CD11b+Gr1+ neutrophil population (data not shown). However, when recipient mice were challenged with non-pathogenic Candida albicans extracts, which are known to trigger neutrophil-dependent inflammatory responses 43, KO but not WT mouse chimera experienced massive alveolar MCA771GA+/neutrophil infiltration (Figure 6A and 6B) and died within 12 h after immunization. In situ detection of enhanced CRAMP and S100A9 production (Figure 6C and 6D) in AKNA KO but not in WT mouse chimeras was indicative of rapid neutrophil mobilization and recruitment to the alveolar compartment. Furthermore, alveolar airspace in KO mice was visibly enlarged compared with that in WT mouse chimeras (Figure 6A and 6B), emphasizing the contribution of AKNA-deficient neutrophils to lung inflammation. Similarly, MMP9 expression was strikingly higher in the KO than in the WT mouse chimeras (Figure 6E), which underscores the involvement of this proteolytic enzyme in the observed alveolar damage. Together, these results suggest that neutrophils are the chief mediators of the pathogen-induced inflammatory hyperreactions underlying the AKNA KO phenotype.
Figure 6.
Alveolar neutrophil infiltration after adoptive bone marrow transplants and C. albicans challenge. (A) HE staining results of paraffin-embedded lung sections obtained from adult mice that received bone marrow transplants from either WT (WT transplants) or AKNA KO (KO transplants) mice. (B) Immunohistochemical results comparing MCA771GA+ neutrophil levels within the alveolar microenvironment of mice that received either WT or KO bone marrow transplants. (C-E) Immunofluorescent histologic results showing CRAMP, S100A9 and MMP9 expression on lung sections from mouse recipients of donor WT or KO bone marrow transplants.
Acute neutrophil responses in KO mice are pathogen-triggered
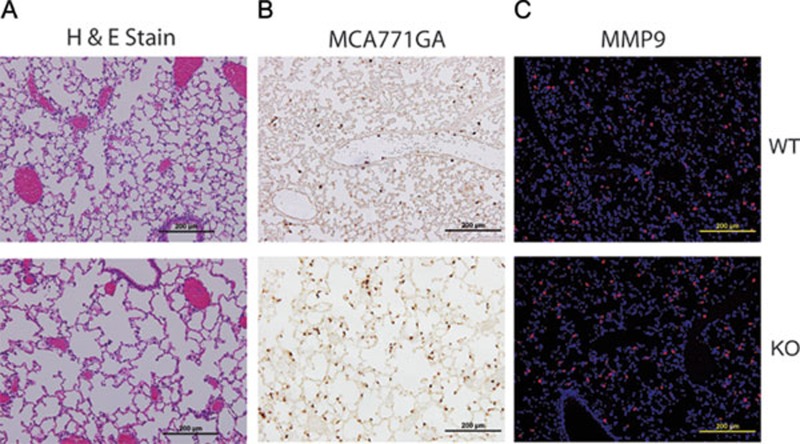
Although most KO mice die suddenly within the first week of life (Figure 1D), some survive past postnatal day 10. However, these surviving KO mice remain exceedingly susceptible to pathogen-induced inflammation. In agreement with these findings, AKNA-deficient mice died in C. albicans challenge experiments and exhibited exacerbated airspace enlargement, whereas their WT littermates were resistant to the pathogen and showed no significant alterations in the alveolar architecture (Figure 7A). The involvement of neutrophils in the acute inflammatory reactions elicited by the pathogen was corroborated by the striking infiltration of MCA771GA+ cells at the sites of major airspace enlargement (Figure 7B). Moreover, the parallel detection of enhanced MMP9 expression within the same histological compartment after the C. albicans treatment was also suggestive of pathogen-elicited, neutrophil-mediated alveolar destruction (Figure 7C). Of note, toll-like receptor-mediated neutrophil activation by the administration of CpG oligonucleotides 44 also led to fulminant inflammation and death of KO mouse survivors but not of WT littermates (data not shown). These results emphasize the contribution of pathogen-driven signals to the inflammatory phenotype of AKNA-deficient mice.
Figure 7.
Neutrophil-mediated airspace destruction in AKNA KO mouse survivors. (A) HE staining shows differences in airway space and alveolar architecture between a 10-week-old AKNA KO mouse survivor and a WT littermate after two sequential ip injections of inactive C. albicans extracts (see Materials and Methods). (B) Immunohistochemical results comparing the level of pathogen-triggered MCA771GA+ neutrophil alveolar infiltration between the KO mouse survivor and the WT littermate. (C) Immunofluorescent histologic results depicting MMP9 levels in the alveoli of the KO mouse survivor and the WT littermate after C. albicans challenge.
