Abstract
OBJECTIVES:
Interleukin-23 (IL-23) has emerged as a new therapeutic target for the treatment of inflammatory bowel disease (IBD). As biomarkers of disease state and treatment efficacy are becoming increasingly important in drug development, we sought to identify efficacy biomarkers for anti-IL-23 therapy in Crohn's disease (CD).
METHODS:
Candidate IL-23 biomarkers, downstream of IL-23 signaling, were identified using shotgun proteomic analysis of feces and colon lavages obtained from a short-term mouse IBD model (anti-CD40 Rag2−/−) treated preventively with monoclonal antibodies (mAbs) to the IL-23 receptor (IL-23R). The biomarkers were then measured in an IBD T-cell transfer model treated therapeutically with a mAb to IL-23 (p19), confirming their association with IBD. To assess the clinical relevance of these markers, we assessed their concentrations in clinical serum, colon tissue, and feces from CD patients.
RESULTS:
We identified 57 proteins up or downregulated in diseased animals that returned to control values when the mice were treated with mAbs to IL-23R. Among those, S100A8, S100A9, regenerating protein 3β (REG), REG3γ, lipocalin 2 (LCN2), deleted in malignant tumor 1 (DMBT1), and macrophage migration inhibitory factor (MIF) mRNA levels correlated with disease score and dose titration of mAbs to IL-23R or IL-23(p19). All biomarkers, except DMBT1, were also downregulated after therapeutic administration of mAbs to IL-23(p19) in a T-cell transfer IBD mouse model. In sera from CD patients, we confirmed a significant upregulation of S100A8/A9 (43%), MIF (138%), pancreatitis-associated protein (PAP, human homolog of REG3β/γ 49%), LCN2 (520%), and CCL20 (1280%), compared with control samples, as well as a significant upregulation of S100A8/A9 (887%), PAP (401%), and LCN2 (783%) in human feces from CD patients compared with normal controls.
CONCLUSIONS:
These studies identify multiple protein biomarkers downstream of IL-23 that could be valuable tools to assess the efficacy of this new therapeutic agent.
INTRODUCTION
Inflammatory bowel disease (IBD) is a gastrointestinal disorder that can manifest as Crohn's disease (CD) and ulcerative colitis.1 Interleukin-23 (IL-23) is a heterodimeric cytokine2 composed of a p40 subunit, shared with IL-12, and a unique p19 subunit. IL-23 acts through a heterodimeric receptor comprising an IL-12Rβ1 chain and a unique IL-23 receptor (IL-23R) chain.3 IL-23(p19)- or IL-23R-deficient mice are resistant to IBD,4 and the inhibition of IL-23 by anti-p19 neutralizing monoclonal antibodies (mAbs) blocks organ-specific autoimmune inflammation.5, 6, 7, 8 Emphasizing the role of the IL-23 pathway in IBD, IL-23(p19), IL-12(p40), and IL-17A expression is elevated in human inflamed gut from CD and ulcerative colitis patients,9, 10 and the p.Arg381Gln allele polymorphism of IL-23R is associated with protection from CD and ulcerative colitis.11 Moreover, that particular mutation is a loss of receptor function that reduces STAT3 phosphorylation upon stimulation with IL-23, and decreases the number of IL-23 responsive T-cells.12 Overall, these accumulating experimental and clinical data have pointed to IL-23 as a new therapeutic target for the treatment of IBD.13
One of the major pitfalls resulting in failure of new therapies has been a lack of efficacy biomarkers in place before starting drug development.14 Because of this, objective and robust target-engagement biomarker confirming that the drug has bound its target in a functional manner, and efficacy biomarkers to guide the drug-dose selection in clinical trials have become an integral part of drug development.14 The most attractive target engagement and efficacy biomarkers are those in which changes occur early after the interaction of the drug with its target, and whose expression reflects the immediate downstream effects of target engagement, confirming a positive effect of therapeutic intervention. This is critical for toxicology studies in which the drug target is a low abundant cytokine, which is undetectable in normal patients: it demonstrates that the absence of adverse effects is not the result of the drug not engaging its target.
Discovery of possible target engagement and efficacy biomarkers can be carried out in animal models in which a treatment group is included. The resulting candidate protein biomarkers can be subsequently verified in a large number of samples with higher throughput techniques, such as western blot or enzyme-linked immunosorbent assay (ELISA) on human plasma/serum or other clinically relevant and accessible samples.
The purpose of the present study was twofold. First, we evaluated the preventive effect of neutralizing mAbs to IL-23R in the anti-CD40 Rag2−/− T-cell-independent IBD mouse model and compared it to the previously described inhibition by mAbs to IL-23(p19).8 This mouse model recapitulates the alterations in the innate immune response that contributes to disease triggering in human IBD. Anti-IL-23R treatment effects on weight loss, inflammation of the colonic lamina propria, and cytokines and chemokines levels were evaluated. Second, we sought to identify efficacy biomarkers for anti-IL-23 therapy in CD using the same animal model. The candidate biomarkers were then confirmed in a T-cell-dependent mouse model of IBD, as T cell are essential in maintaining intestinal inflammation in human disease. Lastly, the mechanism of regulation of the candidate biomarkers was analyzed in vitro using human epithelial cell line, and the biomarkers were confirmed in human-derived samples from normal and CD patients.
