Background: The STAM2/Hrs complex is part of the ESCRT-0 (endosomal sorting complexes required for transport) machinery responsible for cargo sorting to the lysosome.
Results: Binding of the VHS-UIM construct of STAM2 to Lys63-linked diubiquitin is cooperative with a specific structural organization.
Conclusion: Spatial arrangement of the VHS-UIM/Lys63-linked diubiquitin complex probably influences the sorting of Lys63 polyubiquitinated proteins.
Significance: Characterization of the VHS-UIM/Lys63-linked diubiquitin complex is crucial for understanding lysosomal degradation.
Keywords: Endocytosis, Molecular Docking, Nuclear Magnetic Resonance, Protein-Protein Interactions, Ubiquitin, Polyubiquitin
Abstract
As the upstream component of the ESCRT (endosomal sorting complexes required for transport) machinery, the ESCRT-0 complex is responsible for directing ubiquitinated membrane proteins to the multivesicular body pathway. ESCRT-0 is formed by two subunits known as Hrs (hepatocyte growth factor-regulated substrate) and STAM (signal transducing adaptor molecule), both of which harbor multiple ubiquitin-binding domains (UBDs). In particular, STAM2 possesses two UBDs, the VHS (Vps27/Hrs/Stam) and UIM (ubiquitin interacting motif) domains, connected by a 20-amino acid flexible linker. In the present study, we report the interactions of the UIM domain and VHS-UIM construct of STAM2 with monoubiquitin (Ub), Lys48- and Lys63-linked diubiquitins. Our results demonstrate that the UIM domain alone binds monoubiquitin, Lys48- and Lys63-linked diubiquitins with the same affinity and in the same binding mode. Interestingly, binding of VHS-UIM to Lys63-linked diubiquitin is not only avid, but also cooperative. We also show that the distal domain of Lys63-linked diubiquitin stabilizes the helical structure of the UIM domain and that the corresponding complex adopts a specific structural organization responsible for its greater affinity. In contrast, binding of VHS-UIM to Lys48-linked diubiquitin and monoubiquitin is not cooperative and does not show any avidity. These results may explain the better sorting efficiency of some cargoes polyubiquitinated with Lys63-linked chains over monoubiquitinated cargoes or those tagged with Lys48-linked chains.
Introduction
The turnover of many membrane proteins is determined through the endocytic pathway, which can either result in their recycling or lysosomal degradation (1–3). In the case of lysosomal degradation, membrane proteins are sorted into distinctive endosomes known as multivesicular bodies (MVBs).3 These MVBs fuse with lysosomes, resulting in the degradation of their cargoes. Prior to entering the MVB pathway, cargoes have to be properly tagged by a process called ubiquitination. Ubiquitination occurs through the attachment of a single ubiquitin (Ub), a 76-amino acid protein, to a lysine (monoubiquitination) (4) or several lysines (multi-monoubiquitination) (5) of a target protein, as well as by the attachment of polyubiquitin (poly-Ub) chains (6). Interestingly, depending on the type of ubiquitin chain linkages, tagged proteins are committed to different pathways (7). Although monoubiquitination is a sufficient signal to direct cargoes through the MVB pathway (8, 9), in many cases, polyubiquitination by Lys63-linked chains is a more efficient signal for cargo sorting (10–12). Most proteins that are not ubiquitinated follow other pathways and are recycled from the endosome to other cellular compartments (13).
