Abstract
Diabetes is featured by hyperglycemia, which facilitates the formation of advanced glycation end-products (AGEs). AGEs are a causal factor in development of diabetic complications. AGE receptor-1 (AGE-R1) is responsible for detoxification and clearance of AGEs. Type 2 diabetes mellitus is commonly accompanied by non-alcoholic steatohepatitis, which could cause hepatic fibrosis. Little attention has been paid to effects of AGEs on hepatic fibrogenesis. Curcumin, a phytochemical from turmeric, has been reported to inhibit the activation of hepatic stellate cells (HSCs), the major effectors during hepatic fibrogenesis, and to protect against hepatic fibrogenesis in vitro and in vivo. The current study was designed to evaluate effects of AGEs on inducing HSC activation, to assess the role of curcumin in diminishing the AGE effects and to explore the underlying mechanisms. Our results showed that AGEs stimulated HSC activation by inducing cell proliferation and expression of genes relevant to HSC activation, which were abrogated by curcumin. Curcumin induced gene expression of AGE-R1 in passaged HSCs, which might facilitate the attenuation of the stimulatory effects of AGEs on the activation of HSCs. Further experiments revealed that curcumin inhibited the activity of extracellular signal-regulated kinase (ERK) and induced gene expression and the activity of peroxisome proliferator-activated receptor-gamma (PPARγ), leading to the induction of AGE-R1 gene expression. In summary, AGEs stimulated HSC activation. Curcumin eliminated the AGE effects at least partially by inducing AGE-R1 gene expression. The process was mediated by inhibiting ERK activity, inducing gene expression of PPARγ and stimulating its trans-activity.
Keywords: Hyperglycemia, hepatic fibrosis, hepatic stellate cell, gene expression, diabetes
INTRODUCTION
Type 2 diabetes mellitus (T2DM) is featured by insulin-independently elevated levels of blood glucose, i.e. hyperglycemia. Hyperglycemia is a high risk factor for the development of non-alcoholic steatohepatitis (NASH) (1), which is an understudied complication of T2DM. Approximately 15–40% of NASH patients develop hepatic fibrosis (2). Hyperglycemia facilitates the non-enzymatic formation of advanced glycation end-products (AGEs), which are a heterogeneous group of molecules formed by non-oxidative and oxidative reactions of sugars with proteins and/or lipids (3). AGEs accumulate in tissues and circulation during aging, as well as diabetic, chronic renal failure and liver fibrogenesis (4), leading to inflammation and pathogenesis (5). Effects of AGEs are mediated by their receptor system, which could be generally divided into two categories. Receptor for AGEs (RAGE) facilitates oxidative stress (OS), cell growth, and inflammation (6). AGE receptors (AGE-Rs), e.g. AGE-R1 (also called OST-48), are responsible for detoxification and clearance of AGEs (7). In contrast to a dramatic increase in expression of RAGE in diabetes with high levels of AGEs (8), the abundance of AGE-R1 is significantly reduced in diabetic organs, e.g. kidney (9), suggesting a possible inverse relationship between AGEs-mediated cell injury and low expression of AGE-R1. In addition to its participation in AGE removal, AGE-R1 negatively regulates AGE pro-inflammatory signal processing (7).
Hepatic stellate cells (HSCs) are the major effectors during hepatic fibrogenesis (10). During hepatic injury, quiescent HSCs undergo profound phenotypic changes, including enhanced cell proliferation, loss of lipid droplets, de novo expression of α-smooth muscle actin (α-SMA), and excessive production of extracellular matrix, including type I collagen (10). This process is called HSC activation. Freshly-isolated HSCs spontaneously become fully activated in culture (11), mimicking the process seen in vivo. The spontaneous activation of HSCs during cell culture provides a good model for elucidating underlying mechanisms of HSC activation and studying potential therapeutic intervention of the process (10, 12). Few effective medicines are currently available for inhibiting HSC activation and combating hepatic fibrosis, including T2DM- & NASH-associated hepatic fibrosis (13). It is, therefore, of high priority to identify innocuous anti-fibrotic agents. Curcumin, the yellow pigment in curry from turmeric, has received attention as a promising dietary supplement for the protection against fibrogenic insults (14). We recently demonstrated that curcumin inhibited HSC activation and protected the liver from CCl4-caused injury in vitro and in vivo (15–18).
The liver is not only a site for cleaning AGEs, but also a target organ for AGEs. Elevated levels of serum AGEs were observed in patients with NASH (19). However, little attention has been paid to effects of AGEs on HSC activation and on T2DM- & NASH-associated hepatic fibrogenesis. AGEs were reported to induce the proliferation of cultured HSCs (20). The accumulation of AGEs and the reduction of the AGE-R1 abundance may represent important mediators in hyperglycemia-induced HSC activation. However, the underlying mechanism remains largely undefined. The current study was designed to evaluate effects of AGEs on inducing HSC activation, to assess the role of curcumin in inhibiting the effects of AGEs and to explore the underlying mechanisms. Results in this report supported our initial hypothesis that AGEs might stimulate HSC activation, which could be eliminated by curcumin at least partially by inducing AGE-R1 gene expression.
Materials and Methods
AGE preparation and chemicals
AGEs-bovine serum albumin (BSA) were prepared following the protocol described by others (21). In brief, 50 mg/ml of BSA (USB Corp., Cleveland, OH) and 0.5 M of glucose (Sigma-Aldrich Corp. St. Louis, MO) were dissolved in 0.2 M of sodium phosphate buffer (pH 7.4). A BSA control and a glucose control were respectively prepared by dissolving BSA (50 mg/ml) alone, or glucose (0.5 M) alone, in sodium phosphate buffer (pH 7.4) (0.2 M). After sterilization with sterile Acrodisc® syringe filters, the solutions were incubated in the dark at 37 °C for 60 days. Unbound materials were removed by extensive dialysis against phosphate-buffered saline (PBS).
The concentration of AGEs was determined as described by measuring AGEs-specific fluorescence with excitation at 360 nm and emissions at 440 nm (22, 23). The fluorescence of the BSA control was used as a base line. The fluorescence of qualified AGEs-BSA used in our experiments must be at least 70-fold high than that of the BSA control. The quality and eligibility of AGEs were evaluated and confirmed as suggested (24). No contamination with insulin-like growth factor-1 and/or endotoxin was detected. Curcumin (purity >94%) and 15-deoxy-Δ12,14-prostaglandin J2 (PGJ2) were purchased from Sigma (St. Louis, MO). PD68235, a specific PPARγ antagonist, was kindly provided by Pfizer (Ann Arbor, MI) (25). Rosiglitazone (BRL 49653), purchased from Cayman Chemical (Ann Arbor, MI), was dissolved in dimethyl sulfoxide (100 mM). The ERK inhibitor PD98059 was purchased from CalBiochem (La Jolla, CA, USA). Primary antibodies were purchased from Santa Cruz Biotech. Inc. (Santa Cruz, CA), unless otherwise noted, and were previously described (26).
Isolation and culture of HSCs
Male Sprague-Dawley rats (200–250g), purchased from the Harlan Laboratories, Inc. (Indianapolis, IN), were housed in a temperature-controlled animal facility (23°C) with a 12:12h light-dark cycle and allowed free access to regular chew and water ad libitum. HSCs were isolated by the pronase-collagenase perfusion in situ prior to density gradient centrifugation, as we previously described (16). The animal protocol for use of rats was approved by Institutional Animal Care and Use Committee of Saint Louis University. Freshly-isolated HSCs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% of fetal bovine serum (FBS) for 48 hr. Cells were passaged in DMEM with 10% of FBS. Semi-confluent HSCs with 4–9 passages were used in experiments. In some of experiments, cells were cultured in serum-depleted DMEM for 24 hr before treatment, which rendered HSCs more sensitive to exogenous stimuli (26). Cells were subsequently treated and cultured in serum-depleted media, which excluded the interference from other factors in FBS.
Determination of cell proliferation in vitro
Cell growth was colorimetrically determined by using the non-radioactive cell proliferation assay kit (i.e. MTS assays) (Promega, Madison, WI), following the protocol provided by the manufacturer (16).
Western blotting analyses
Preparation of whole cell extracts, SDS-PAGE, transblotting and subsequent immuno-reactions were conducted as we previously described (16). β-actin, or β-tubulin, was used as an invariant control for equal loading. Densities of bands in Western blotting analyses were normalized with the internal invariable control. Levels of target protein bands were densitometrically determined by using Quantity One® 4.4.1 (Bio-Rad, Hercules, CA). Variations in the density were expressed as fold changes compared to the control in the blot.
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA was treated with DNase I prior to the synthesis of the first strand of cDNA. Real-time PCR were performed as we previously described using SYBR Green Supermix (16). mRNA levels were expressed as fold changes after normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as described by Schmittgen et al (27). The following primers were used for determining the level of rat AGE-R1 mRNA by real-time PCR: (F) 5′-GCT CTG ATA TCG GTG ACC CT-3′, (R) 5′-TCG TAG TTG TGG TGG TCG AT-3′. Other primers used in this study have been described in our prior reports (26).
Plasmids and transient transfection assays
The cDNA expression plasmids pa-ERK and pdn-ERK, respectively containing a full length of cDNA fragment encoding the constitutively active form of ERK (a-ERK), or the dominant-negative form of ERK (dn-ERK), were previously described and used (28). The PPARγ activity luciferase reporter plasmid pPPRE-TK-Luc was also previously described (16). The luciferase reporter plasmid pAGE-R1-Luc was generated by subcloning a fragment (−3838/+67 bp) of the murine AGE-R1 gene promoter into HindIII/MluI sites of pGL3-Basic vector. Semi-confluent HSCs in 6-well cell culture plates were transiently transfected with a total of 3 – 4.5 μg DNA per well, using the LipofectAMINE® reagent (Invitrogen Corp. Carlsbad, CA), as we previously described (16). Each sample was in triplicate in every experiment. Transfection efficiency was normalized by co-transfection of the β-galactosidase reporter plasmid pSV-β-gal (0.5 μg/well) (Promega, Madison, WI). β-galactosidase activities were measured by using a chemiluminescence assay kit (Tropix, Bedford, MA). Luciferase activities were expressed as relative unit after normalization with β-galactosidase activities per microgram of protein. Results were combined from at least three independent experiments.