METHODS
Anti-CD40 IBD mouse model
C57BL/6 Rag2−/− (recombinase-activating gene 2 deficient) mice (Taconic Farm, Hudson, NY) were used at >8–12 weeks of age. Animals were housed under specific pathogen-free conditions and kept in microisolators with filtered air at Schering-Plough Biopharma (Palo Alto, CA). Conventional animal chow and water were provided ad libitum. One day before disease induction, mice (N=4 per group) were injected subcutaneously with anti-IL-23R mAbs (Schering-Plough Biopharma, clone 21A4), at 60 mg/kg, or with an IgG1 control antibody (clone 27F11) at 60 mg/kg. At day 0, mice were administered intravenously with 50 μg of anti-CD40 (IgG2a, monoclonal Ab FGK45) to induce IBD.15 The naïve group did not receive any treatment. Mice were killed by carbon dioxide asphyxiation at day 1, 3, 5, or 7. Plasma was collected for ELISA and multiplex serum analysis. Colons were dissected and the preformed feces were extracted and snap-frozen for further analysis. Two centimeter of colon, proximal to the cecum, was collected and treated as follows: (1) fixed in 10% neutral buffered formalin, paraffin-embedded and tissue sections stained with hematoxylin and eosin to evaluate tissue pathology; (2) snap-frozen in liquid nitrogen for further RNA extraction; or (3) placed in ice-cold, calcium-, magnesium- and phenol red-free Hank's balanced salt solution (HBSS) for the separation of epithelial cells from lamina propria. All animal procedures were approved by the Institutional Animal Care and Use Committee of Schering-Plough Biopharma, in accordance with guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care.
T-cell transfer model
C3H.SCID mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and housed in the University of Alabama animal facility. Cecal bacterial antigens (CBA) and CBA-specific T cells were prepared as previously described.16 A total of 1 × 106 cecal bacterial antigen-specific T cells were injected into groups of 3–5 C3H.SCID mice intravenously. Adoptive transfer of anti-CD3 activated T cells was used as a control. Anti-IL-23p19 mAb or control mAb was administered intraperitoneally (100 μg/mouse) on the same day as cell transfer and weekly thereafter for the preventive study and from week 4–8, weekly, in the therapeutic study. Two months later or at earlier time points based on the requirement of experiments, the mice were killed and histopathology examined. In addition, total RNA was recovered from the colon of each mouse for real-time reverse-transcription PCR.
All studies were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
ELISA and luminex
Tumor necrosis factor-α (TNF-α) and IL-17A were measured using multiplex serum analysis (Millipore Corporation, Billerica, MA). S100A8/A9 (Calprotectin) ELISA kit was from Hycult Biotechnology (Uden, The Netherlands) and the pancreatitis-associated protein (PAP) kit from Dynabio SA. (Marseille, France). Mouse IL-22 and human migration inhibitory factor (MIF), lipocalin 2 (LCN2), and CCL20/MIP-3α were Quantikine kits from R&D systems (Minneapolis, MN). For human feces, phosphate-buffered saline (containing protease inhibitors “Complete” from Roche, Penzberg, Germany) was added to feces (w/w). Samples were vortexed, incubated under agitation for 2 h at 4 °C, and spun 20 min at 2,000 g Supernatant was collected and spun at 16,000 g for 15 min at 4 °C. Clear supernatant was collected and stored at −80 °C until ELISA analysis.
Shotgun proteomic analysis
Epithelial cell and lamina propria separation
Colons were opened longitudinally, rinsed in ice cold HBSS calcium-, magnesium- and phenol red-free, cut in pieces and incubated under agitation for 20 min at 37 °C in cell dissociation buffer (HBSS calcium-, magnesium- and phenol red-free supplemented with 5 mM EDTA and 10 mM HEPES). Samples were vortexed for 1 min and the supernatant containing the epithelial cells was spun at 2,000 g. Cell pellets were stored at −80 °C, until protein extraction. This epithelial cell enrichment process was repeated twice. Cell pellets and remaining colons (LP) were stored at −80 °C, until protein extraction.
Cell culture
HT-29 primary human colonic epithelial cells (ATCC, St Cloud, MN) were grown in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were starved overnight in serum-free RPMI before stimulation with cytokines. Human IL-17A, IL-22, and TNF-α (R&D Systems) were added to the culture at 20 ng/ml in fresh serum-free medium. Human IL-23 (Schering-Plough Biopharma) was added at 50 ng/ml. Cells were then incubated for 24–48 h, at 37 °C. Supernatants were harvested and analyzed by ELISA. For RNA extraction, cells were rinsed in phosphate-buffered saline and treated with the RNeasy Mini kit (Qiagen, Valencia, CA).
Western blots
For feces, EC, and LP, proteins were extracted in lysis buffer (25 mM Tris pH 8.8/0; 4% SDS). Thirty micrograms of proteins from EC and LP were labeled with Cy5 (1 μl, diluted 1/20) for 20 min on ice. For feces, the quantity of proteins labeled was based on the estimation of protein concentration done by SyproRuby staining (cf. shotgun proteomic analysis). Proteins were separated on SDS–polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene difluoride. Membranes were scanned using a Typhoon 9,400 imager (GE Healthcare, Piscataway, NJ) to assess the amount loaded in each lane. After blocking in 5% milk in TBST (10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature, membranes were probed with the different antibodies at 1 μg/ml, incubated with appropriate HRP-linked secondary antibodies, and signal was detected using the ECL plus kit from GE Healthcare. Band intensities were estimated with Image J 1.39 (NIH). MIF (clone FL-115) and DMBT1 (deleted in malignant tumor 1; Clone P-20) were from Santa Cruz Biotechnology (Santa Cruz, CA); S100A8 (AF3059) and LCN2 (AF1857) antibodies from R&D Systems; and REG3γ antibodies were from Schering-Plough Biopharma.
Real-time reverse-transcription PCR
DNase-treated total RNA was reverse-transcribed using Superscript II (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Primers were designed with Primer Express (PE Biosystems, Foster City, CA) or obtained commercially from Applied Biosystems (Carlsbad, CA). The real-time PCR were performed as described.17
Clinical samples
Serum samples were obtained from Bioreclamation (East Meadow, NY), which works directly with Institutional Review Boards to organize and collect specimens from disease state patients. Colon biopsies were obtained from the Mayo Clinic (Rochester, MN; see Supplementary Table S2 for patient characteristics). The study was approved by the Institutional Review Board of the Mayo Clinic, and all patients included provided written informed consent.