Exon 19-21 targeting/AKNA KO and exon 3 deletion/AKNA KO2 mirror phenotypes
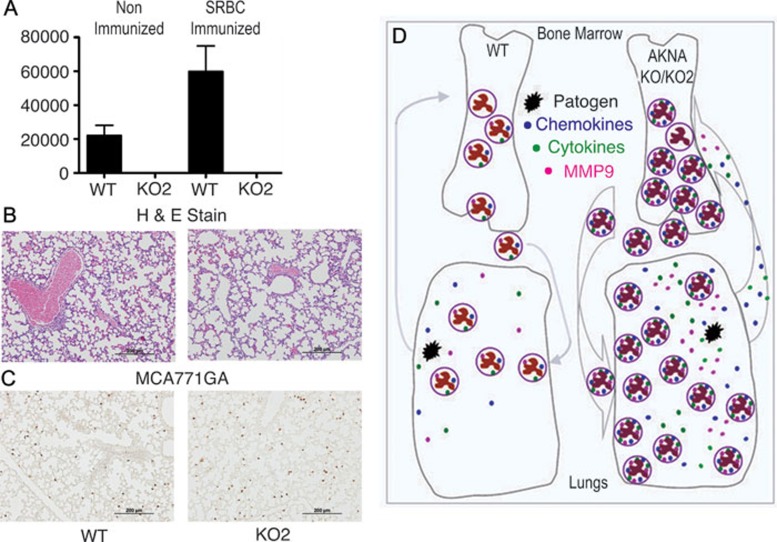
The emerging concept from the data presented so far is that the deletion of mouse AKNA's putative C-terminus ALM sequence appears to be sufficient to cause dysregulation of functionally related genes, which reciprocally manifests in a lethal inflammatory phenotype. However, the overall AKNA transcription rate in KO mice was reduced but not totally abolished (Supplementary information, Figure S1B), so the increased protease, cytokine and chemokine expression could reflect a failure of exon 19-21-truncated AKNA KO proteins to attain gene regulatory functions. Independent assessment of in vivo AKNA physiopathology was needed to test this hypothesis. Hence, a new mouse line with complete gene inactivation was generated and studied. The new AKNA line (AKNA KO2) was produced by the targeted disruption of exon 3, which is the largest coding exon and may include an N-terminus AT-hook motif. After confirmation of the AKNA KO2 genotype, the line was bred for 10 generations into a pure C57BL/6 genetic background. In remarkable similarity to mice with the exon 19-21 AKNA deficiency, AKNA KO2 mice died prematurely and most did not survive beyond 10 days of age. As a corroborative measure, RNA from C57BL/6 WT and AKNA KO2 mouse neonates was extracted and analyzed by qRT-PCR to compare AKNA expression. As shown in Figure 8A, AKNA expression in WT animals was at the expected levels and this increased by nearly 3-fold after activation of T cell-dependent immunity 45. In contrast, AKNA transcription in KO2 mice was completely abolished, with or without immunization (Figure 8A). Similar to the phenotype observed in the AKNA KO mouse line, KO2 mice died of pathogen-induced inflammation, manifested by prominent airspace enlargement (Figure 8B). Furthermore, the involvement of neutrophils at the onset of AKNA KO2 inflammatory reactions was substantiated by the conspicuous MCA771GA+ cell infiltration (Figure 8C).
Figure 8.
Loss of AKNA expression in KO2 mice and resemblance to AKNA KO pathology. (A) The qRT-PCR results showing AKNA expression levels in the spleen of WT and KO2 mice without (left column set) or with (right column set) sheep red blood cell immunizations (n = 6/condition, P < 0.05). Bars represent ± SEM. (B) Results of HE-stained lung sections that compare airway space and alveolar morphology between WT and AKNA KO2 mice at postnatal day 7. (C) Immunohistochemical results showing the level of alveolar MCA771GA+ neutrophil infiltration in WT and AKNA KO2 mice. (D) A hypothesis proposing that AKNA tightly regulates the expression of cytokines, chemokines and proteolytic enzymes to temper the degree of neutrophil responses to infections, in a manner that ensures the elimination of pathogens without irreparable tissue damage. This means that under physiological conditions, when AKNA expression is intact (WT mice), neutrophil mobilization from the bone marrow and cytokine, chemokine and protease activity are at the levels required to neutralize and destroy pathogens (schema on the left). By contrast, when either defective function or total loss of AKNA expression (KO and KO2 mice) occurs, exacerbated neutrophil reactions could ensue (schema on the right) and cause death. These reactions include enhanced autocrine and/or paracrine cytokine/chemokine/MMP9-mediated neutrophil activation, proliferation, circulation, alveolar infiltration and severe tissue damage. The hypothetical AKNA function is supported by the lethal phenotypes observed in AKNA KO and KO2 mice and by the death of mice transplanted with AKNA-deficient but not with WT bone marrow after the challenge with C albicans.