Statistical analyses
Percentages in differences were calculated in the formula: [(# in target HSCs − # in compared HSCs)/# in compared HSCs] × 100% (n≥3). Differences between means were evaluated using an unpaired two-sided Student’s test (p<0.05 considered as significant). Where appropriate, comparisons of multiple treatment conditions with control were analyzed by analysis of variance with the Dunnett’s test for post hoc analysis.
RESULTS
AGEs stimulated HSC proliferation and induced expression of genes closely relevant to HSC activation
To determine effects of AGEs on inducing HSC activation, passaged HSCs were cultured in serum-depleted DMEM for 24 hr, which restored some of quiescent phenotypes and made HSCs more sensitive to exogenous stimuli (26). Serum-starved HSCs were stimulated with AGEs-BSA, or BSA, at 0–200μg/ml in serum-depleted media for 24 hr. Subsequent culture in serum-depleted DMEM eliminated the interference from growth factors in FBS. As shown in Fig. 1A by cell proliferation assays, AGEs-BSA dose-dependently increased the number of viable cells. For instance, compared with the untreated control (the 1st black column), AGEs-BSA at 100μg/ml significantly increased the number of viable HSCs by 95%. However, BSA by itself had no such stimulatory effect on the number of viable HSCs (white columns). These results suggested that AGEs stimulated cell proliferation of HSCs in vitro.
Figure 1. AGEs dose-dependently stimulated HSC activation in vitro.
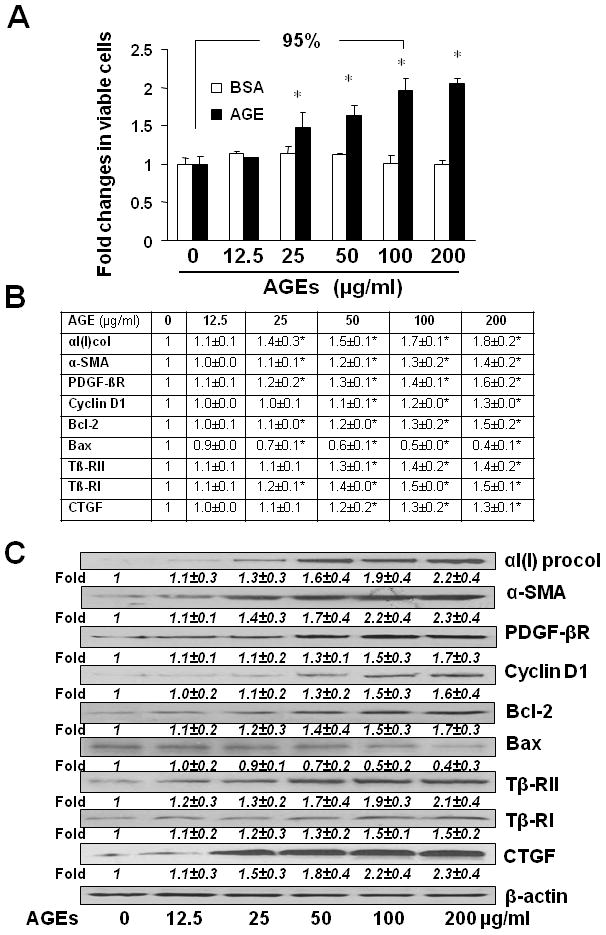
HSCs were serum-starved in DMEM for 24 hr prior to the stimulation with AGEs-BSA, or BSA, at indicated doses in serum-depleted media for additional 24 hr. (A) Cell proliferation was determined by colorimetric MTS assays. Results were expressed as fold changes in the number of viable cells, compared with the untreated control (mean± s. d., n=3) (the corresponding 1st column). The percentage in difference was calculated in the formula: [(# in target HSCs − # in compared HSCs)/# in compared HSCs] x 100%. *p<0.05 vs. the untreated control (the 1st column). (B) real-time PCR analyses. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. the untreated control (the corresponding 1st column). (C) Western blotting analyses. Representatives were from three independent experiments. β-actin was used as an internal control for equal loading. Italic numbers beneath blots were fold changes (means ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
Further experiments by real-time PCR (Fig. 1B) and Western blotting analyses (Fig. 1C) indicated that AGEs-BSA stimulated gene expression of αI(I) collagen and α-SMA, two unique markers for activated HSCs. In addition, AGEs-BSA induced expression of genes relevant to cell growth, including pro-mitogenic PDGF-βR, cyclin D1 and anti-apoptotic Bcl-2, while suppressed gene expression of pro-apoptotic Bax. Furthermore, AGEs-BSA induced expression of genes relevant to pro-fibrogenesis, including the type I and II TGF-β receptors (Tβ-RI/II) and connective tissue growth factor (CTGF). In contrast, BSA by itself had no such dose-dependent effects on the regulation of expression of the genes (data not shown). These results collectively suggested that AGEs induced the activation of HSCs in vitro. Since AGEs-BSA at 100 μg/ml was enough for inducing HSC activation, this concentration was chosen for the following experiments.
Curcumin eliminated the effects of AGEs on the induction of HSC activation in vitro
Curcumin by itself has shown its effects on the regulation of expression of genes relevant to the activation of HSCs in vitro and in vivo, including αI(I) collagen (15, 16), α-SMA (15, 16), TGF-βI/II (17), CTGF (29, 30), Bax and BcL-2 (17), etc. To assess the role of curcumin in attenuating the stimulatory effects of AGEs on the activation of HSCs, serum-starved HSCs were treated with or without (w/wt) AGEs-BSA (100μg/ml) in the presence of curcumin at indicated concentrations (0–30 μM) in serum-depleted media for 24 hr. As shown in Fig. 2A by cell proliferation assays, AGEs increased, as expected, the number of viable cells by 92% (the 2nd column), compared with the untreated control (the 1st column). Curcumin dose-dependently eliminated the effect of AGEs. For instance, compared with AGEs alone (the 2nd column), curcumin at 20 μM diminished the effect of AGEs and reduced the number of viable cells by 33% (the 4th column).
Figure 2. Curcumin eliminated the effects of AGEs on the induction of HSC activation.
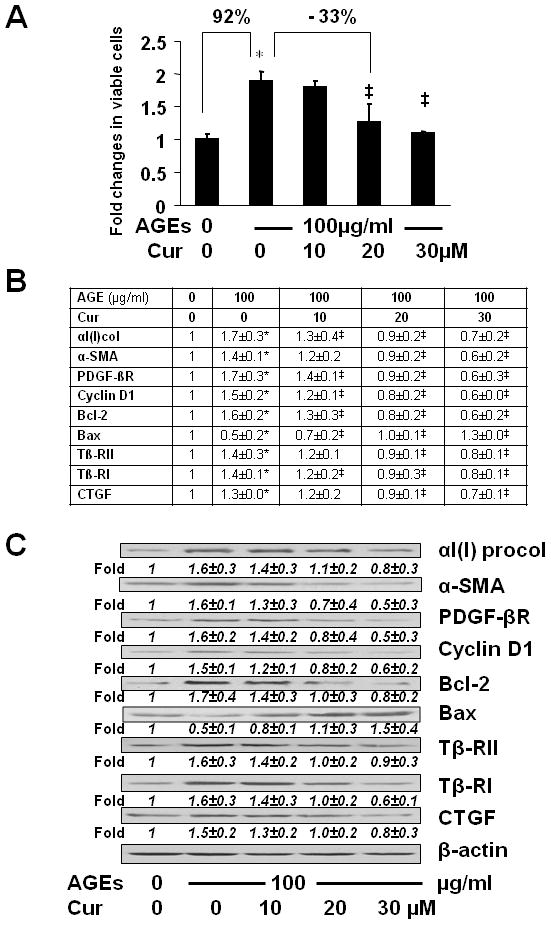
Serum-starved HSCs were treated with or without AGEs-BSA at 100 μg/ml plus or minus curcumin at indicated concentrations in serum-depleted media for 24 hr. (A) Cell proliferation was determined by MTS assays. Results were expressed as fold changes in the number of viable cells, compared with the untreated control (mean± s. d., n=3). *p<0.05 vs. the untreated control (the 1st column); ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). The percentage in difference was calculated in the formula: [(# in target HSCs − # in compared HSCs)/# in compared HSCs] x 100%. (B) real-time PCR analyses. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. the untreated control; ‡p<0.05 vs. cells treated with AGEs alone. (C) Western blotting analyses. Representatives were from three independent experiments. β-actin was used as an internal control for equal loading. Italic numbers beneath blots were fold changes (means ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
Further experiments of real-time PCR (Fig. 2B) and Western blotting analyses (Fig. 2C) revealed that curcumin eliminated the effect of AGEs on regulating expression of genes relevant to the activation of HSCs by reducing gene expression of αI(I) collagen, α-SMA, PDGF-βR, cyclin D1, Bcl-2, Tβ-RI/II and CTGF, as well as by inducing gene expression of Bax, at both levels of mRNA and protein. Taken together, these results demonstrated that curcumin eliminated the effects of AGEs on the induction of HSC activation in vitro.