Statistics
Weight, cytokine, and western-blot intensity were analyzed with Prism 3 software (GraphPad Software, La Jolla, CA). P values were calculated with a Mann–Whitney non-paired t-test. The multivariate discriminant analysis was performed with JMP (SAS, Cary, NC).
RESULTS
Proteomic profiling in the anti-CD40 induced colitis Rag2−/− mice
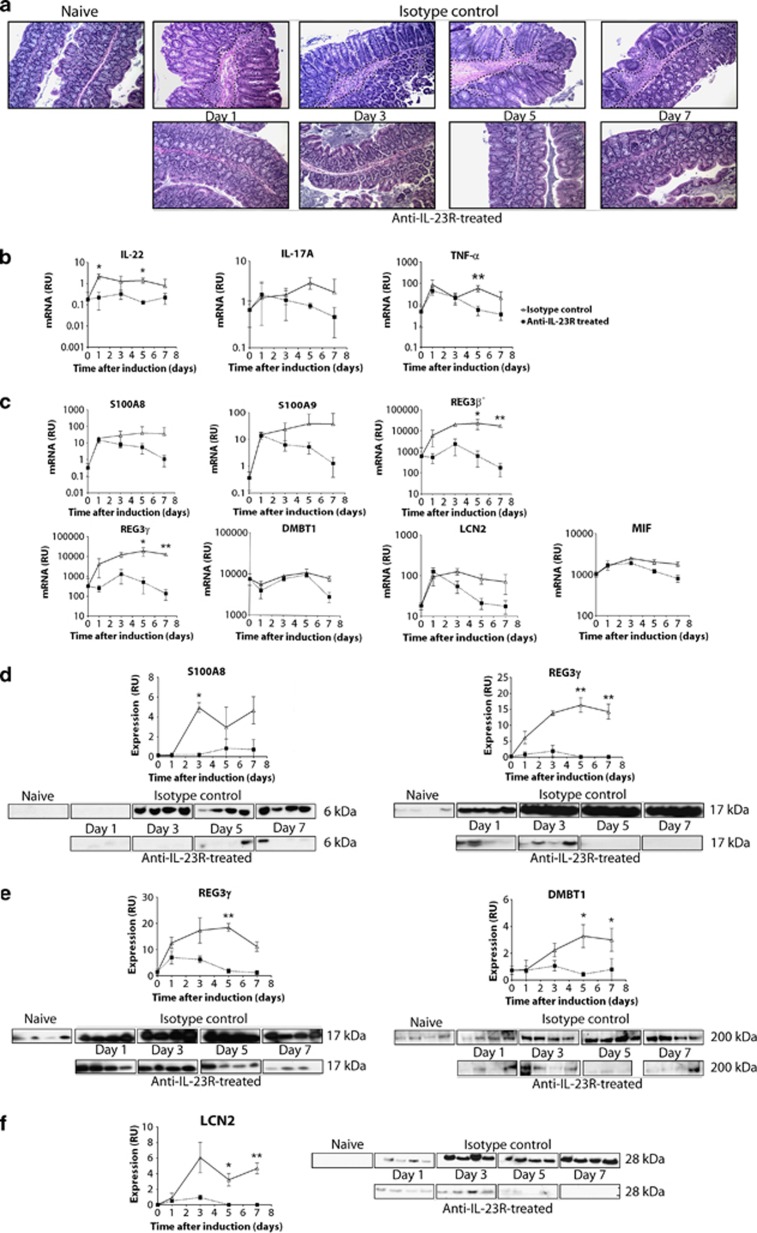
One day after a preventive treatment with either anti-IL-23R mAb or an isotype control, autoimmune colitis was induced in Rag2−/− mice by direct activation of inflammatory dendritic cells and macrophages with a CD40 agonist that mimics activated T cells (Supplementary Figure S1).8 Isotype-control-treated animals showed intense histological signs of colitis from day 1 to day 7, which were associated with epithelial hyperplasia and leukocyte infiltration within the lamina propria. Treatment with a mAb to IL-23R completely prevented this intestinal inflammation (Figure 1a). Consistent with leukocyte infiltration, control animals treated with an isotype mAb had increased colon expression of innate immune cell markers such as CD11b, lactoferrin, and neutrophil elastase, and a marked increase of pro-inflammatory cytokines such as IL-6 and interferon-γ (Supplementary Figure S2A). Treatment with mAbs to IL-23R prevented these increases. Anti-CD40 treatment also rapidly induced IL-22, IL-17A, and TNF-α gene expression in colon tissue (Figure 1b), and treatment with mAbs to IL-23R blocked these increases locally. IL-22 protein concentrations were also increased in the plasma 1 day after the challenge with the CD40 agonist, and this increase was inhibited by treatment with mAbs to IL-23R. In contrast, TNF-α plasma concentrations were unaffected by the treatment (Supplementary Figure S2B). Systemic production of IL-17A was undetectable.
Figure 1.
Candidate biomarker gene expression and protein concentration in mouse colon tissue or feces of a T-cell-independent model. One day before disease induction, Rag2−/− mice were injected subcutaneously with isotype control (at 60 mg/kg) or anti-interleukin(IL)-23R neutralizing antibodies (at 60 mg/kg). On the day of disease induction, mice were injected with 50 μg of anti-CD40 mAbs intravenously to induce immune-mediated colitis (see Methods). Mice were killed at days 1, 3, 5, and 7 (N=4 per group). (a) Hematoxylin and eosin staining of proximal colon. The dashed lines indicate mucosal inflammation. Real-time reverse-transcription PCR analysis of (b) IL-22, IL-17A, and tumor necrosis factor-α (TNF-α), (c) S100A8, S100A9, REG3β, REG3γ, DMBT1, LCN2, and MIF transcripts in mouse proximal colon. Results are presented relative to ubiquitin transcripts (RU, relative units). (d) Western blot analysis of S100A8 and REG3γ in feces, (e) REG3γ and DMBT1 in colonic epithelial cells, and (f) LCN2 in colonic lamina propria. Data are representative of two experiments. Mean value is shown±s.d. *P<0.05, **P<0.01. DMBT1, deleted in malignant tumor 1; LCN2, lipocalin 2; MIF, migration inhibitory factor; REG, regenerating protein.