Discussion
Multicellular development, maturation and function require the timely activation or inhibition of a myriad of genes controlled by central regulatory mechanisms 1, 2. Although homeobox proteins have been established as master gene transcription factors 46, DNA architecture-modulating AT-hook proteins constitute an evolutionarily conserved family with analogous global gene-regulation functions 1, 2, 4, 6, 9. In accordance with this notion, AT-hook proteins bind a broad spectrum of AT-rich sequence-containing promoters 5 and regulate pathway-specific gene expression networks 3, 8, 47, 48. The present in vivo studies aimed to assess the function of AKNA, a transcription factor with a theoretical N-terminus AT-hook and C-terminus ALM. We found that targeting AKNA exons 19-21, which could produce truncated and non-functional AKNA proteins (AKNA KO), resulted in severe alveolar damage and neonatal death, mediated by neutrophil inflammatory reactions that included a coordinated activation of MMP9, IL-1β, IFN-γ, CRAMP and S100A9 gene expression (Figures 3,4, 5, 6). Although no evidence is presented nor are claims being made in support of bona fide ALM gene-regulation capacity, MMP9, IL-1β, IFN-γ, NGP, CRAMP and S100A9 promoters do possess regulatory AT-rich sequences 49, 50, 51, 52, 53. We also found that disruption of exon 3 (AKNA KO2), which completely annulled AKNA expression, reproduced the neutrophil-mediated inflammation, alveolar damage and sudden neonatal death seen in exon 19-21/AKNA deficient mice. Although we cannot definitively conclude that ALM and/or AT-hook deletions directly contribute to the respective AKNA KO and KO2 mouse phenotypes, our results support the increasing evidence for the involvement of canonical AT-hook HMGA1 in signaling inflammatory responses 54 and underscore the significance of the enhanced myeloproliferation (including neutrophils) previously observed in HMGA1-deficient mice 55.
Given that AKNA deficiency results in the concurrent increase of MMP9, IL-1β and IFN-γ, NGP, CRAMP and S100A9 gene expression, we speculate that AKNA functions as a multi-faceted transcriptional repressor that can coordinately temper pathway-specific gene transcription. Our finding that AKNA has the capacity to downregulate MMP9 promoter-driven gene reporter expression (Figure 5C), while not conclusive, provides support for AKNA's repression function. In keeping with the targeting of theoretical AT-hook and ALM sequences and the enhanced gene expression resulting from AKNA deficiency, IL-1β promoter includes palindromic A/T-rich sequences that seem to be responsible for the gene's constitutive repression 47, 50. Moreover, consistent with the notion of multifaceted gene regulation and in support of the provocative link between A/T-rich regulatory elements and gene promoter inhibition, non-HMG AT-hook PATZ and RNF4 proteins have been shown to cooperate to mediate broad transcriptional repression 8.
Without endorsing any specific mechanism of action, we hypothesize that AKNA's function is necessary to regulate the magnitude of pathogen-elicited neutrophil activation, proliferation and tissue infiltration by coordinately restricting autocrine/paracrine cytokine, chemokine and proteolytic MMP9 levels. This implies that when AKNA is productively expressed, as in WT mice, neutrophil reactions are increased to neutralize and destroy pathogens (Figure 8D, left). However, loss of AKNA expression (AKNA KO and KO2) could lead to exacerbated neutrophil activation and irreparable tissue damage (Figure 8D, right). This hypothesis is tune with the enhanced MMP9, IL-1β, IFN-γ, NGP, CRAMP and S100A9 expression and the resulting lethal syndrome associated with AKNA deficiency, in which transcriptional repression is seemingly lost. The finding that bone marrow transplants from AKNA KO, but not WT donors, reproduced MMP9/cytokine/chemokine-dependent pulmonary inflammation and alveolar destruction in healthy mouse recipients provides additional support to AKNA's regulatory function and emphasizes the involvement of Gr1+CD11b+ neutrophils in the pathogenesis of KO and KO2 mice.
Because neonatal death of AKNA KO and KO2 mice is exacerbated in the C57BL/6 background, our data additionally stress the importance of genetic susceptibility to acute neutrophil-mediated inflammatory reactions. Hence, in the context of a linkage with strain in mice and potentially with race in humans, we consider two hypothetical scenarios in AKNA deficiency: (1) acute and fatal hyper-reactions in genetically prone populations 56, 57 and (2) subtle states of inflammation in populations with no inherent genetic susceptibility that create a microenvironment that facilitates the initiation and progression of degenerative and/or neoplastic disorders 58.
Although our in vivo studies assessed only the function of mouse AKNA, the evolutionary parallels between human and mouse physiopathology are noteworthy. For instance, human and mouse AKNA genes are respectively located within fragile chromosome 9 FRA9E and 4 FRA4C2 regions, and these loci are known to be frequently altered in inflammatory diseases and cancer 24, 58. This suggests that, in humans, AKNA gene lesions could be a determining risk factor for how neutrophil-mediated inflammatory reactions resolve into acute syndromes and/or neoplastic disorders 59. Although we make no parallel between AKNA and HMG proteins, we speculate that given the evidence of HMGA1 direct contribution to neoplastic transformation 60, 61, 62, the shared involvement in inflammation 54, and the linkage of SNP at human AKNA AT-hook domain with higher risk for cervical cancer 23, physiopathological cooperation is at least plausible.