The machinery responsible for committing ubiquitinated cargoes to the MVB pathway is the ESCRT machinery (14), whose most upstream component is ESCRT-0 that is composed of the STAM/Hrs complex (15) in mammalian cells (Vps27/Hse1 complex in yeast) (16, 17). Recognition of the ubiquitin signal is achieved by modular motifs called ubiquitin-binding domains (UBDs) (18). Both STAM and Hrs harbor two UBDs. The VHS (19) and UIM (ubiquitin-interacting motif) (20) domains of STAM, which are connected by a 20-amino acid linker, are key players in ubiquitin recognition and cargo sorting. Deletion of either the VHS or UIM domains of STAM gives rise to a partial loss of function of ESCRT-0 (19, 21). These results highlight the importance of both the VHS and UIM domains, and it has recently been proposed that ESCRT-0 as well as the VHS-UIM domains of STAM1 bind avidly to long polyubiquitin chains and show a moderate degree of selectivity for Lys63- over Lys48-linked chains (21). However, the molecular details of the recognition of Lys63-linked diubiquitin (Lys63-Ub2) chains by the ESCRT machinery remain poorly understood, and structural studies are required to decipher the underlying mechanisms. Because ESCRT-0 appears to be primarily responsible for cargo clustering (22), we are seeking to understand if any particular structural organization forms the basis for the polyubiquitin chain selectivity of ESCRT-0 and, more specifically, of the VHS-UIM fragment of STAM2. To address this question, we compared interactions of the UIM domain and the VHS-UIM construct of STAM2 with monoubiquitin (mono-Ub), Lys48- and Lys63-Ub2. Here we show that the UIM domain binds mono-Ub, Lys48- and Lys63-Ub2 with the same affinity and binding mode, in striking contrast with our previous findings for the VHS domain (23). However, the situation is somewhat different when the UIM and the VHS domains are connected. Our results indicate that interaction of the VHS-UIM construct with Lys63-Ub2, but not with mono-Ub or Lys48-Ub2, is cooperative. In addition, site-directed paramagnetic spin labeling data allow modeling of the VHS-UIM/Lys63-Ub2 complex and clearly demonstrate that each domain of STAM2 VHS-UIM interacts with a specific Ub unit in Lys63-Ub2.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
STAM2(1–188) (VHS-UIM) and STAM2(162–187) (UIM) were purified as previously described for STAM2(4–156) (VHS) (23). The TEV cleavage reaction left N-terminal extensions GAMG for STAM2(1–188) and GAMGM for STAM2(162–187). Ubwt, UbT12CD77, UbT12CK63R, UbK63R, UbK48R, UbT12CK48R, and UbD77 were produced as described in Refs. 24–26. Diubiquitins were produced as described in Ref. 25 and segmentally isotope-labeled as detailed in Refs. 24, 27, and 28. The UIMT193Y variant was engineered using site-directed mutagenesis.
NMR Experiments
To be consistent with our previous study related to VHS (23), the ensemble of NMR experiments was acquired at 288 K where the NMR samples were exchanged into a buffer containing 20 mm sodium phosphate at pH 6.8, 10% D2O, 0.02% (w/v) NaN3. Assignment and spin relaxation experiments for UIM and VHS-UIM have been carried out on a Varian Inova Unity 600 equipped with a triple-resonance probe and on a Bruker Avance 600 equipped with TXI cryoprobe or TCI cryoprobe.
Protein Resonance Assignment
Backbone resonance assignments for VHS-UIM were obtained using a combination of the following experiments: [15N,1H]-HSQC, CBCA(CO)NH, HNCA, HNCACB, and three-dimensional NOESY-[15N,1H]-HSQC, whereas the backbone resonance assignments for UIM were obtained using three-dimensional NOESY-[15N,1H]-HSQC and three-dimensional TOCSY-[15N,1H]-HSQC. Data were processed with NMRpipe (29) and analyzed with CARA (30) starting with the assignment list of the STAM2 VHS domain (23). The 3JHN-Hα coupling constants for the UIM domain were derived from 1H,15N-transverse relaxation optimized spectroscopy (TROSY) spectra (31) acquired with 8192 complex points in the F2 dimension (1H). For consistency with our previous study of the VHS domain (23), the UIM and VHS-UIM sequences were renumbered according to the STAM2-VHS structure (PDB code 1X5B) by adding 7 to the STAM2 sequence (Uniprot accession number O75886). The complete VHS-UIM backbone assignment is available from BioMagResBank under the accession number 18185.