To identify protein biomarkers secreted in the lumen of the gut that were modulated in the disease state and returned to control values by treatment with mAbs to IL-23R, we performed a shotgun proteomic analysis18 of proteins extracted from colon lavages and feces. Before mass spectrometry analysis, proteins were separated by SDS–polyacrylamide gel electrophoresis to decrease the complexity of the samples. Numerous proteins appeared in multiple bands, at different molecular weights (e.g., DMBT1, carbonic anhydrase 1, neutrophilic elastase), suggesting a possible degradation and/or various post-translational modifications in the colon lumen. Using this approach, we identified peptides from ∼130 unique proteins, 57 of which were modulated in the disease state but remained at control values by treatment with mAbs to IL-23R (Supplementary Table S1). These proteins belonged to different functional families: as enzymes (25%), peptidases (10%), kinases (9%), transporters (5%) and others (51%). Among them, S100A8, S100A9, Reg3β, Reg3γ, DMBT1, MIF, and LCN2 were selected for further validation. Our selection criteria included an up- or downregulation profile in the disease state compared with naïve animals, a reversion by the anti-IL-23R treatment, a known association with IBD pathology and/or IL-23 biology, and availability of reagents to quantify them.
Candidate biomarkers in a T-cell-independent mouse model of model
As our shotgun proteomic analysis is a semi-quantitative approach, we verified the differences observed in naïve, diseased, and treated by mAbs to IL-23R- or IL-23(p19) mice using real-time reverse-transcription PCR analysis of colon tissue. S100A8, S100A9, REG3β, REG3γ, LCN2, and MIF gene expression levels were upregulated at day 1 and sustained at a high level until day 7 in isotype-control-treated animals (Figure 1c). Treatment by mAbs to IL-23R prevented the upregulation of REG3β and REG3γ at day 1, whereas it only showed effects at day 3 for S100A8, S100A9, LCN2, and MIF (Figure 1c). DMBT1 transcripts were significantly downregulated at day 7 in the mice treated by mAbs to IL-23R compared with the one treated by the isotype control (Figure 1c). The upregulation of S100A8 and Reg3γ proteins could be confirmed by western blot in feces from diseased animals compared with mice treated by mAbs to IL-23R (Figure 1d), suggesting that in addition to S100A8/A9 (calprotectin), a well-known fecal marker of IBD,19 Reg3γ is also stable enough to be measured in feces. Reg3γ and DMBT1 were upregulated in colon epithelial cells during disease (Figure 1e) but were undetectable in the lamina propria. On the other hand, LCN2, which was increased in diseased animals, could only be detected in the lamina propria (Figure 1f). Protein concentration of all the candidate biomarkers were maintained at control values in mice treated by mAbs to IL-23R.
In addition, these biomarker transcripts were monitored in dose titration of mAbs to IL-23R and IL-23(p19), and all showed a dose-dependent response with either mAbs treatment in mouse proximal colonic tissue (Supplementary Figure S3A and Supplementary Figure S4A). Three mg/kg of mAbs to IL-23R decreased the expression of S100A8, S100A9, MIF, REG3γ, and LCN2 transcripts to control values, whereas 30 mg/kg were needed for REG3β and DMBT1. Remarkably, the disease score measured in the proximal colon was 0 at both 3 and 30 mg/kg, reflecting complete protection from the disease as measured histologically (Supplementary Figure S3B). The mAbs to IL-23(p19) had a lower affinity than the mAbs to IL-23R (in house data), so animals were treated with larger doses of mAbs to IL-23(p19) to achieve complete disease protection. In all, 120 mg/kg of mAbs to IL-23(p19) decreased the amount of expressed mRNA of all the selected biomarkers to control values (Supplementary Figure S4A), as well as completely protecting the mice from colonic disease as assessed histologically (Supplementary Figure S4B).
Candidate biomarkers in a T-cell-dependent mouse model of colitis
The anti-CD40 model is a T-cell-independent model in which the innate immune system is activated to induce colitis. As T cells are also involved in human disease, we assessed the validity of the candidate biomarkers in the T-cell-dependent colitis mouse model established by Elson et al.6 In these mice, adoptive transfer of Bir14 CD4+ T cells to SCID recipients, colitis is established over several weeks, allowing us to evaluate candidate biomarker gene expression in the colon with both prophylactic and therapeutic treatments by mAbs to IL-23(p19); (Figure 2a). Control recipient mice receiving anti-CD3 activated T cells did not develop colitis,6 whereas mice receiving pathogenic T cells developed intense colonic inflammation, which was associated with an increase of IL-17A, IL-22, and TNF-α gene expression (Figure 2a). As in the anti-CD40 IBD model, S100A8, S100A9, REG3β and γ, LCN2, and MIF transcripts increased with colitis, and treatment by mAbs to IL-23(p19) mAbs administered on the same day as cell transfer prevented this upregulation (Figure 2a). Therapeutic administration of mAbs to IL-23(p19) from week 4 after disease induction also downregulated our set of biomarkers (Figure 2b), except for DMBT1, which was upregulated in diseased animals but unaffected at the transcript level by treatment.
Figure 2.
Expression profiles of the candidate biomarkers in a T-cell-dependent mouse colitis model: preventive and therapeutic treatment with anti-interleukin(IL)-23(p19). (a) Real-time reverse-transcription PCR (real-time RT-PCR) of transcript levels in colon tissue. Bir14 CD4+ T cells or control anti-CD3T cells were transferred intravenously into C3H.SCID mice, and anti-IL-23p19 mAb or control mAb was administered intraperitoneally (100 μg/mouse) on the same day of cell transfer and weekly thereafter. All mice were killed 8 weeks after cell transfer. (b) Bir14 CD4+ T cells or control anti-CD3T cells were transferred into C3H/HeJ SCID mice intravenously. Group 1 was killed after 4 weeks to assess that a colitis was present. Then, groups 2 and 3 were administered either isotype control or anti-IL-23(p19) mAbs at 100 μg/mouse weekly for 4 weeks. Real-time RT-PCR of the markers was performed twice with groups of 3 and 4 mice each time. Results are presented relative to ubiquitin transcripts (RU, relative units). Bars indicate the median values. *P<0.05, **P<0.01 and ***P<0.001.