Curcumin abrogated the inhibitory effect of AGEs and induced gene expression of AGE-R1 in passaged HSCs
To elucidate the underlying mechanism by which curcumin eliminated the effects of AGEs on the induction of HSC activation, we assumed that one of the mechanisms for AGEs to induce HSC activation was to suppress gene expression of AGE-R1, which could be attenuated by curcumin by inducing gene expression of AGE-R1. To test the assumption, serum-starved HSCs were stimulated with AGEs at indicated doses in serum-depleted media for 24 hr w/wt the presence of curcumin (0–30 μM). As shown in Fig. 3A & B respectively by real-time PCR and Western blotting analyses, AGEs dose-dependently suppressed gene expression of AGE-R1. For instance, compared with the untreated control (the 1st column and lane), AGEs at 100 μg/ml significantly reduced AGE-R1 at both levels of transcript and protein by approximately 70% (the 5th column and lane).
Figure 3. Curcumin abrogated the effect of AGEs and induced gene expression of AGE-R1 in cultured HSCs.
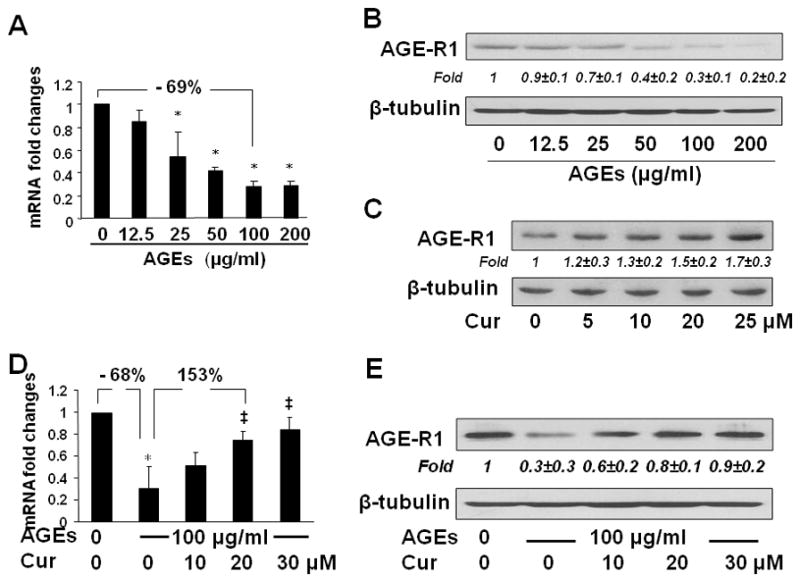
Serum-starved HSCs were treated with AGEs at 0–200 μg/ml (A & B), or with AGEs at 100 μg/ml plus curcumin at indicated concentrations (D & E), in serum-depleted media for 24 hr. On the other hand, passaged HSCs were treated with curcumin at indicated concentrations in DMEM with 10% FBS for 24 hr (C).
A & D: Real-time PCR analyses. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. the untreated control (the 1st column), ‡p<0.05 vs. the cells treated with AGEs alone (the 2nd column).
B, C, & E: Western blotting analyses. Representatives were from three independent experiments. β-tubulin was used as an internal control for equal loading. Italic numbers beneath blots were fold changes (means ± s. d.) in the densities of the bands compared with the control without treatment in the blot (n=3), after normalization with the internal invariable control.
To evaluate the role of curcumin in regulating expression of AGE-R1, passaged HSCs were treated with curcumin at various concentrations in DMEM with 10% FBS for 24 hr. Western blotting analyses indicated that curcumin by itself increased the abundance of AGE-R1 in HSCs in a dose-dependent manner (Fig. 3C). Additional experiments revealed that curcumin dose-dependently attenuated the effect of AGEs by increasing gene expression of AGE-R1 demonstrated by real-time PCR (Fig. 3D) and Western blotting analyses (Fig. 3E). For example, compared with AGEs alone (the 2nd column and lane), curcumin at 20 μM diminished the effect of AGEs and increased the levels of mRNA and protein by more than 150% (the 4th column and lane). Prior studies have shown that serum-starvation partially restored phenotypic features of quiescent HSCs, including reduced levels of αI(I) collagen and α-SMA (26). However, HSCs in Fig. 3C were cultured in regular DMEM with 10% of FBS, which inhibited gene expression of AGE-R1 in HSCs. The difference in media conditions prior to the treatment could cause a difference in the basal levels of AGE-R1 in the un-treated controls in Fig. 3C verse B and E. Take together, our results demonstrated that curcumin eliminated the inhibitory effect of AGEs and induced gene expression of AGE-R1 in passaged HSCs.
The inhibition of ERK activity diminished the effect of AGEs and induced gene expression of AGE-R1 in cultured HSCs
To start to elucidate the mechanisms by which curcumin eliminated the effect of AGEs on inhibiting gene expression of AGE-R1 in HSCs, we presumed that AGEs stimulated the activity of ERK in HSCs, leading to the suppression of gene expression of AGE-R1, and that curcumin induced gene expression of AGE-R1 by inhibiting the activity of ERK in HSCs. We have shown that curcumin by itself dose-dependently reduces the level of phosphorylated ERK in cultured HSCs (31). Our prior experiments showed that the acute activation of ERK could reach its peak within 20–30 minutes in passaged HSCs after the exposure to stimuli, which was attenuated and inhibited by curcumin (26, 32). To test our presumption, serum-starved HSCs were treated with AGEs at indicated doses in serum-depleted media for 30 minutes w/wt the pretreatment with curcumin (0–30 μM) for 1 hr. Whole cell extracts were prepared for analyzing levels of phosphorylated ERK. As shown in Fig. 4A by Western blotting analyses, AGEs caused a dose-dependent increase in the level of phosphorylated ERK, indicating that AGEs indeed induced the activation of ERK in HSCs in vitro. The inductive effect of AGEs on the level of phosphorylated ERK was diminished by curcumin in a dose-dependent manner (Fig. 4B).
Figure 4. The inhibition of ERK activity diminished the effect of AGEs and induced gene expression of AGE-R1 in cultured HSCs.
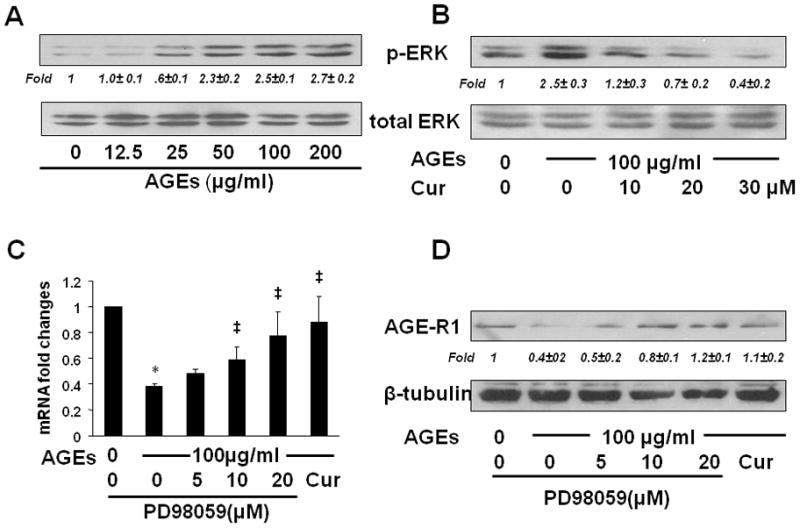
(A & B) Serum-starved HSCs were treated with AGEs at indicated doses in serum-depleted media for 30 minutes with or without the pretreatment with curcumin (0–30 μM) for 1 hr. Whole cell extracts were prepared for analyzing levels of phosphorylated ERK by Western blotting analyses. Total ERK was used as an internal control for equal loading. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control. Representatives were from three independent experiments.
(C & D) Serum-starved HSCs were pretreated with or without the selective ERK inhibitor PD98059 (0–20 μM) or curcumin (20 μM) for 1 hr prior to the exposure to AGEs (100 μg/ml) for additional 24 hr. Total RNA and whole cell extracts were prepared from the cells. (C) real-time PCR assays. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. cells with no treatment (the 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). (D) Western blotting analyses. β-tubulin was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
To evaluate the role of the activation of ERK in regulating gene expression of AGE-R1, serum-starved HSCs were pretreated w/wt the selective ERK inhibitor PD98059 (0–20 μM) or curcumin (20 μM) for 1 hr prior to the exposure to AGEs (100 μg/ml) for additional 24 hr. Total RNA and whole cell extracts were prepared from the cells. As shown by real-time PCR (Fig. 4C) and Western blotting analyses (Fig. 4D), compared with the untreated control (the 1st column and lane), AGEs reduced, as expected, the levels of mRNA and protein of AGE-R1 (the 2nd column and lane). The inhibition of ERK activity by PD98059, mimicking the role of curcumin (the last column and lane), dose-dependently eliminated the inhibitory effect of AGEs and elevated the contents of AGE-R1 mRNA and protein in the cells (the 3rd – 5th columns and lanes). These results collectively supported our presumption and suggested that the stimulation of ERK activity by AGEs might lead to the suppression of gene expression of AGE-R1, which could be abrogated by curcumin by inhibiting the activity of ERK in HSCs.