In conclusion, our combined data support the working hypothesis that AKNA deficiency reflects a loss of global gene repression functions, in which a dysregulation of genes essential to control the degree of neutrophil responses leads to systemic inflammation, severe tissue damage and sudden death. These data additionally emphasize the relevance of mechanisms that may be involved in the initiation and progression of complex pathologies such as anaphylaxis and tumorigenesis 23, 56, 57, 59. Although our studies are by no means exhaustive, the combined data from AKNA KO and KO2 mice concur with our hypothesis that AKNA has a regulatory function on gene transcription networks, which we suggest is linked to the mechanisms that control the degree of neutrophil responses to pathogen invasion (Figure 8D). Future comprehensive research is needed to substantiate the resemblance between mouse and human AKNA physiopathology, their mechanisms of action and their influence on systemic inflammation.
Materials and Methods
Mice
To generate AKNA KO and KO2 mouse lines, exons 19-21 or large exon 3 of the mouse AKNA gene on chromosome 4 were respectively replaced by homologous recombination in 129 embryonic stem cells. Before any pathophysiologic analyses, both AKNA/AT-hook and AKNA KO mouse lines were independently backcrossed until mice were 99% of pure C57BL/6 genetic background. All mice under study were housed under specific pathogen-free conditions and experiments were conducted according to Institutional Animal Care and Use Committee guidelines. WT littermates were routinely used as controls. AKNA KO and AKNA KO2 genotype experiments were performed by Southern blotting and qPCR, whereas AKNA expression was monitored by qRT-PCR.
Lung suspension cell isolation
After mice were euthanized lungs were completely exsanguinated by flushing with phosphate-buffered saline (PBS), aseptically removed and minced into small pieces. The dissected tissue was incubated in RPMI (Invitrogen, Carlsbad, CA, USA) medium containing 1 mg/ml Type-D collagenase and 0.5 mg/ml DNase Type I (Roche Diagnostics, Mannheim, Germany) for 1 h at 37 °C. Digested lungs were further disrupted by gently pushing the tissue through a 70-μm nylon sieve. Cell suspensions were washed twice with RPMI and then washed with PBS containing 0.5% bovine serum albumin, counted and analyzed by flow cytometry.
Alveolar chord length
Measurements of alveolar airspace via chord-length measurements were determined on hematoxylin-eosin (HE)-stained lung sections as previously described 63. Then eight independent fields at 20× magnification from 6 WT and KO mouse pairs were assessed. Averaged results were used to calculate the mean alveolar chord length.
Flow cytometry
Multiple bone marrow, spleen, peripheral blood and lung cell preparations from WT and AKNA KO mice were concomitantly labeled with fluorescently conjugated FITC/anti-mouse CD11b, APC/anti-mouse Gr-1 and PerCP/anti-mouse CD45. After gating CD45+ cells, quantitative GR1+CD11b+ phenotypes were compared on a Moflo flow cytometer (Cytomation, Inc., Fort Collins, CO, USA) equipped with a 488 nm Argon laser (Enterprise II, Coherent Inc., Palo Alto, CA, USA) and a 647 nm Argon/Krypton laser (Innova 70 Spectrum, Coherent). Individually stained control cells were used to set the compensation. A total of 500 000 events were acquired for the final analysis. Histogram results and statistical plots were obtained using the FlowJo software (Tree Star, Inc. Ashland, OR, USA).
Immunoblot
Proteins lysates from WT and KO lungs were resolved on 10% SDS-PAGE gels, electroblotted and reacted with polyclonal anti-MMP9 IgG (Abcam, Cambridge, MA, USA) overnight at 4 °C. After intense washing, reactions were revealed with HRP-conjugated anti-rabbit IgG (Abcam) and chemilluminiscence (Pierce, Rockford, IL, USA) autoradiography according to the manufacturer's instructions. Blots were subsequently stripped and re-probed with rat monoclonal anti-Tubulin (Abcam), followed by identical detection methods, using HRP-conjugated anti-Rat IgG (Cell Signaling Technology, Danvers, MA, USA) and chemilluminiscence reactions (Pierce). Band scans from four independent WT and KO immunoblots were quantitatively analyzed by chemilluminiscence imaging software (Alpha Innotech, Santa Clara, CA, USA) and the results were plotted.
Immunohistochemistry
Formalin-fixed, paraffin-embedded lung tissue sections were reacted with neutrophil-specific rat anti-mouse monoclonal antibody MCA771GA (ABD Serotec, Oxford, UK) at 1:500 dilution. Reactions were developed by HRP-conjugated anti-rat IgG in combination with 3,3′-Diaminobenzidine, a chromogenic substrate. Sections were counterstained with methyl green (Vector laboratories, Burlington, CA, USA). Double-blind counts of MCA771GA+ cells were performed by 3 individuals on 5 independent 20× magnification fields from 4 WT and KO mouse pairs.