Candidate biomarker reguation by IL-22, IL-17A, and TNF-α in HT-29 human colonic epithelial cells
IL-17A and IL-22 are directly downstream of IL-23.17 The major cell types in the gut mucosa responsive to IL-17A and IL-22 stimulation are colonic epithelial cells.20 Therefore, to assess the relationship of the candidate IBD biomarkers to IL-23 biology, we stimulated human HT-29 colonic epithelial cells with IL-22 and IL-17A. As TNF-α is a powerful activator of epithelial cells and anti-TNF-α treatment is the standard therapy for CD21, 22 we also used TNF-α alone or in combination with IL-17A or IL-22. IL-23 alone was not able to modulate any of the markers (Figure 3), consistent with the absence of IL-23R in HT-29 cells (Supplementary Figure S5C). The combination of IL-22 and IL-17A modestly increased S100A8 and S100A9 transcripts and adding TNF-α to IL-22 and IL-17A led to a substantial synergistic effect (Figure 3a). The combination of IL-22 and IL-17A was the most potent in increasing DMBT1 expression (Figure 3a).
Figure 3.
Regulation of the candidate biomarkers by interleukin (IL)-17A, IL-22 and tumor necrosis factor-α (TNF-α) in HT-29 human colonic epithelial cells. Human HT29 colonic epithelial cells were incubated with IL-23 (50 ng/ml), IL-22, IL-17A, and/or TNF-α at 20 ng/ml for 24–48 h. (a) Expression levels were measured by real-time reverse-transcription PCR (real-time RT-PCR) analysis. Results are shown as expression relative to ubiquitin mRNA levels (RU, relative units). (b) The protein accumulation was measured in cell supernatant by enzyme-linked immunosorbent assay (ELISA). Error bars represent s.d.; data are representative of 3 experiments. DMBT1, deleted in malignant tumor 1; LCN2, lipocalin 2; PAP, pancreatitis-associated protein.
The combination of IL-22 and IL-17A moderately increased PAP (the human homolog of Reg3β/γ) protein concentration (Figure 3b) and the addition of TNF-α further increased its expression at 24 h and 48 h after treatment (Figure 3b). In contrast to our results in the mouse model, LCN2 expression was detected in human colonic epithelial cells and synergistically accumulated in the cell supernatant after treatment with the combination of IL-22 and IL-17A, IL-17A and TNF-α, as well as in the presence of IL-22, IL-17A, and TNF-α (Figure 3b).
CCL20/MIP-3α is markedly elevated in colonic epithelial cells from IBD patients, and this elevation can be induced by TNF-α.23 We confirmed that TNF-α alone increased CCL20/MIP-3α, but a significant synergistic effect was observed upon adding IL-22, IL-17A, and TNF-α simultaneously after 24 and 48 h of stimulation (Figure 3b).
IL-17F, IL-21, and IL-26 are also secreted by Th-17 cells,17 and IL-21R and IL-26R are expressed by HT-29 cells.24, 25 We tested the ability of these three cytokines to increase the expression of the candidate biomarkers in the presence of IL-17A, IL-22, and TNF-α. None of them was essential for biomarker expression (Supplementary Figure S5).
Taken together, these results show that S100A8/A9, PAP, LCN2, DMBT1, and CCL20/MIP-3α are regulated by IL-17A, IL-22, and TNF-α in human epithelial cells.
Expression of potential IL-23-associated biomarkers in patients with Crohn's disease
To translate our findings from the mouse IBD models to human disease, we measured by ELISA the serum concentration of our biomarkers in normal subjects and patients with active CD (see Supplementary Table S2 for patient characteristics). Several biomarkers were significantly upregulated in CD patients, including S100A8/A9 (P<0.001, 43% upregulation), MIF (P<0.001, 138% upregulation), and PAP (P<0.05, 49% upregulation; Figure 4a).26, 27, 28 Of note, we also demonstrated that LCN2 (P<0.001, 520% upregulation) and CCL20 (P<0.001, 1280% upregulation) were increased in sera from active CD patients compared with normal controls (Figure 4a). Further, we showed that the transcripts of S100A8/A9, CCL20/MIP-3α, DMBT1, PAP, and LCN2 were similarly upregulated in biopsies from the inflamed colon of patients with CD, and markedly decreased in biopsy samples from patients in remission (Figure 4b). As previously described, we confirmed that IL-22, IL-17A, and TNF-α expression was upregulated in colon tissue from patients with active CD29, 30, 31 and downregulated in the remission group, but only IL-22 was significantly decreased (Supplementary Figure S6). We also confirmed in human feces that S100A8/A9 was upregulated in CD patients (P<0.01, 887%) compared with controls and showed for the first time that PAP (P<0.05, 401%) and LCN2 (P<0.01, 783%) were also statistically upregulated in CD (Figure 4c).
Figure 4.
Expression of potential interleukin(IL)-23-associated biomarkers in patients with Crohn's disease. (a) Serum concentrations were measured by enzyme-linked immunosorbent assay (ELISA) in normal control subjects (N=18) and Crohn's disease (CD) patients (N=17). Bars indicate the median values. (b) Real-time reverse-transcription PCR for the expression of the candidate biomarkers in colon tissue biopsies from normal control subjects (N=6–7), CD active (N=6–7), and remission patients (N=6). Results are presented relative to ubiquitin transcripts (RU, relative units). Bars indicate the median values. *P<0.05, **P<0.01, and ***P<0.001. (c) Fecal concentrations were measured by ELISA in normal control subjects (N=20) and CD patients (N=11). Bars indicate the median values. *P<0.05, **P<0.01. DMBT1, deleted in malignant tumor 1; LCN2, lipocalin 2; MIF, migration inhibitory factor; PAP, pancreatitis-associated protein.