Relaxation Measurements and Analysis
Relaxation measurements including 15N longitudinal (R1) and transverse (R2) relaxation rates and the 15N-1H cross-relaxation rates, via steady-state 15N{1H}NOE, were performed as previously described (32, 33). NMR spectra were recorded with spectral widths of 2000 Hz in the 15N dimension and 9600 Hz in the 1H dimension. For the R1 measurements, we used 12 relaxation delays: 4, 20, 40, 80 (twice), 240, 480, 800 (twice), 1200, 1400, 1600, 1800, and 2000 ms with a recycling delay of 4 s. The R2 CPMG measurements were performed with transverse relaxation periods of 4, 12, 24, 32 (twice), 40, 48, 56, 80, 120 (twice), 160, and 200 ms and a relaxation delay of 4 s. For 15N{1H}NOE experiments, two-dimensional spectra were recorded with and without presaturation of amide protons (34, 35). The relaxation delay was set to 6 s to allow the bulk water magnetization to return as close as possible to its equilibrium value. All NMR data were processed with NMRpipe (29) and SPARKY (36), and the relaxation rates were extracted using RelaxFit (32). For complexes, the relaxation experiments were performed with a concentration of 200 μm for the protein and a saturating concentration of its corresponding ligand.
NMR Titration Studies
Interaction surfaces on 15N-mono-Ub, 15N-Ub2, 15N-UIM, and 15N-VHS-UIM were characterized by means of chemical shift perturbations (CSPs) where a series of 1H,15N-HSQC experiments were recorded upon addition of the (unlabeled) binding partner. To avoid possible aggregation of the proteins, we started from a 15N-labeled protein at 200 μm concentration and added an increasing volume of a concentrated stock of unlabeled ligand protein until reaching saturation. To derive the corresponding binding constant, spectral perturbations were quantified as the combined amide CSPs: Δδ = [(ΔδH)2 + (ΔδN/5)2]1/2. The equations used to derive the dissociation constants for the 1:1 or 2:1 stoichiometry models can be found in Ref. 27; the formula used to calculate the effective concentration can be found in the supplemental Materials.
Site-directed Spin Labeling
A paramagnetic spin label, 1-oxy-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)methanesulfonate (MTSL), was covalently attached to Lys63-Ub2 or Lys48-Ub2 on the proximal or the distal UbT12C at position 12 through the cysteine side chain, as described previously (28). The paramagnetic relaxation enhancement (PRE) effect was measured as a ratio of protein signal intensities in the 1H,15N NMR spectra obtained when MTSL was in the oxidized (paramagnetic) and the reduced (diamagnetic) states (28). The oxidized state spectrum was obtained for 15N-VHS-UIM in the presence of unlabeled Lys63-Ub2 or Lys48-Ub2 with MTSL attached to the proximal or the distal Ub; the reduced state spectrum was obtained after adding 3 molar eq of ascorbate to that sample. The position of the unpaired electron in MTSL with respect to the protein was reconstructed by fitting the observed PREs using program SLfit (37). The corresponding equations are summarized in supplemental Materials.
Homology Modeling of UIM
The three-dimensional structures of the UIM domain alone and the UIM part in the VHS-UIM construct were obtained by homology modeling. As seen from the pairwise alignment (supplemental Fig. S2A), amino acid sequences of STAM2-UIM and Vps27-UIM1 share 55% identity and 70% similarity. We used the UIM1 domain (38) of Vps27 (PDB code 1Q0V) to model the structure of the UIM domain as well as the UIM part of the VHS-UIM construct. The NMR structure of the VHS domain of STAM2 (PDB code 1X5B) was used to model the VHS part of VHS-UIM. Models were generated by using the Modeler program (39). After alignment of the query and template sequences with Align2D, they were used as input in Modeler. A total of 10 structures were generated for UIM as well as VHS-UIM (supplemental Fig. S2B).