The alterations in the activity of ERK resulted in the changes in the gene promoter activity and the abundance of AGE-R1 in cultured HSCs
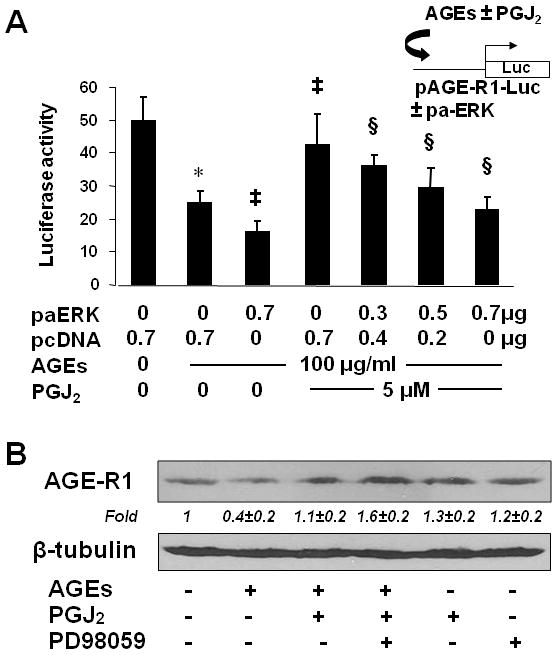
To verify the role of the ERK activity in mediating the effect of AGEs on regulating gene expression of AGE-R1, HSCs in six-well culture plates were co-transfected with a DNA mixture, including 2 μg of the AGE-R1 gene promoter luciferase reporter plasmid pAGE-R1-Luc, 0.5 μg of pSV-β-gal and 0.7 μg of the cDNA expression plasmid pa-ERK encoding the constitutively active form of ERK at various doses plus the empty vector pcDNA, or 1.5 μg of the cDNA expression plasmid pdn-ERK encoding the dominant negative form of ERK (dn-ERK) at various doses plus the empty vector pcDNA. The latter was used to ensure equal amount of total DNA in transfection assays. After recovery, cells were serum-starved for 4 hr prior to the treatment w/wt AGEs (100 μg/ml) in the presence or absence of curcumin (20 μM) in serum-depleted media for additional 24 hr. Results from luciferase activity assays in Fig. 5A & B demonstrated that compared with the untreated control (the corresponding 1st column), AGEs significantly reduced, as expected, the luciferase activity in the cells, indicating a reduction in the promoter activity of AGE-R1 gene (the corresponding 2nd column). Curcumin apparently eliminated the inhibitory effect of AGEs on the gene promoter activity AGE-R1 (the corresponding 3rd column). It was further observed that forced expression of exogenous active ERK cDNA dose-dependently diminished the role of curcumin and reduced luciferase activities in the cells (the 4th–6th columns in Fig. 5A). Compared with the control (the 1st column), the co-transfection of pa-ERK alone, as a control, significantly reduced the luciferase activity in the cells (the last column in Fig. 5A). These results confirmed that the increase in ERK activity could reduce the gene promoter activity of AGE-R1 in HSCs. In contrast, forced expression of exogenous dn-ERK cDNA diminished the effect of AGEs and increased, like curcumin (the 3rd column in Fig. 5B), luciferase activities in a dose-dependent manner (the 4th – 6th columns in Fig. 5B), indicating that the blockade of the ERK signaling pathway by dn-ERK abrogated the effect of AGEs on the inhibition of the promoter activity of AGE-R1 gene.
Figure 5. The alterations in the activity of ERK resulted in the changes in the gene promoter activity and the abundance of AGE-R1 in cultured HSCs.
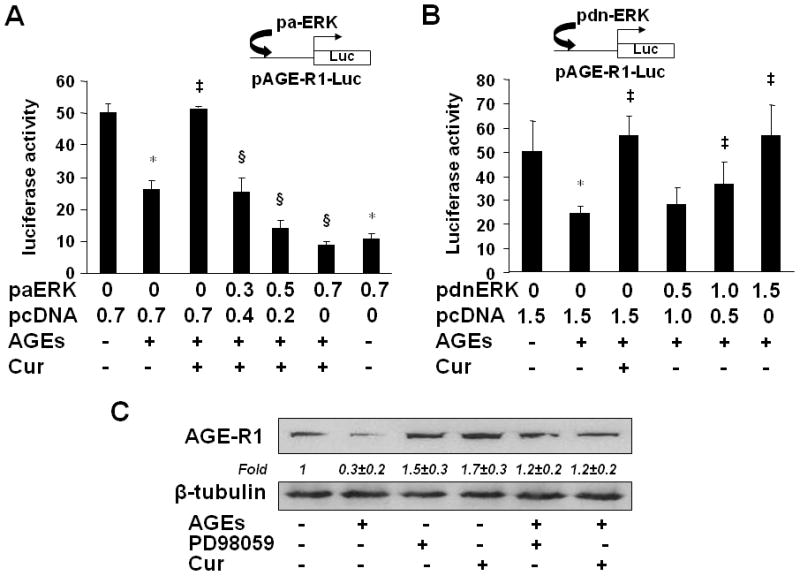
(A & B) HSCs were co-transfected with the AGE-R1 promoter luciferase reporter plasmid pAGE-R1-Luc and the cDNA expression plasmid pa-ERK encoding constitutively active ERK (A), or pdn-ERK encoding dominant negative ERK (B), at indicated doses. After recovery, cells were serum-starved for 4 hr prior to the treatment with or without AGEs (100 μg/ml) in the presence or absence of curcumin (20 μM) in serum-depleted media for additional 24 hr. Luciferase activity assays were conducted (n=6). *p<0.05 vs. cells with no treatment (the 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). §p<0.05 vs. cells treated with AGEs plus curcumin (the 3rd column). The floating schema denoted the plasmids pAGE-R1-Luc and pa-ERK, or pdn-ERK in use for co-transfection.
(C) Serum-starved HSCs were treated with AGEs (100 μg/ml) in the presence of curcumin (20 μM), or the ERK selective inhibitor PD98059 (10 μM), in serum-depleted media for 24 hr. Western blotting analyses were conducted. β-tubulin was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
To further confirm the role of the curcumin-caused inhibition of ERK in the up-regulation of expression of AGE-R1, serum-starved HSCs were treated with AGEs (100 μg/ml) in the presence of curcumin (20 μM), or the ERK selective inhibitor PD98059 (10 μM) in serum-depleted media for 24 hr. Western blotting analyses indicated that compared with the untreated control (the 1st lane), AGEs reduced, as expected, the abundance of AGE-R1 in HSCs (the 2nd lane). It is of interest to observe that the ERK inhibitor PD98059 (the 5th lane) mimicked the role of curcumin (the 5th lane) in the attenuation of the inhibitory effect of AGEs and in the elevation of the level of AGE-R1 in HSCs. PD98059 (the 3rd lane) or curcumin (the 4th lane) alone showed a slight effect on the elevation of the level of AGE-R1 in serum-starved HSCs, likely because of no enough stimuli in serum-depleted media. Taken together, our results indicated that the alterations in the activity of ERK resulted in the changes in the gene promoter activity of AGE-R1 and in the abundance of AGE-R1 in cultured HSCs. Our results also revealed a critical role of the curcumin-caused inhibition of ERK in the up-regulation of expression of AGE-R1.
AGEs suppressed gene expression of PPARγ and reduced its trans-activity in HSCs, which were diminished by curcumin
PPARγ is highly expressed in quiescent HSCs in the normal liver (33). However, the level of PPARγ and its activity are dramatically reduced during the process of HSC activation in vitro and in vivo (33). We previously reported that curcumin induced gene expression of PPARγ in vitro and in vivo (15–17), which was required for curcumin to inhibit HSC activation in vitro (16, 17). To further elucidate the underlying mechanisms by which curcumin eliminated the effect of AGEs and induced gene expression of AGE-R1 in HSCs, we assumed that AGEs suppressed gene expression of PPARγ in HSCs, which was abolished by curcumin. To test the assumption, serum-starved HSCs were stimulated w/wt AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media for 24 hr. Total RNA and whole cell extracts were prepared from the cells. As shown by real-time PCR (Fig. 6A) and Western blotting analyses (Fig. 6B), compared with the untreated control (the 1st column and lane), AGEs significantly reduced gene expression of PPARγ (the 2nd column and lane). The inhibitory effect of AGEs was dose-dependently diminished by curcumin (the 3rd – 5th columns and lanes).
Figure 6. AGEs suppressed gene expression of PPARγ and reduced its trans-activity in HSCs, which were diminished by curcumin.
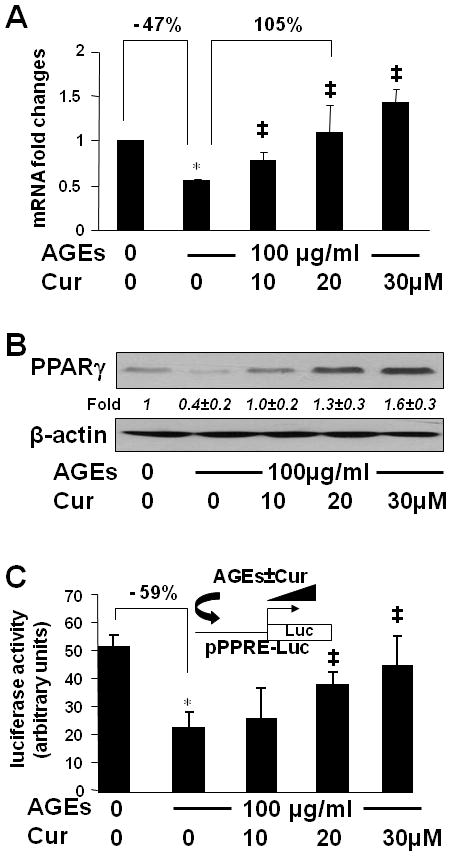
(A & B) Serum-starved HSCs were stimulated with or without AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media for 24 hr. Total RNA and whole cell extracts were prepared. (A). real-time PCR assays. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. cells with no treatment (the 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). (B) Western blotting analyses. β-actin was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
(C) HSCs were transfected with the PPARγ activity luciferase reporter plasmid pPPRE-Luc. After recovery, cells were serum-starved for 4 hr prior to the treatment with or without AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media with PGJ2 (5 μM) for 24 hr. Luciferase activity assays were conducted (n=6). *p<0.05 vs. cells with no treatment (the 1st column), ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). The floating schema denoted the plasmid pPPRE-Luc in use for transfection and the application of AGEs w/wt curcumin to the system.