Serum and fecal values for all individuals were entered into JMP statistical software without any information about the disease status. JMP then predicted the disease status for each individual. In serum, based on LCN2 measurements alone, four individuals were misclassified out of 35 (88% confidence), whereas based on LCN2 and PAP measurements taken together, two individuals were misclassified (94% confidence). But most importantly, when LCN2, PAP, and CCL20 measurements were all taken into consideration, only one individual was misclassified therefore representing a 97% confidence and emphasizing the power of a biomarker panel over single analyte measurement (Supplementary Table S3). In addition, in feces, the measurement of S100A8/A9, LCN2, and PAP showed a confidence of 94% in discriminating between normal and CD patients (94% confidence, Supplementary Table S3).
DISCUSSION
IL-23R inactivation blocks IBD
In this study, we used the anti-CD40-induced mouse model for IBD to search for efficacy biomarkers of treatment by mAbs to IL-23R. Anti-CD40 treatment induced a strong activation of inflammatory dendritic cells and macrophages, resulting in colon inflammation, epithelial cell hyperplasia, and myeloid cell accumulation in the lamina propria. We demonstrated that IL-23R inactivation by neutralizing mAbs completely prevented the development of colitis. Our results are consistent with those of Uhlig et al.,8 who showed that CD40-induced inflammation was prevented by treatment with mAbs to IL-23(p19). Overall, no significant difference was observed on the prevention of weight loss, inflammation of the colonic laminia propria, and cytokines, and chemokines levels blocking IL-23R versus p19 itself.
IL-22 and IL-17
IL-22 was upregulated 24 h after colitis was triggered by CD40 stimulation in our model, and anti-IL23R treatment completely prevented this in the serum and the colon tissue, suggesting that IL-23 signaling is indispensable in promoting IL-22 production. IL-22 serum concentrations are increased in CD patients and correlate with disease activity and IL-23R genotype status32 and correspondingly, we also demonstrated that IL-22 transcripts increase during active disease, and revert back to control values in patients in remission. However, IL-22 protein is often undetectable in serum from healthy individuals and not systematically detectable in CD patients, making this proximal target-engagement biomarker hard to use as an index of disease clinically. IL-17A, another cytokine directly downstream of our target, is a hallmark of CD, but we showed that IL-17A does not vary consistently with CD and remission states. Moreover, IL-17A is also difficult to detect in serum from CD patients, therefore limiting its utility in the clinic.33 Nevertheless, IL-17A can be detected in the feces of patients with active CD, but at a very low levels.33
Antimicrobial proteins S100A8, S100A9, REG3β and γ, LCN2, and DMBT1
We demonstrated that the concentrations of the antimicrobial proteins S100A8, S100A9, REG3β, REG3γ, and LCN2 were induced in the colon tissue of diseased mice with T-cell-independent and T-cell-dependent colitis, and that treatment by mAbs to IL-23R prevented these upregulations, likely through the downregulation of IL-22, IL-17A, and TNF-α. We showed in vitro that, in human colonic epithelial cell line HT-29, stimulation with IL-22, IL-17A, and/or TNF-α led to upregulation of the expression of these markers, supporting the role of the IL-23/TH17 pathway as a key component of mucosal immunity.20
We showed that several antimicrobial proteins were candidate biomarkers for IL-23 blockage efficacy. S100A8 and A9, termed calprotectin when complexed together, are calcium-binding proteins expressed in abundance by activated neutrophils or epithelial cells that exhibit chemoattractant properties.34 Calprotectin measurement by ELISA in feces has been approved by the FDA (Food and Drug Administration) to assess the level of gut inflammation in IBD and monitor response to therapy (http://www.accessdata.fda.gov/cdrh_docs/pdf5/k050007.pdf). S100A8/A9 also served as a control for the different analysis methods used in the present study.
REG3β and REG3γ in mouse (PAP in human), which belong to the secreted C-type lectin protein family, are induced during gut bacterial colonization, and could participate in epithelial cell repair and protection during the onset of inflammation.28, 35, 36 PAP, previously shown to be upregulated in gut biopsies and serum from CD patients compared with controls, could act as an anti-inflammatory molecule by inhibiting NF-κB, which controls cytokine production and adhesion molecules in inflamed tissue.28 Notably, REG protein expression in the colon is IL-22 dependent through STAT3 signaling.37, 38 Moreover, STAT3 and other downstream mediators of IL-23R such as JAK2 have been identified as CD susceptibility genes.39 Here, we show that PAP expression is downregulated in CD patients in remission, and statistically upregulated in feces from CD patients, making it a potential biomarker for monitoring anti-IL-23 therapy.
LCN2, another antimicrobial protein expressed by immune cells and colonic epithelial cells, exerts a bacteriostatic effect by sequestering small iron-binding molecules synthesized by bacteria as a mean of iron acquisition.40 LCN2 overexpression is part of an IBD-specific gene signature established in patient gut biopsies.41, 42 Here, we show that LCN2 is differentially expressed in the serum and feces from CD patients when compared with controls, and that its transcript levels vary with disease activity in colon biopsies from patients in remission.
DMBT1 belongs to the superfamily of scavenger receptor cysteine-rich genes. It mediates anti-inflammatory effects during intestinal inflammation by aggregating Gram-positive and -negative bacteria,43 and by possibly decreasing NF-κB signaling. Furthermore, DMBT1 gene expression is upregulated in gut biopsies from CD patients, and a deletion variant of DMBT1 may have a role in the pathogenesis of CD.44 DMBT1 is a target for the intracellular pathogen receptor NOD2, which is the major CD susceptibility gene. We showed that the blockade of IL-23 signaling decreased DMBT1 gene expression only in the T-cell-independent model. In human samples, we confirmed DMBT1 upregulation in colon biopsies from CD patients compared with controls and demonstrated its downregulation with therapy. However, we failed to detect DMBT1 in mouse feces by western blot analysis, and it could not be measured in serum due to the lack of reagents, restricting its practical use.