Immunofluorescent staining
Formalin-fixed, paraffin-embedded lung tissue sections were reacted overnight at 4 °C with the following primary antibodies: polyclonal rabbit anti-MMP9 antibody (Abcam Inc, Cambridge, MA, USA) at 1:1 000 dilution; polyclonal rabbit anti-CRAMP antibody (a generous gift from Dr Richard L Gallo, University of California, San Diego, USA) at 1:1 000 dilution; and polyclonal goat anti-S100A9 antibody (Santa Cruz Biotechnology Inc, Santa Barbara, CA, USA) at 1:100 dilution. Primary antibody reactions were then revealed by incubating the slides for 1 h at room temperature with fluorescence-conjugated antibodies (Molecular Probes, Eugene, OR, USA) as follows: Alexa 594 (red fluorescence) anti-rabbit IgG was used to reveal both MMP9 and CRAMP expression and Alexa 488 (green fluorescence) anti-donkey IgG for S100A9 detection. All dilutions for these reactions were 1:5 000 in PBS/0.1% Tween-20 buffer. Hoechst 33342 (Molecular Probes) served as a nuclear counterstain. The slides were mounted in universal mount solution (Research Genetics, Huntsville, AL, USA) and images were captured with an Olympus BX41 microscope (Center Valley, PA, USA).
Reporter assay
Hela cells were seeded into 24-well plates (1 × 105 cells/well) and incubated in RPMI 1640 medium. After 24 h, cells in each well were transfected using Lipofectamine 2000 (Invitrogen) with 200 ng of MMP9 promoter-driven luciferase reporter 42, generously provided by Dr Douglas D Boyd (Department of Cancer Biology, The University of Texas M.D. Anderson Cancer Center, USA) or 5 ng of control Renilla luciferase reporter pRL-tk-luc (Promega, Madison, WI, USA) in conjunction with either empty vector PCDNA3.1 (800 ng) or PCDNA3.1-AKNA plasmid (800 ng). After 48 h of transfection, dual luciferase assays were performed according to the manufacturer's instructions (Promega) and luciferase activity was measured using a single-photon-channel scintillation counter (Beckman, Fullerton, CA, USA).
Adoptive bone marrow cell transfers and mouse immunizations
Bone marrow cells were harvested from WT and KO mice. A total of 5 × 106 bone marrow cells from either WT or KO donors (CD45.2) were independently injected into cohorts of lethally irradiated (1 000 cGy) WT mouse recipients (CD45.1) by tail vein injections. Twelve weeks after cell transplants, separate groups of WT and AKNA KO chimera mice received (in 7-day intervals) intraperitoneal (ip) and intravenous (iv, via the tail-vein) 100-μl injections of formalin-fixed C. albicans extracts (equivalent to 1 × 107 cells/mouse), followed by C. albicans extract (100 μl) intranasal (in) instillations (under anesthesia and according to institutional guidelines) 7 days after iv injections. All WT chimera mice survived the C. albicans challenge, whereas most AKNA/AT-hook KO chimera mice died within 24 h after the second or third in application. Lungs of both WT (euthanized) and KO (dead after antigen challenge) chimera mice were collected post mortem for histology and immunostaining.
Elastin staining
Paraffin-embedded lung sections were stained overnight with a mixture containing 5 ml of Harts Elastin stock solution (Elastin Products Company Inc, Owensville, MO, USA), 50 ml of 70% ethanol and 1 ml of 12N HCl. Sections were then washed for 30 min in ddH2O and counterstained with 2.5% Tartrazine.
In situ apoptosis detection
Sections were stained using the TdT in situ Apoptosis Detection Kit (TACS Blue Label–TA4626, R&D Systems, Minneapolis, MN, USA) according to the manufacturer's method and counterstained with Nuclear Fast Red. Positive controls were obtained by treating serial sections with TACS-nuclease according to the manufacturer's instructions.
Assessment of mRNA expression by qRT-PCR
Total RNA from lungs of 3 WT and AKNA KO or KO2 mouse pairs was prepared for comparative gene expression analyses. The qRT-PCR was performed by standard procedures using SYBR Green/Rox PCRmaster mix (Sabioscience, Frederick, MD, USA) in a 7500 fast real time PCR system (Applied BioSystems, Carlsbad, CA, USA). The qRT-PCR primers for TNF-α (PPM03113), IL-13 (PPM03021A), IL-1β (PPM03109E), VEGF (PPM03041E), TGF-β (PPM02001A), IFNγ (PPM03121A), β-Actin (PPM02945A), AKNA-1439 (PPM63382A) and AKNA-3194 custom-ordered with sequence coordinates comprising the putative AT-hook-like region (PPMC0002A) were from SABioscience. Other forward and reverse primers used were as follows:
MMP9 forward: 5′-AGTGGACGCGACCGTAGTTG-3′, reverse: 5′-GCCACCAGGAACAGGGTGTA-3′ CRAMP forward: 5′-GCTGTGGCGGTCACTATCAC-3′, reverse: 5′-TGTCTAGGGACTGCTGGTTGA-3′ S100A9 forward: 5′-ATACTCTAGGAAGGAAGGACACC-3′, reverse: 5′-TCCATGATGTCATTTATGAGGGC-3′ NGP forward: 5′-AGACCTTTGTATTGGTGGTGGC-3′, reverse: 5′-GGTTGTATGCCTCTATGGCTCTA-3′ MMP12 forward: 5′-GAGTCCAGCCACCAACATTAC-3′, reverse: 5′-GCGAAGTGGGTCAAAGACAG-3′ TIMP-1 forward: 5′-GCAACTCGGACCTGGTCATAA-3′, reverse: 5′-CGGCCCGTGATGAGAAACT-3′.