To evaluate the effect of AGEs on the trans-activity of PPARγ, HSCs were transfected with the PPARγ activity luciferase reporter plasmid pPPRE-Luc. After recovery, cells were serum-starved for 4 hr prior to the treatment w/wt AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media with PGJ2 (5 μM) for additional 24 hr. The exogenous PPARγ agonist PGJ2 was added because there was no PPARγ agonist in serum-depleted media. Results from luciferase activity assays indicated that compared with the untreated control (the 1st column), AGEs reduced luciferase activity by 59% (the 2nd column), suggesting a significant reduction in the trans-activity of PPARγ in the cells. The inhibitory effect of AGEs was dose-dependently eliminated by curcumin (the 3rd –5th columns). These results collectively demonstrated that AGEs suppressed gene expression of PPARγ and reduced its trans-activity in HSCs, which were diminished by curcumin by inducing gene expression of PPARγ.
The activation of PPARγ eliminated the inhibitory effect of AGEs and stimulated gene expression of AGE-R1 in HSCs in vitro
To evaluate the role of the activation of PPARγ in regulating the gene promoter activity of AGE-R1, HSCs were transfected with the AGE-R1 gene promoter luciferase reporter plasmid pAGE-R1-Luc. After recovery, cells were serum-starved for 4 hr prior to the treatment w/wt AGEs (100 μg/ml) in the presence or absence of the natural PPARγ agonist PGJ2, or the synthesized PPARγ agonist rosiglitazone (Rosi), at 0–10 μM in serum-depleted media for additional 24 hr. Results from luciferase activity assays in Fig. 7A demonstrated that compared with the untreated control (the corresponding 1st column), AGEs significantly reduced, as expected, luciferase activity in the cells (the corresponding 2nd column). The inhibitory effect of AGEs was dose-dependently abrogated by either PGJ2 or rosiglitazone (the corresponding 3rd – 5th columns), suggesting that the activation of PPARγ eliminated the inhibitory effect of AGEs on the gene promoter activity of AGE-R1 in passaged HSCs.
Figure 7. The activation of PPARγ eliminated the inhibitory effect of AGEs and stimulated gene expression of AGE-R1 in HSCs in vitro.
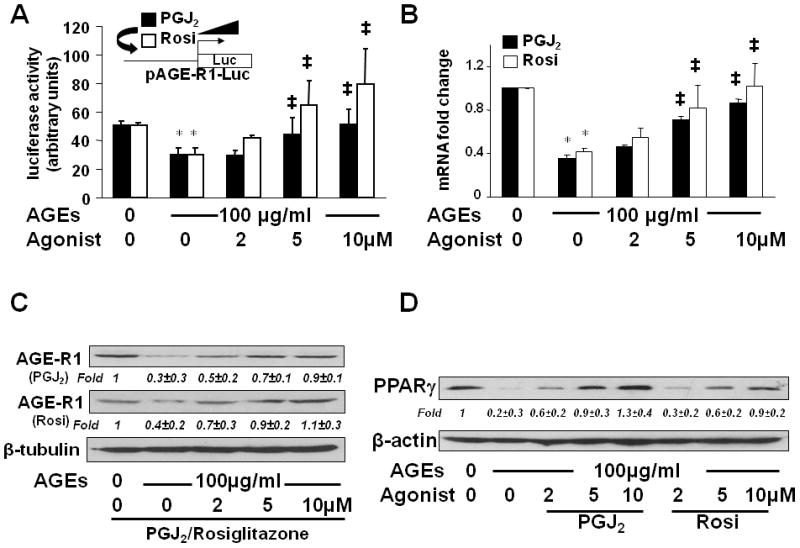
(A) HSCs were transfected with the AGE-R1 promoter luciferase reporter plasmid pAGE-R1-Luc. After recovery, cells were serum-starved for 4 hr prior to the treatment with or without AGEs (100 μg/ml) in the presence of PGJ2, or rosiglitazone (Rosi), at 0–10 μM in serum-depleted media for 24 hr. Luciferase activity assays were conducted (n=6). *p<0.05 vs. cells with no treatment (the corresponding 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the corresponding 2nd column). The floating schema denoted pAGE-R1-Luc in use for transfection and the application of PGJ2, or Rosi, to the system.
(B, C & D) Serum-starved HSCs were stimulated with or without AGEs (100 μg/ml) in the presence of PGJ2, or Rosi, at 0–10 μM in serum-depleted media for 24 hr. Total RNA and whole cell extracts were prepared. (B). real-time PCR assays. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. cells with no treatment (the corresponding 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the corresponding 2nd column). (C & D) Western blotting analyses. β-tubulin, or β-actin, was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
To verify the role of the activation of PPARγ in inducing gene expression of AGE-R1, serum-starved HSCs were stimulated w/wt AGEs (100 μg/ml) in the presence of the PPARγ agonist PGJ2, or rosiglitazone (Rosi) at 0–10 μM in serum- depleted media for 24 hr. Results from real-time PCR (Fig. 7B) and Western blotting analyses (Fig. 7C) demonstrated that the activation of PPARγ by PGJ2 or rosiglitazone eliminated the inhibitory effect of AGEs and dose-dependently induced gene expression of AGE-R1 at the levels of transcript and protein in the cells (the corresponding 3rd – 5th columns and lanes).
Additional experiments were conducted to address the question whether the stimulation of the AGE-R1 gene expression in HSCs by PGJ2 or rosiglitazone corresponded to an increase in PPARγ. HSCs were treated w/wt AGEs (100 μg/ml) in the presence of PGJ2, or rosiglitazone at 0–10 μM in serum-depleted media for 24 hr. Western blotting analyses in Fig. 7D demonstrated that PGJ2 or rosiglitazone, mimicking the role of curcumin (Fig. 6B), dose-dependently eliminated the effect of AGEs on the level of PPARγ in HSCs in vitro. To our knowledge, the role of the PPARγ agonist PGJ2, or rosiglitazone, in inducing gene expression of PPARγ in HSCs has never been reported. Taken together, our results demonstrated that the activation of PPARγ eliminated the inhibitory effect of AGEs and stimulated gene expression of AGE-R1 in HSCs in vitro.
The stimulation of the trans-activity of PPARγ played a critical role in the curcumin-caused elimination of the effect of AGEs on the inhibition of gene expression of AGE-R1 in HSCs in vitro
To further evaluate the role of the activation of PPARγ in the curcumin-caused elimination of the inhibitory effect of AGEs on gene expression of AGE-R1, serum-starved HSCs were pretreated w/wt the PPARγ antagonist PD68235 (20 μM) for 30 min prior to the exposure to AGEs (100 μg/ml), or curcumin (20 μM), or both, in serum-depleted media with PGJ2 (5 μM) for additional 24 hr. Total RNA and whole cell extracts were prepared for real-time PCR (Fig. 8A) and Western blotting analyses (Fig. 8B). Compared with the untreated control (the 1st column and lane), AGEs significantly reduced the levels of mRNA and protein of AGE-R1 in the cells (the 2nd column and lane). Curcumin, as expected, dramatically eliminated the effect of AGEs and increased the contents of mRNA and protein of AGE-R1 (the 3rd column and lane). It was of interest to observe that the blockade of PPARγ activation by the pretreatment with the PPARγ antagonist PD68235 (PD) apparently attenuated the role of curcumin in eliminating the inhibitory effect of AGEs (the last column and lane), suggesting a critical role of the activity of PPARγ in the process. PD68235 alone had no apparent effect (the 4th column and lane). These results collectively indicated that the stimulation of the trans-activity of PPARγ played a critical role in the curcumin-caused elimination of the effect of AGEs on the inhibition of gene expression of AGE-R1 in HSCs in vitro.
Figure 8. The stimulation of the trans-activity of PPARγ played a critical role in the curcumin-caused elimination of the effect of AGEs on inhibiting gene expression of AGE-R1 in HSCs in vitro.
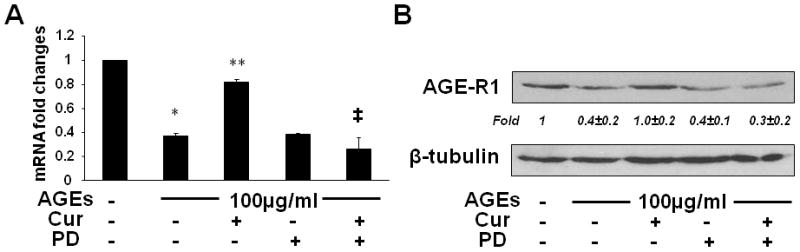
Serum-starved HSCs were pretreated w/wt the PPARγ antagonist PD68235 (PD) (20 μM) for 30 min prior to the exposure to AGEs (100 μg/ml), or curcumin (20 μM), or both, in serum-depleted media with PGJ2 (5 μM) for additional 24 hr. Total RNA and whole cell extracts were prepared. (A). real-time PCR assays. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. cells with no treatment (the 1st column); **p<0.05 vs. cells treated with AGEs alone (the 2nd column); ‡p<0.05 vs. cells treated with both AGEs and curcumin (the 3rd column). (B) Western blotting analyses. β-tubulin was used as an internal control for equal loading. Representatives were shown from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
ERK and PPARγ acted sequentially in the attenuation of the effect of AGEs on the regulation of the expression of AGE-R1 in HSCs
To elucidate the relationship between the ERK signaling pathway and the activation of PPARγ in regulating gene expression of AGE-R1, passaged HSCs in six-well plates were co-transfected with a total of 4.5 μg of a DNA mixture per well, including 2 μg of the promoter activity luciferase reporter plasmid pAGE-R1-Luc, 0.5μg of pSV-β-gal and 2 μg of the cDNA expression plasmid pa-ERK, encoding constitutively active ERK, at indicated doses plus the empty vector pcDNA. The latter was used to ensure an equal amount of total DNA in transfection assays. After overnight recovery from transfection, cells were serum-starved for 4 hr prior to the treatment w/wt AGEs (100 μg/ml) in the presence or absence of PGJ2 (5 μM) in serum-depleted media for additional 24 hr. As shown in Fig. 9, compared to the untreated control (the 1st column), AGEs significantly reduced, as expected, luciferase activity in cells transfected with pAGE-R1-Luc (the 2nd column). Forced expression of constitutively active ERK potentiated the inhibitory effect of AGEs (the 3rd column). On the other hand, the activation of PPARγ with PGJ2 dramatically diminished the inhibitory effect of AGEs (the 4th column). It was of interest to observe that forced expression of constitutively active ERK eliminated the role of PGJ2 and reduced luciferase activities in a dose-dependent manner (the 4th – 7th columns), indicating that the activation of the ERK signaling pathway counteracted the role of PPARγ activation in stimulating the promoter activity of AGE-R1. This result suggested a sequential relationship between ERK and PPARγ in regulating the promoter activity of AGE-R1 in HSCs.