MIF and CCL20
In addition to these antimicrobial proteins, MIF, a pro-inflammatory cytokine, was upregulated in mouse colon tissue.45 MIF plasma concentration is upregulated in CD patients, and anti-MIF treatment suppresses the established colitis in a mouse T-cell transfer model.27 We confirmed higher MIF serum concentration in CD patients compared with normals, although the difference was not as significant as it was for calprotectin, PAP, LCN2, or CCL20.
CCL20 transcript was not upregulated in our mouse models but has been previously shown to be overexpressed in colonic biopsies from CD patients and can regulate the chemotaxis of T cells and dendritic cells in IBD.46 CCR6, the receptor of CCL20, has been recenthy identified as a CD susceptibility gene.39 Treatment of colonic explants with antibody to TNF-α in vitro reduced CCL20 secretion in tissues from healthy patients but not in tissue from CD patients.46 Our study results could partially explain the latter as we show that IL-17A and IL-22 are also able to control CCL20 expression. We confirmed the upregulation of CCL20 in colon biopsies from CD patients compared with normals and showed a restoration to control values in patients in remission. In addition, we validated the upregulation of CCL20 in serum from CD patients.
Overall remarks
We also demonstrated that in both a T-cell-independent and -dependent mouse model of IBD, all the markers track with IL-23 therapy both in a prophylactic and importantly, a therapeutic manner. This highlights their potential value in monitoring anti-IL-23 therapy, especially since they are also increased in the inflamed colons of CD patients and decreased in patients in remission. Last, we found that the concentrations of most of the markers was actually controlled by TNF-α, IL-22, and IL-17A in the gut, suggesting that the therapeutic inhibition of IL-23 signaling, which would lead to a decrease in IL-17A, IL-22, and TNF-α, could be more effective than the inhibition of TNF-α alone.
Conclusion
In conclusion, proteomic analysis of fecal proteins in disease and treatment mouse models allowed us to uncover a number of potential disease- and target-engagement biomarkers. Taken together, we confirmed that calprotectin, PAP, and MIF concentrations are higher in the serum of CD patients than in healthy individuals, and we added two CD-associated serum biomarkers to this list: LCN2 and CCL20. We showed also for the first time that PAP and LCN2 were statistically upregulated in human feces from CD patients. We believe that the measurement of these markers as panels in both serum and stool will be helpful tool to evaluate the response to anti-IL-23 therapy.
Study Highlights

Acknowledgments
We thank the Bioanalytics and Lead Identification group for their advice with the ELISA, the Protein Chemistry and Analytics group for the monoclonal antibody preparations (anti-IL-23(p19) and anti-IL-23R), Lorena Taricani and Dave Parry for their advice on anti-REG3γ polyclonal antibody generation, and Jeanine Mattson and Rick Cheung for their critical comments.
Guarantor of the article: Maribel Beaumont, PhD.
Specific author contributions: Study concept and design, implementation, analysis and interpretation, drafting of the manuscript: Corinne Cayatte; study concept and design, technical and material support: Barbara Joyce-Shaikh; technical and material support: Felix Vega, Jeffrey Grein, Erin Murphy, Wendy M. Blumenschein, Smiley Chen, and Maria-Christina Malinao; study concept and design: Katia Boniface and Brent S. McKenzie; pathological analysis and interpretation: Robert Pierce; critical revision: Charles O. Elson and Rene de Waal Malefyt; statistics: Beth Basham; critical revision and scientific input on study concept: Edward P. Bowman and Daniel Cua; clinical coordinator and sample acquisition: William A. Faubion; study concept and design, study supervision: Terrill K. McClanahan; study supervision: Robert A. Kastelein; study concept and design, critical revision, and study supervision: Maribel Beaumont.
Financial support: This work was supported by Merck & Co., with the exception of the T-cell transfer data that was funded by NIH Grant DK071176.