Statistical analyses
P values were obtained using an unpaired two-tailed t-test, where P ≤ 0.05 was taken to indicate statistical significance. Error bars indicate SEM.
Acknowledgments
We thank Dr Shao-Cong Sun and Dr Yong-Jun Liu (The University of Texas MD Anderson Cancer Center, USA) for their critiques and advice. We also thank Dr Douglas D Boyd (The University of Texas MD Anderson Cancer Center, USA) and Dr Richard L Gallo (University of California, San Diego, USA) for their generosity in providing the MMP9 promoter/Luciferase reporter and the anti-CRAMP antibody, respectively. The present work was supported by grants AI056125-01 and AI065796-01 from the National Institutes of Health (NIH) and by NCI CCSG Core grant CA106672. MRM was supported by the training grant T32 CA009598-15 from the NIH and WM was supported by the Odyssey Program and The Cockrell Foundation Award for Scientific Achievement at The University of Texas MD Anderson Center Cancer.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
AKNA Genotype and expression analyses.
Comparative assessment of alveolar airspace.
Lung Elastin deposit, cytokine expression and apoptosis.
Circulating lymphocytes and lung cytokine expression.
References
- Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev. 2009;19:437–443. doi: 10.1016/j.gde.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Ng KH, Yu H, Ito T. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 2009;7:e1000251. doi: 10.1371/journal.pbio.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- Aulner N, Monod C, Mandicourt G, et al. The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation. Mol Cell Biol. 2002;22:1218–1232. doi: 10.1128/MCB.22.4.1218-1232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekdal C, Sjottem E, Johansen T. The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J Biol Chem. 2000;275:40288–40300. doi: 10.1074/jbc.M006978200. [DOI] [PubMed] [Google Scholar]
- Fedele M, Benvenuto G, Pero R, et al. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J Biol Chem. 2000;275:7894–7901. doi: 10.1074/jbc.275.11.7894. [DOI] [PubMed] [Google Scholar]
- Siddiqa A, Sims-Mourtada JC, Guzman-Rojas L, et al. Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature. 2001;410:383–387. doi: 10.1038/35066602. [DOI] [PubMed] [Google Scholar]
- Torrungruang K, Alvarez M, Shah R, et al. DNA binding and gene activation properties of the Nmp4 nuclear matrix transcription factors. J Biol Chem. 2002;277:16153–16159. doi: 10.1074/jbc.M107496200. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Thomas JO, Travers AA. HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- Cui T, Leng F. Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: a SELEX study. Biochemistry. 2007;46:13059–13066. doi: 10.1021/bi701269s. [DOI] [PubMed] [Google Scholar]
- Whitson RH, Tsark W, Huang TH, Itakura K. Neonatal mortality and leanness in mice lacking the ARID transcription factor Mrf-2. Biochem Biophys Res Commun. 2003;312:997–1004. doi: 10.1016/j.bbrc.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Sutherland HG, Newton K, Brownstein DG, et al. Disruption of Ledgf/Psip1 results in perinatal mortality and homeotic skeletal transformations. Mol Cell Biol. 2006;26:7201–7210. doi: 10.1128/MCB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Maezawa S, Koiwai K, Hayano T, Koiwai O. Identification of functional domains in TdIF1 and its inhibitory mechanism for TdT activity. Genes Cells. 2007;12:941–959. doi: 10.1111/j.1365-2443.2007.01105.x. [DOI] [PubMed] [Google Scholar]
- Senthilkumar R, Mishra RK. Novel motifs distinguish multiple homologues of Polycomb in vertebrates: expansion and diversification of the epigenetic toolkit. BMC Genomics. 2009;10:549. doi: 10.1186/1471-2164-10-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BR, Li Y, Wang L, et al. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims-Mourtada JC, Bruce S, McKeller MR, et al. The human AKNA gene expresses multiple transcripts and protein isoforms as a result of alternative promoter usage, splicing, and polyadenylation. DNA Cell Biol. 2005;24:325–338. doi: 10.1089/dna.2005.24.325. [DOI] [PubMed] [Google Scholar]
- Thye T, Burchard GD, Nilius M, Muller-Myhsok B, Horstmann RD. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am J Hum Genet. 2003;72:448–453. doi: 10.1086/367714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landvik NE, Hart K, Skaug V, et al. A specific interleukin-1B haplotype correlates with high levels of IL1B mRNA in the lung and increased risk of non-small cell lung cancer. Carcinogenesis. 2009;30:1186–1192. doi: 10.1093/carcin/bgp122. [DOI] [PubMed] [Google Scholar]