Figure 9. ERK and PPARγ acted sequentially in the attenuation of the effect of AGEs on the regulation of the expression of AGE-R1 in HSCs.
(A) Passaged HSCs in six-well plates were co-transfected with a total of 4.5 μg of a DNA mixture per well, including 2 μg of the promoter activity luciferase reporter plasmid pAGE-R1-Luc, 0.5μg of pSV-β-gal and 2 μg of the cDNA expression plasmid pa-ERK, encoding constitutively active ERK, at indicated doses plus the empty vector pcDNA. The latter was used to ensure an equal amount of total DNA in transfection assays. After overnight recovery, cells were serum-starved for 4 hr prior to the treatment w/wt AGEs (100 μg/ml) in the presence or absence of the PPARγ agonist PGJ2 (5 μM) in serum-depleted media for additional 24 hr. Luciferase activity assays were conducted (n=6). *p<0.05 vs. cells with no treatment (the 1st column); ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column); §p<0.05 vs. cells transfected with no pa-ERK, but treated with both AGEs and PGJ2 (the 4th column). The floating schema denoted pAGE-R1-Luc in use w/wt pa-ERK for co-transfection and the application of AGEs w/wt PGJ2 to the system.
(B) serum-starved HSCs were treated with AGEs (100 μg/ml) and the PPARγ agonist PGJ2 (5 μM) plus or minus the ERK selective inhibitor PD98059 (10 μM) in serum-depleted media for 24 hr. Western blotting analyses were conducted. β-tubulin was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
To further elucidate the relationship between ERK and PPARγ in the attenuation of the inhibitory effect of AGEs on the regulation of gene expression of AGE-R1, serum-starved HSCs were treated with AGEs (100 μg/ml) and the PPARγ agonist PGJ2 (5 μM) plus or minus the ERK selective inhibitor PD98059 (10 μM) in serum-depleted media for 24 hr. Western blotting analyses in Fig. 9B revealed that compared with the untreated cells (the 1st lane), PGJ2 by itself increased the level of AGE-R1 (the 5th lane). AGEs significantly reduced the level of AGE-R1 in HSCs (the 2nd lane), which was dramatically attenuated, as expected, by the activation of PPARγ by PGJ2 (the 3rd lane). It was further shown that the presence of PD98059 strengthened the role of PGJ2 in the attenuation of the inhibitory effect of AGEs on the expression of AGE-R1 in HSCs (the 4th lane). Taken together, our results suggested a sequential relationship between upstream ERK and downstream PPARγ in the attenuation of the effect of AGEs on the regulation of the expression of AGE-R1 in HSCs.
DISCUSSION
The development of T2DM is coupled with the increase in the levels of many detrimental factors, some of which stimulate the activation of HSCs, including hyperinsulinemia (26), hyperleptinemia (34), dyslipidemia (35, 36) and hyperglycemia (37). Our present study provided the evidence that AGEs, whose formation is facilitated and stimulated by hyperglycemia, were an additional stimulus for inducing HSC activation. AGE-R1 has been reported to have functions in detoxification and clearance of AGEs (7). The current study was designed to evaluate effects of AGEs on inducing HSC activation, to assess the role of curcumin in diminishing the AGE effects and to explore the underlying mechanisms. We observed that AGEs induced the activation of HSCs and suppressed gene expression of AGE-R1 in activated HSCs. The phytochemical curcumin eliminated the effects of AGEs and induced gene expression of AGE-R1 likely by inhibiting ERK activity and stimulating the trans-activity of PPARγ.
AGEs elicit their effects via receptors. While AGE-R1 and other receptors, including CD36 and Scr-II, assist AGE clearance, RAGE facilitates pro-inflammation (7, 38). The AGEs-RAGE-OS axis is involved in diabetic complications (3). A dramatic increase in RAGE expression is found in diabetic patients with high levels of plasma AGEs (8, 39). On the other hand, the AGE-R1 abundance is significantly reduced in diabetic kidney (9), suggesting a possible inverse relationship between AGEs-mediated cell injury and low expression of AGE-R1. Over-expression of AGE-R1 reduces basal levels of AGEs and OS, enhances resistance to hyperglycemia and protects against inflammation in vivo (40). Inhibition of AGE formation, blockade of AGEs-RAGE interaction, suppression of RAGE expression, interruption of its signaling and induction of AGE-R1 expression are, thus, novel therapeutic strategies for treatment of diabetic complications (41). To our knowledge, this study is the first report to observe that AGEs suppressed gene expression of AGE-R1 in passaged HSCs, which could be eliminated by curcumin. Prior studies suggested that AGE-R1 might be a negative regulator in the inflammatory response to AGEs in mesangial cells (7). It was also reported that an increase in the dietary AGE contents reduced the ratio of AGE-R1 to RAGE in cells, leading to an increase in oxidative stress and organ damage, and a reduction in life span (42). In addition to the induction of gene expression of AGE-R1 in HSCs, our preliminary results also suggested that curcumin inhibited gene expression of RAGE in HSCS in vitro (data not shown). Additional experiments are ongoing in our lab to elucidate molecular mechanisms by which curcumin divergently regulated gene expression of RAGE and AGE-R1 in HSCs.
Oxidative stress induces HSC activation and hepatic fibrogenesis (10). Studies have suggested that AGE-R1 is not only a scavenger receptor which facilitates uptake and degradation of AGEs, but also a receptor mediating the attenuation of oxidative stress (43). Therefore, the curcumin-caused induction of gene expression of AGE-R1 could attenuate oxidative stress in passaged HSCs, leading to the inhibition of the AGEs-induced activation of HSCs. On the other hand, curcumin itself is a potent antioxidant, whose antioxidant capacity is 100-fold stronger than that of vitamin E/C (44), likely by inducing gene expression of glutamate-cysteine ligase, a key rate-limiting enzyme in de novo synthesis of glutathione (18). It is plausible to assume that in addition to the induction of AGE-R1 gene expression, curcumin itself could attenuate oxidative stress, which also facilitates the inhibition of the AGEs-induced activation of HSCs.
As demonstrated in the report, AGEs inhibited gene expression of AGE-R1, which could be diminished by the activation of PPARγ by PGJ2 or rosiglitazone (Fig. 7C). It was explained that a portion of the stimulatory role of PGJ2 might be used to counteract the inhibitory effect of AGEs. Therefore, the effect of PGJ2 alone on elevating the level of AGE-R1 was more prominent than that of the treatment with both PGJ2 and AGEs (Fig. 9B). On the other hand, our results could not exclude other possible mechanisms by which the activation of PPARγ by PGJ2 elevated the abundance of AGE-R1, including increasing the protein stability and/or mRNA half-life of AGE-R1 in HSCs in vitro.
Our results in this report indicated that AGEs stimulated the activation of ERK, suppressed gene expression of PPARγ and reduced its trans-activation activity in activated HSCs in vitro. Our results also suggested a sequential relationship between upstream ERK and downstream PPARγ in the attenuation of the effect of AGEs on the regulation of the expression of AGE-R1 in HSCs. Our observations were in agreement with prior other reports (45, 46). The activation of mitogen-activated protein kinase (MAPK) signaling pathways stimulates the phosphorylation of PPARγ, resulting in a reduction in the trans-activation activity of the nuclear transcription factor (45, 46). We demonstrated that curcumin inhibited the activity of ERK and induced gene expression of PPARγ in passaged HSCs (16, 17), both of which attenuated the inhibitory effects on the promoter activity of AGE-R1 in HSCs. It is, therefore, understandable that curcumin could eliminate the effect of AGEs and induce gene expression of AGE-R1 in HSCs. It is worth mentioning that the pharmacological approach, i.e. using the selective ERK inhibitor, might not be the best, but a proper one, to address the questions studied in this report. Additional experiments are necessary to elucidate the underlying molecular mechanisms of the PPARγ-dependent ERK signaling pathway in regulating the promoter activity of AGE-R1 in HSCs. However, it is noteworthy that our results did not exclude possible PPARγ-independent effects of the ERK pathway on inhibiting the promoter activity of AGE-R1 in HSCs.
The toxicity of curcumin to cultured HSCs was previously evaluated (16). Based on results from lactate dehydrogenase release assays, trypan blue exclusion assays and a rapid recovery of cell proliferation after withdrawal of curcumin, it was concluded that curcumin up to 100 μM was not toxic to cultured HSCs. Curcumin at 20 μM was used in most of our in vitro experiments. The systemic bioavailability of curcumin is relatively low (47). Curcumin concentrations in human plasma can reach up to 2 μM following oral intake of very high amounts of curcumin (48). Few reports could be found regarding serum levels of the AGE-proteins in human populations with or without diabetes. Among these limited studies, the levels of serum AGEs in human were not consistent (49–53). AGEs at 100 μg/ml was used in most of experiments in this project. The same dose of human glycated albumin was also used to examine its effects on insulin signaling in L6 skeletal muscle cells (24). The concentration of AGEs used in our experiments was determined by measuring AGEs-specific fluorescence with excitation at 360 nm and emissions at 440 nm (22, 23). N-carboxymethyl-lysine (CML) was a major AGE among them (22, 23). The level of CML was previously observed to be higher in diabetic patients than non-diabetic controls (51). It is noteworthy that because the in vivo system is multi-factorial, directly extrapolating in vitro conditions and results, e. g. effective concentrations, to the in vivo system, or vice versa, might be misleading.