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Maloy KJ. The interleukin-23/interleukin-17 axis in intestinal inflammation. J Intern Med. 2008;263:584–590. doi: 10.1111/j.1365-2796.2008.01950.x. [DOI] [PubMed] [Google Scholar]
- Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- Parham C, Chirica M, Timans J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- Yen D, Cheung J, Scheerens H, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Langrish CL, McKenzie B, et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest. 2006;116:1317–1326. doi: 10.1172/JCI25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson CO, Cong Y, Weaver CT, et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–2370. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- Hue S, Ahern P, Buonocore S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlig HH, McKenzie BS, Hue S, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Giese T, Ludwig B, et al. Expression of interleukin-12-related cytokine transcripts in inflammatory bowel disease: elevated interleukin-23p19 and interleukin-27p28 in Crohn's disease but not in ulcerative colitis. Inflamm Bowel Dis. 2005;11:16–23. doi: 10.1097/00054725-200501000-00003. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Okamoto S, Hisamatsu T, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn's disease. Gut. 2008;57:1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidasheva S, Trifari S, Phillips A, et al. Functional Studies on the IBD Susceptibility Gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS One. 2011;6:e25038. doi: 10.1371/journal.pone.0025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman EP, Chackerian AA, Cua DJ. Rationale and safety of anti-interleukin-23 and anti-interleukin-17A therapy. Curr Opin Infect Dis. 2006;19:245–252. doi: 10.1097/01.qco.0000224818.42729.67. [DOI] [PubMed] [Google Scholar]
- Kola I. The state of innovation in drug development. Clin Pharmacol Ther. 2008;83:227–230. doi: 10.1038/sj.clpt.6100479. [DOI] [PubMed] [Google Scholar]
- Rolink A, Melchers F, Andersson J. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- Cong Y, Weaver CT, Lazenby A, Elson CO. Colitis induced by enteric bacterial antigen-specific CD4+ T cells requires CD40-CD40 ligand interactions for a sustained increase in mucosal IL-12. J Immunol. 2000;165:2173–2182. doi: 10.4049/jimmunol.165.4.2173. [DOI] [PubMed] [Google Scholar]
- Wilson NJ, Boniface K, Chan JR, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- Bhat VB, Choi MH, Wishnok JS, Tannenbaum SR. Comparative plasma proteome analysis of lymphoma-bearing SJL mice. J Proteome Res. 2005;4:1814–1825. doi: 10.1021/pr0501463. [DOI] [PubMed] [Google Scholar]
- von Roon AC, Karamountzos L, Purkayastha S, et al. Diagnostic precision of fecal calprotectin for inflammatory bowel disease and colorectal malignancy. Am J Gastroenterol. 2007;102:803–813. doi: 10.1111/j.1572-0241.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Valdez P. IL-22 in mucosal immunity. Mucosal Immunol. 2008;1:335–338. doi: 10.1038/mi.2008.26. [DOI] [PubMed] [Google Scholar]
- Van Deventer SJ. Tumour necrosis factor and Crohn's disease. Gut. 1997;40:443–448. doi: 10.1136/gut.40.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuch PL, Hanauer SB. Treatment of inflammatory bowel disease: a review of medical therapy. World J Gastroenterol. 2008;14:354–377. doi: 10.3748/wjg.14.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JH, Keates S, Bassani L, et al. Colonic epithelial cells are a major site of macrophage inflammatory protein 3alpha (MIP-3alpha) production in normal colon and inflammatory bowel disease. Gut. 2002;51:818–826. doi: 10.1136/gut.51.6.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso R, Fina D, Peluso I, et al. A functional role for interleukin-21 in promoting the synthesis of the T-cell chemoattractant, MIP-3alpha, by gut epithelial cells. Gastroenterology. 2007;132:166–175. doi: 10.1053/j.gastro.2006.09.053. [DOI] [PubMed] [Google Scholar]
- Dambacher J, Beigel F, Zitzmann K, et al. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207–1217. doi: 10.1136/gut.2007.130112. [DOI] [PubMed] [Google Scholar]
- Leach ST, Yang Z, Messina I, et al. Serum and mucosal S100 proteins, calprotectin (S100A8/S100A9) and S100A12, are elevated at diagnosis in children with inflammatory bowel disease. Scand J Gastroenterol. 21;2007:1–11. doi: 10.1080/00365520701416709. [DOI] [PubMed] [Google Scholar]
- de Jong YP, Abadia-Molina AC, Satoskar AR, et al. Development of chronic colitis is dependent on the cytokine MIF. Nat Immunol. 2001;2:1061–1066. doi: 10.1038/ni720. [DOI] [PubMed] [Google Scholar]
- Gironella M, Iovanna JL, Sans M, et al. Anti-inflammatory effects of pancreatitis associated protein in inflammatory bowel disease. Gut. 2005;54:1244–1253. doi: 10.1136/gut.2004.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh A, Zhang Z, Inatomi O, et al. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 2005;129:969–984. doi: 10.1053/j.gastro.2005.06.071. [DOI] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand S, Beigel F, Olszak T, et al. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- Schmechel S, Konrad A, Diegelmann J, et al. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008;14:204–212. doi: 10.1002/ibd.20315. [DOI] [PubMed] [Google Scholar]
- Holtta V, Klemetti P, Sipponen T, et al. IL-23/IL-17 immunity as a hallmark of Crohn's disease. Inflamm Bowel Dis. 2008;14:1175–1184. doi: 10.1002/ibd.20475. [DOI] [PubMed] [Google Scholar]
- Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Fukushima K, Naito H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- Moucadel V, Soubeyran P, Vasseur S, et al. Cdx1 promotes cellular growth of epithelial intestinal cells through induction of the secretory protein PAP I. Eur J Cell Biol. 2001;80:156–163. doi: 10.1078/0171-9335-00148. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Pickert G, Neufert C, Leppkes M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott KM, Mori K, Li JY, et al. Dual action of neutrophil gelatinase-associated lipocalin. J Am Soc Nephrol. 2007;18:407–413. doi: 10.1681/ASN.2006080882. [DOI] [PubMed] [Google Scholar]
- Csillag C, Nielsen OH, Vainer B, et al. Expression of the genes dual oxidase 2, lipocalin 2 and regenerating islet-derived 1 alpha in Crohn's disease. Scand J Gastroenterol. 2007;42:454–463. doi: 10.1080/00365520600976266. [DOI] [PubMed] [Google Scholar]
- Galamb O, Gyorffy B, Sipos F, et al. Inflammation, adenoma and cancer: objective classification of colon biopsy specimens with gene expression signature. Dis Markers. 2008;25:1–16. doi: 10.1155/2008/586721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstiel P, Sina C, End C, et al. Regulation of DMBT1 via NOD2 and TLR4 in intestinal epithelial cells modulates bacterial recognition and invasion. J Immunol. 2007;178:8203–8211. doi: 10.4049/jimmunol.178.12.8203. [DOI] [PubMed] [Google Scholar]
- Renner M, Bergmann G, Krebs I, et al. DMBT1 confers mucosal protection in vivo and a deletion variant is associated with Crohn's disease. Gastroenterology. 2007;133:1499–1509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Metz CN, Bucala R. Role of macrophage migration inhibitory factor in the regulation of the immune response. Adv Immunol. 1997;66:197–223. doi: 10.1016/s0065-2776(08)60598-2. [DOI] [PubMed] [Google Scholar]
- Kaser A, Ludwiczek O, Holzmann S, et al. Increased expression of CCL20 in human inflammatory bowel disease. J Clin Immunol. 2004;24:74–85. doi: 10.1023/B:JOCI.0000018066.46279.6b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.