- Savas S, Liu G. Genetic variations as cancer prognostic markers: review and update. Hum Mutat. 2009;30:1369–1377. doi: 10.1002/humu.21078. [DOI] [PubMed] [Google Scholar]
- Perales G, Burguete-Garcia AI, Dimas J, et al. A polymorphism in the AT-hook motif of the transcriptional regulator AKNA is a risk factor for cervical cancer. Biomarkers. 15:470–474. doi: 10.3109/1354750X.2010.485332. [DOI] [PubMed] [Google Scholar]
- Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neptune ER, Podowski M, Calvi C, et al. Targeted disruption of NeuroD, a proneural bHLH factor, impairs distal lung formation and neuroendocrine morphology in the neonatal lung. J Biol Chem. 2008;283:21160–21169. doi: 10.1074/jbc.M708692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipple TE, Welty SE, Nelin LD, Hansen JM, Rogers LK. Alterations of the thioredoxin system by hyperoxia: implications for alveolar development. Am J Respir Cell Mol Biol. 2009;41:612–619. doi: 10.1165/rcmb.2008-0224OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhuang F, Liu YH, et al. TGF-beta receptor II in epithelia versus mesenchyme plays distinct roles in the developing lung. Eur Respir J. 2008;32:285–295. doi: 10.1183/09031936.00165407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson PM, Vandivier RW, Douglas IS. Cell death, remodeling, and repair in chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2006;3:713–717. doi: 10.1513/pats.200605-104SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Gao XP, Ramchandran R, et al. Nonmuscle myosin light-chain kinase mediates neutrophil transmigration in sepsis-induced lung inflammation by activating beta2 integrins. Nat Immunol. 2008;9:880–886. doi: 10.1038/ni.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Murray AR, et al. Increased accumulation of neutrophils and decreased fibrosis in the lung of NADPH oxidase-deficient C57BL/6 mice exposed to carbon nanotubes. Toxicol Appl Pharmacol. 2008;231:235–240. doi: 10.1016/j.taap.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Schymeinsky J, Mocsai A, Walzog B. Neutrophil activation via beta2 integrins (CD11/CD18): molecular mechanisms and clinical implications. Thromb Haemost. 2007;98:262–273. [PubMed] [Google Scholar]
- Laubitz D, Larmonier CB, Bai A, et al. Colonic gene expression profile in NHE3-deficient mice: evidence for spontaneous distal colitis. Am J Physiol Gastrointest Liver Physiol. 2008;295:G63–G77. doi: 10.1152/ajpgi.90207.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DG, Huang X, Kaminski N, et al. Loss of integrin alpha(v)beta6-mediated TGF-beta activation causes Mmp12-dependent emphysema. Nature. 2003;422:169–173. doi: 10.1038/nature01413. [DOI] [PubMed] [Google Scholar]
- Shukla A, Barrett TF, Nakayama KI, et al. Transcriptional up-regulation of MMP12 and MMP13 by asbestos occurs via a PKCdelta-dependent pathway in murine lung. FASEB J. 2006;20:997–999. doi: 10.1096/fj.05-4554fje. [DOI] [PubMed] [Google Scholar]
- Nair RR, Solway J, Boyd DD. Expression cloning identifies transgelin (SM22) as a novel repressor of 92-kDa type IV collagenase (MMP-9) expression. J Biol Chem. 2006;281:26424–26436. doi: 10.1074/jbc.M602703200. [DOI] [PubMed] [Google Scholar]
- Shapiro SD, Goldstein NM, Houghton AM, et al. Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am J Pathol. 2003;163:2329–2335. doi: 10.1016/S0002-9440(10)63589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen U, Whitsett JA, Wert SE, Tichelaar JW, Bry K. Interleukin-1beta causes pulmonary inflammation, emphysema, and airway remodeling in the adult murine lung. Am J Respir Cell Mol Biol. 2005;32:311–318. doi: 10.1165/rcmb.2004-0309OC. [DOI] [PubMed] [Google Scholar]
- O'Donnell LC, Druhan LJ, Avalos BR. Molecular characterization and expression analysis of leucine-rich alpha2-glycoprotein, a novel marker of granulocytic differentiation. J Leukoc Biol. 2002;72:478–485. [PubMed] [Google Scholar]
- Gallo RL, Kim KJ, Bernfield M, et al. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem. 1997;272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J Immunol. 2005;174:6257–6265. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- Newton RA, Hogg N. The human S100 protein MRP-14 is a novel activator of the beta 2 integrin Mac-1 on neutrophils. J Immunol. 1998;160:1427–1435. [PubMed] [Google Scholar]
- Yan C, Wang H, Boyd DD. KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha -induced block of p65/p50 nuclear translocation. J Biol Chem. 2001;276:1164–1172. doi: 10.1074/jbc.M008681200. [DOI] [PubMed] [Google Scholar]