Prior studies showed the role of curcumin in inhibiting the AGEs-induced increase in NF-kB and AP-1 activity, VEGF mRNA up-regulation, and the resultant increase in DNA synthesis in microvascular endothelial cells (54). The influence of curcumin on the level of AGEs and the cross-linking of collagen in diabetic rats were studied (55). A correlation between the level of AGEs and collagen cross-linking was noted. It was further found that accelerated accumulation of AGE-collagen in diabetic animals was prevented by curcumin. The preventive effect of curcumin on the advanced glycation and cross-linking of collagen was pronounced. Results from this report demonstrated the role of curcumin administration in the prevention of AGE-induced complications of diabetes mellitus (55). Additional in vivo experiments are necessary to verify the in vitro observed protective role of curcumin in attenuating the AGE effects on the activation of HSCs.
In conclusion, results in this report supported our initial hypothesis and demonstrated that AGEs stimulated HSC activation, which was eliminated by curcumin at least partially by inducing AGE-R1 gene expression. This process was likely mediated by inhibiting ERK activity, inducing gene expression of PPARγ and sequentially stimulating its trans-activity. These results were summarized in Fig. 10. The authors admit that the evidence presented in this report is somewhat indirect. Experiments with high expression of AGE-R1 in HSCs by stable transfection with exogenous AGE-R1 cDNA, and knockdown expression of AGE-R1 by RNA interference technology, including small interfering RNA, would provide more direct evidence to the involvement and role of AGE-R1 in mediating the curcumin inhibition of AGE-induced HSC activation. These experiments are ongoing in our laboratory. It bears emphasis that our results do not exclude any other mechanisms by which curcumin eliminates the stimulatory effects of AGEs on the activation of HSCs and induces gene expression of AGE-R1 in HSCs. It remains unknown how the curcumin-caused inhibition of ERK and stimulation of PPARγ activity could result in the induction of gene expression of AGE-R1 in HSCs. Additional experiments are necessary to explore the underlying molecular mechanisms. The results from this study provide novel insight into effects of diabetes-associated AGEs on inducing HSC activation and hepatic fibrogenesis, the roles and underlying mechanisms of curcumin in abrogating the stimulatory effects of AGEs.
Figure 10. A simplified action model of AGEs and curcumin in regulating the activation of HSCs.
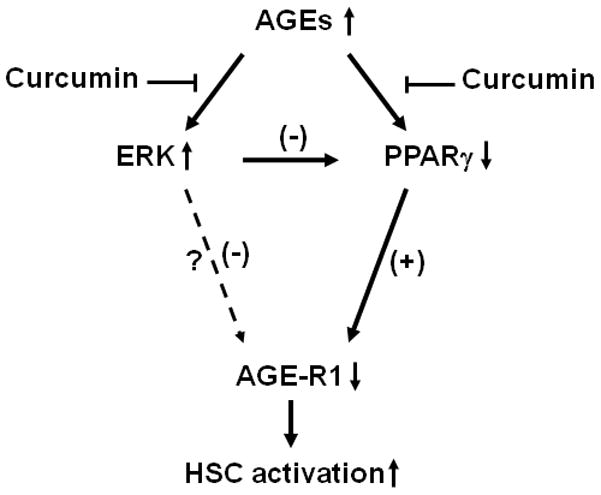
AGEs stimulated HSC activation, which was eliminated by curcumin by inducing expression of AGE-R1 gene. The process was likely mediated by inhibiting ERK activity, inducing gene expression of PPARγ and sequentially stimulating its trans-activity. “↑” or “↓” indicates the effect of AGEs.
Acknowledgments
The work was supported by the grant DK 47995 from NIH/NIDDK to A. Chen.
ABBREVIATIONS
- AGEs
advanced glycation end-products
- AGE-R1
AGE receptor-1
- α-SMA
alpha-smooth muscle actin
- BSA
bovine serum albumin
- CTGF
connective tissue growth factor
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HSCs
hepatic stellate cells
- NASH
non-alcoholic steatohepatitis
- PDGF-βR
platelet-derived growth factor-beta receptor
- PPARγ
peroxisome proliferator-activated receptor-gamma
- RAGE
the receptor for AGEs
- TGF-βR
transforming growth factor-beta receptor
References
- 1.Tsochatzis E, Papatheodoridis GV, Manesis EK, Kafiri G, Tiniakos DG, Archimandritis AJ. Metabolic syndrome is associated with severe fibrosis in chronic viral hepatitis and non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2008;27(1):80–89. doi: 10.1111/j.1365-2036.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- 2.Clark JM. The epidemiology of nonalcoholic fatty liver disease in adults. J Clin Gastroenterol. 2006;40 (Suppl 1):S5–10. doi: 10.1097/01.mcg.0000168638.84840.ff. [DOI] [PubMed] [Google Scholar]
- 3.Bierhaus A, Humpert PM, Morcos M, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83(11):876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 4.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54(6):1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 5.Libby P, Plutzky J. Diabetic macrovascular disease: the glucose paradox? Circulation. 2002;106(22):2760–2763. doi: 10.1161/01.cir.0000037282.92395.ae. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt AM, Yan SD, Yan SF, Stern DM. The biology of the receptor for advanced glycation end products and its ligands. Biochim Biophys Acta. 2000;1498(2–3):99–111. doi: 10.1016/s0167-4889(00)00087-2. [DOI] [PubMed] [Google Scholar]
- 7.Lu C, He JC, Cai W, Liu H, Zhu L, Vlassara H. Advanced glycation endproduct (AGE) receptor 1 is a negative regulator of the inflammatory response to AGE in mesangial cells. Proc Natl Acad Sci U S A. 2004;101(32):11767–11772. doi: 10.1073/pnas.0401588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett J, Schmidt AM, Yan SD, et al. Survey of the distribution of a newly characterized receptor for advanced glycation end products in tissues. Am J Pathol. 1993;143(6):1699–1712. [PMC free article] [PubMed] [Google Scholar]
- 9.Li YM, Mitsuhashi T, Wojciechowicz D, et al. Molecular identity and cellular distribution of advanced glycation endproduct receptors: relationship of p60 to OST-48 and p90 to 80K-H membrane proteins. Proc Natl Acad Sci U S A. 1996;93(20):11047–11052. doi: 10.1073/pnas.93.20.11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134(6):1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman SL, Rockey DC, McGuire RF, Maher JJ, Boyles JK, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15(2):234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- 12.Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol. 2006;21 (Suppl 3):S84–87. doi: 10.1111/j.1440-1746.2006.04584.x. [DOI] [PubMed] [Google Scholar]
- 13.Calamita G, Portincasa P. Present and future therapeutic strategies in non-alcoholic fatty liver disease. Expert Opin Ther Targets. 2007;11(9):1231–1249. doi: 10.1517/14728222.11.9.1231. [DOI] [PubMed] [Google Scholar]
- 14.O’Connell MA, Rushworth SA. Curcumin: potential for hepatic fibrosis therapy? Br J Pharmacol. 2008;153(3):403–405. doi: 10.1038/sj.bjp.0707580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, Zheng S, Lin J, Ryerse J, Chen A. Curcumin protects the rat liver from CCl4-caused injury and fibrogenesis by attenuating oxidative stress and suppressing inflammation. Mol Pharmacol. 2008;73(2):399–409. doi: 10.1124/mol.107.039818. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Fu Y, Chen A. Activation of peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of curcumin on rat hepatic stellate cell growth. Am J Physiol Gastrointest Liver Physiol. 2003;285(1):G20–30. doi: 10.1152/ajpgi.00474.2002. [DOI] [PubMed] [Google Scholar]
- 17.Zheng S, Chen A. Activation of PPARgamma is required for curcumin to induce apoptosis and to inhibit the expression of extracellular matrix genes in hepatic stellate cells in vitro. Biochem J. 2004;384(Pt 1):149–157. doi: 10.1042/BJ20040928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng S, Yumei F, Chen A. De novo synthesis of glutathione is a prerequisite for curcumin to inhibit hepatic stellate cell (HSC) activation. Free Radic Biol Med. 2007;43(3):444–453. doi: 10.1016/j.freeradbiomed.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyogo H, Yamagishi S, Iwamoto K, et al. Elevated levels of serum advanced glycation end products in patients with non-alcoholic steatohepatitis. J Gastroenterol Hepatol. 2007;22(7):1112–1119. doi: 10.1111/j.1440-1746.2007.04943.x. [DOI] [PubMed] [Google Scholar]
- 20.Iwamoto K, Kanno K, Hyogo H, et al. Advanced glycation end products enhance the proliferation and activation of hepatic stellate cells. J Gastroenterol. 2008;43(4):298–304. doi: 10.1007/s00535-007-2152-7. [DOI] [PubMed] [Google Scholar]
- 21.Makita Z, Vlassara H, Cerami A, Bucala R. Immunochemical detection of advanced glycosylation end products in vivo. J Biol Chem. 1992;267(8):5133–5138. [PubMed] [Google Scholar]
- 22.Monnier VM, Kohn RR, Cerami A. Accelerated age-related browning of human collagen in diabetes mellitus. Proc Natl Acad Sci U S A. 1984;81(2):583–587. doi: 10.1073/pnas.81.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrobel K, Wrobel K, Garay-Sevilla ME, Nava LE, Malacara JM. Novel analytical approach to monitoring advanced glycosylation end products in human serum with on-line spectrophotometric and spectrofluorometric detection in a flow system. Clin Chem. 1997;43(9):1563–1569. [PubMed] [Google Scholar]
- 24.Miele C, Riboulet A, Maitan MA, et al. Human glycated albumin affects glucose metabolism in L6 skeletal muscle cells by impairing insulin-induced insulin receptor substrate (IRS) signaling through a protein kinase C alpha-mediated mechanism. J Biol Chem. 2003;278(48):47376–47387. doi: 10.1074/jbc.M301088200. [DOI] [PubMed] [Google Scholar]
- 25.Camp HS, Chaudhry A, Leff T. A novel potent antagonist of peroxisome proliferator-activated receptor gamma blocks adipocyte differentiation but does not revert the phenotype of terminally differentiated adipocytes. Endocrinology. 2001;142(7):3207–3213. doi: 10.1210/endo.142.7.8254. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Zheng S, Chen A. Curcumin attenuates the effects of insulin on stimulating hepatic stellate cell activation by interrupting insulin signaling and attenuating oxidative stress. Lab Invest. 2009;89(12):1397–1409. doi: 10.1038/labinvest.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285(2):194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 28.Chen A, Beno DW, Davis BH. Suppression of stellate cell type I collagen gene expression involves AP-2 transmodulation of nuclear factor-1-dependent gene transcription. J Biol Chem. 1996;271(42):25994–25998. doi: 10.1074/jbc.271.42.25994. [DOI] [PubMed] [Google Scholar]
- 29.Chen A, Zheng S. Curcumin inhibits connective tissue growth factor gene expression in activated hepatic stellate cells in vitro by blocking NF-kappaB and ERK signalling. Br J Pharmacol. 2008;153(3):557–567. doi: 10.1038/sj.bjp.0707542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng S, Chen A. Curcumin suppresses the expression of extracellular matrix genes in activated hepatic stellate cells by inhibiting gene expression of connective tissue growth factor. Am J Physiol Gastrointest Liver Physiol. 2006;290(5):G883–893. doi: 10.1152/ajpgi.00450.2005. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zheng S, Lin J, Zhang QJ, Chen A. The interruption of the PDGF and EGF signaling pathways by curcumin stimulates gene expression of PPARgamma in rat activated hepatic stellate cell in vitro. Lab Invest. 2007;87(5):488–498. doi: 10.1038/labinvest.3700532. [DOI] [PubMed] [Google Scholar]
- 32.Lin J, Chen A. Activation of peroxisome proliferator-activated receptor-gamma by curcumin blocks the signaling pathways for PDGF and EGF in hepatic stellate cells. Lab Invest. 2008;88(5):529–540. doi: 10.1038/labinvest.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukamoto H, She H, Hazra S, Cheng J, Miyahara T. Anti-adipogenic regulation underlies hepatic stellate cell transdifferentiation. J Gastroenterol Hepatol. 2006;21 (Suppl 3):S102–105. doi: 10.1111/j.1440-1746.2006.04573.x. [DOI] [PubMed] [Google Scholar]
- 34.Tang Y, Zheng S, Chen A. Curcumin eliminates leptin’s effects on hepatic stellate cell activation via interrupting leptin signaling. Endocrinology. 2009;150(7):3011–3020. doi: 10.1210/en.2008-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang Q, Chen A. Curcumin suppresses expression of low-density lipoprotein (LDL) receptor, leading to the inhibition of LDL-induced activation of hepatic stellate cells. Br J Pharmacol. 2009;157(8):1354–1367. doi: 10.1111/j.1476-5381.2009.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Q, Chen A. Curcumin eliminates oxidized LDL roles in activating hepatic stellate cells by suppressing gene expression of lectin-like oxidized LDL receptor-1. Lab Invest. 2009;89(11):1275–1290. doi: 10.1038/labinvest.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Chen A. Curcumin diminishes the impacts of hyperglycemia on the activation of hepatic stellate cells by suppressing membrane translocation and gene expression of glucose transporter-2. Mol Cell Endocrinol. 2011;333:160–171. doi: 10.1016/j.mce.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sourris KC, Forbes JM. Interactions between advanced glycation end-products (AGE) and their receptors in the development and progression of diabetic nephropathy - are these receptors valid therapeutic targets. Curr Drug Targets. 2009;10(1):42–50. doi: 10.2174/138945009787122905. [DOI] [PubMed] [Google Scholar]
- 39.Tanji N, Markowitz GS, Fu C, et al. Expression of advanced glycation end products and their cellular receptor RAGE in diabetic nephropathy and nondiabetic renal disease. J Am Soc Nephrol. 2000;11(9):1656–1666. doi: 10.1681/ASN.V1191656. [DOI] [PubMed] [Google Scholar]
- 40.Torreggiani M, Liu H, Wu J, et al. Advanced glycation end product receptor-1 transgenic mice are resistant to inflammation, oxidative stress, and post-injury intimal hyperplasia. Am J Pathol. 2009;175(4):1722–1732. doi: 10.2353/ajpath.2009.090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagishi S, Nakamura K, Matsui T, Ueda S, Fukami K, Okuda S. Agents that block advanced glycation end product (AGE)-RAGE (receptor for AGEs)-oxidative stress system: a novel therapeutic strategy for diabetic vascular complications. Expert Opin Investig Drugs. 2008;17(7):983–996. doi: 10.1517/13543784.17.7.983. [DOI] [PubMed] [Google Scholar]
- 42.Vlassara H, Uribarri J, Cai W, Striker G. Advanced glycation end product homeostasis: exogenous oxidants and innate defenses. Ann N Y Acad Sci. 2008;1126:46–52. doi: 10.1196/annals.1433.055. [DOI] [PubMed] [Google Scholar]
- 43.Cai W, He JC, Zhu L, Chen X, Striker GE, Vlassara H. AGE-receptor-1 counteracts cellular oxidant stress induced by AGEs via negative regulation of p66shc-dependent FKHRL1 phosphorylation. Am J Physiol Cell Physiol. 2008;294(1):C145–152. doi: 10.1152/ajpcell.00350.2007. [DOI] [PubMed] [Google Scholar]
- 44.Sreejayan, Rao MN. Curcuminoids as potent inhibitors of lipid peroxidation. J Pharm Pharmacol. 1994;46(12):1013–1016. doi: 10.1111/j.2042-7158.1994.tb03258.x. [DOI] [PubMed] [Google Scholar]
- 45.Camp HS, Tafuri SR. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J Biol Chem. 1997;272(16):10811–10816. doi: 10.1074/jbc.272.16.10811. [DOI] [PubMed] [Google Scholar]
- 46.Camp HS, Tafuri SR, Leff T. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology. 1999;140(1):392–397. doi: 10.1210/endo.140.1.6457. [DOI] [PubMed] [Google Scholar]
- 47.Sharma RA, McLelland HR, Hill KA, et al. Pharmacodynamic and pharmacokinetic study of oral Curcuma extract in patients with colorectal cancer. Clin Cancer Res. 2001;7(7):1894–1900. [PubMed] [Google Scholar]
- 48.Garcea G, Jones DJ, Singh R, et al. Detection of curcumin and its metabolites in hepatic tissue and portal blood of patients following oral administration. Br J Cancer. 2004;90(5):1011–1015. doi: 10.1038/sj.bjc.6601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butscheid M, Schafer C, Brenner S, et al. Unchanged serum levels of advanced glycation endproducts in patients with liver disease. Naunyn Schmiedebergs Arch Pharmacol. 2007;375(6):401–406. doi: 10.1007/s00210-007-0171-9. [DOI] [PubMed] [Google Scholar]
- 50.Scharnagl H, Stojakovic T, Winkler K, Rosinger S, Marz W, Boehm BO. The HMG-CoA reductase inhibitor cerivastatin lowers advanced glycation end products in patients with type 2 diabetes. Exp Clin Endocrinol Diabetes. 2007;115(6):372–375. doi: 10.1055/s-2007-973830. [DOI] [PubMed] [Google Scholar]
- 51.Basta G, Sironi AM, Lazzerini G, et al. Circulating soluble receptor for advanced glycation end products is inversely associated with glycemic control and S100A12 protein. J Clin Endocrinol Metab. 2006;91(11):4628–4634. doi: 10.1210/jc.2005-2559. [DOI] [PubMed] [Google Scholar]
- 52.Schiel R, Franke S, Appel T, et al. Improvement in quality of diabetes control and concentrations of AGE-products in patients with type 1 and insulin-treated type 2 diabetes mellitus studied over a period of 10 years (JEVIN) J Diabetes Complications. 2003;17(2):90–97. doi: 10.1016/s1056-8727(02)00203-9. [DOI] [PubMed] [Google Scholar]
- 53.Ghanem AA, Elewa A, Arafa LF. Pentosidine and N-carboxymethyl-lysine: biomarkers for type 2 diabetic retinopathy. Eur J Ophthalmol. 2011;21(1):48–54. doi: 10.5301/ejo.2010.4447. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto T, Yamagishi S, Inagaki Y, et al. Angiogenesis induced by advanced glycation end products and its prevention by cerivastatin. Faseb J. 2002;16(14):1928–1930. doi: 10.1096/fj.02-0030fje. [DOI] [PubMed] [Google Scholar]
- 55.Sajithlal GB, Chithra P, Chandrakasan G. Effect of curcumin on the advanced glycation and cross-linking of collagen in diabetic rats. Biochem Pharmacol. 1998;56(12):1607–1614. doi: 10.1016/s0006-2952(98)00237-8. [DOI] [PubMed] [Google Scholar]