- Fulurija A, Ashman RB, Papadimitriou JM. Neutrophil depletion increases susceptibility to systemic and vaginal candidiasis in mice, and reveals differences between brain and kidney in mechanisms of host resistance. Microbiology. 1996;142:3487–3496. doi: 10.1099/13500872-142-12-3487. [DOI] [PubMed] [Google Scholar]
- Knuefermann P, Baumgarten G, Koch A, et al. CpG oligonucleotide activates Toll-like receptor 9 and causes lung inflammation in vivo. Respir Res. 2007;8:72. doi: 10.1186/1465-9921-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae S, Asano M, Horai R, Sakaguchi N, Iwakura Y. IL-1 enhances T cell-dependent antibody production through induction of CD40 ligand and OX40 on T cells. J Immunol. 2001;167:90–97. doi: 10.4049/jimmunol.167.1.90. [DOI] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Mouri F, Tsukada J, Mizobe T, et al. Intracellular HMGB1 transactivates the human IL1B gene promoter through association with an Ets transcription factor PU.1. Eur J Haematol. 2008;80:10–19. doi: 10.1111/j.1600-0609.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- Grant MA, Baron RM, Macias AA, et al. Netropsin improves survival from endotoxaemia by disrupting HMGA1 binding to the NOS2 promoter. Biochem J. 2009;418:103–112. doi: 10.1042/BJ20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Kita M, Seiki M. v-Src activates the expression of 92-kDa type IV collagenase gene through the AP-1 site and the GT box homologous to retinoblastoma control elements. A mechanism regulating gene expression independent of that by inflammatory cytokines. J Biol Chem. 1993;268:23460–23468. [PubMed] [Google Scholar]
- Lebedeva TV, Singh AK. Constitutive activity of the murine IL-1 beta promoter is regulated by a transcriptional repressor. Biochim Biophys Acta. 1997;1353:32–38. doi: 10.1016/s0167-4781(97)00040-7. [DOI] [PubMed] [Google Scholar]
- Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- Elloumi HZ, Holland SM. Complex regulation of human cathelicidin gene expression: novel splice variants and 5'UTR negative regulatory element. Mol Immunol. 2008;45:204–217. doi: 10.1016/j.molimm.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken W, Lekstrom-Himes JA, Sorg C, Manitz MP. Molecular analysis of the mouse S100A9 gene and evidence that the myeloid specific transcription factor C/EBPepsilon is not required for the regulation of the S100A9/A8 gene expression in neutrophils. J Cell Biochem. 2001;80:606–616. [PubMed] [Google Scholar]
- Resar LM. The high mobility group A1 gene: transforming inflammatory signals into cancer. Cancer Res. 2010;70:436–439. doi: 10.1158/0008-5472.CAN-09-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele M, Fidanza V, Battista S, et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006;66:2536–2543. doi: 10.1158/0008-5472.CAN-05-1889. [DOI] [PubMed] [Google Scholar]
- Nagarkar DR, Wang Q, Shim J, et al. CXCR2 is required for neutrophilic airway inflammation and hyperresponsiveness in a mouse model of human rhinovirus infection. J Immunol. 2009;183:6698–6707. doi: 10.4049/jimmunol.0900298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blood-Siegfried J. The role of infection and inflammation in sudden infant death syndrome. Immunopharmacol Immunotoxicol. 2009;31:516–523. doi: 10.3109/08923970902814137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AK, Rondini EA. Review paper: the role of inflammation in mouse pulmonary neoplasia. Vet Pathol. 2009;46:369–390. doi: 10.1354/vp.08-VP-0217-B-REV. [DOI] [PubMed] [Google Scholar]
- Ardi VC, Kupriyanova TA, Deryugina EI, Quigley JP. Human neutrophils uniquely release TIMP-free MMP-9 to provide a potent catalytic stimulator of angiogenesis. Proc Natl Acad Sci USA. 2007;104:20262–20267. doi: 10.1073/pnas.0706438104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LJ, Maher JF, Bunton TE, Resar LM. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60:4256–4261. [PubMed] [Google Scholar]
- Wood LJ, Mukherjee M, Dolde CE, et al. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol. 2000;20:5490–5502. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sumter TF, Bhattacharya R, et al. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64:3371–3375. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- Blackburn MR, Volmer JB, Thrasher JL, et al. Metabolic consequences of adenosine deaminase deficiency in mice are associated with defects in alveogenesis, pulmonary inflammation, and airway obstruction. J Exp Med. 2000;192:159–170. doi: 10.1084/jem.192.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AKNA Genotype and expression analyses.
Comparative assessment of alveolar airspace.
Lung Elastin deposit, cytokine expression and apoptosis.
Circulating lymphocytes and lung cytokine expression.