RESULTS
Mapping the Interaction of UIM with Mono-Ub, Lys48- and Lys63-Ub2
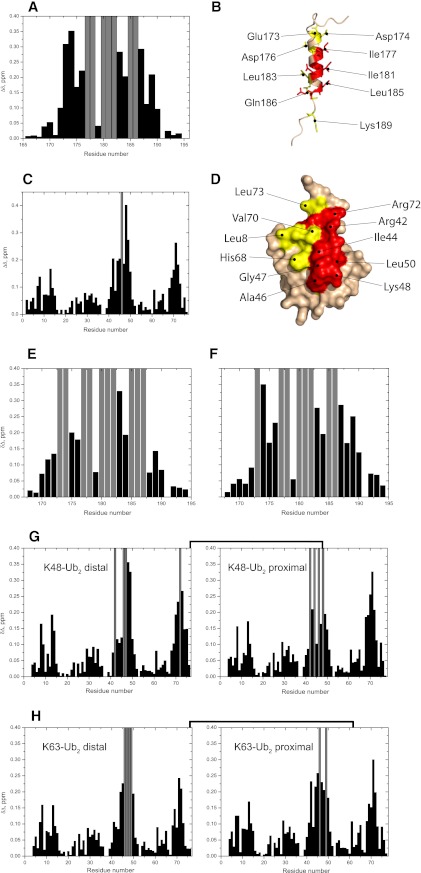
To compare Ub/poly-Ub binding properties of the isolated UIM domain and the tandem VHS-UIM construct, we first mapped the interactions between UIM and mono-Ub or Ub2 by means of NMR CSPs. To obtain information from both sides of the complex, we monitored signal shifts in 1H,15N-HSQC spectra of 15N-labeled UIM, mono-Ub, or Ub2 upon addition of the corresponding unlabeled binding partner. As supported by the 1H,15N-TROSY and three-dimensional 15N-dispersed NOESY spectra (see supplemental Figs. S1 and S2), the UIM domain forms an α-helix. Chemical shift index (40) predicts an α-helical region from Lys172 to Glu187. Upon titration of mono-Ub into 15N-UIM, several residues experienced significant CSPs, whereas some showed a strong decrease (up to 70% already at the beginning of titration) in signal intensity indicative of intermediate exchange (Fig. 1A). Specifically, strong signal attenuations were detected for the hydrophobic residues Ile177, Ala178, Ala180, Ile181, and Leu185. These residues form a stretch along one face of the α-helix of UIM, flanked on both sides by negatively charged residues Glu173, Asp174, Asp176, and Glu187, which show significant CSPs (Fig. 1B). These perturbed residues are in good agreement with those perturbed on other UIM domains upon binding to mono-Ub (38, 41). On the mono-Ub side, the CSPs cluster around the canonical hydrophobic patch of Ub, specifically including residues Leu8, Ile44, Ala46, Leu50, Val70, and Leu73, and also including Arg42, Lys48, Gly47, His68, and Arg72 (Fig. 1, C and D).
FIGURE 1.
NMR mapping of the interface between UIM and mono-Ub, Lys48- or Lys63-Ub2. Shown are CSPs observed in UIM at the end point of titration with mono-Ub (A), Lys48-Ub2 (E), and Lys63-Ub2 (F), and in mono-Ub (C), Lys48-Ub2 (G), and Lys63-Ub2 (H) upon saturation with UIM. Gray bars indicate residues experiencing intermediate exchange on the NMR time scale, resulting in strong attenuations of their signals in the 1H,15N-HSQC spectra. B and D, mapping of the residues affected by UIM/mono-Ub binding on the three-dimensional structure of UIM (B) and Ub (D) (the UIM structure was obtained by homology modeling, see “Experimental Procedures”). In UIM (B), residues in intermediate exchange are colored red, whereas residues with significant CSPs (Δδ > 0.2) are colored yellow. In Ub (D), residues with significant CSPs (Δδ > 0.2) and/or intermediate exchange are colored red and residues with 0.2 ≥ Δδ > 0.1 are colored yellow.
We performed similar studies to examine and compare UIM binding to Lys48-Ub2 and Lys63-Ub2. To enable studies of UIM interactions with each individual Ub unit in the chain, the Ub2 constructs were segmentally 15N-labeled (see “Experimental Procedures”) on either the proximal Ub (that carries the free C terminus) or the distal Ub (whose C terminus is linked to Lys48 or Lys63 on the proximal Ub). Our NMR data (Fig. 1, E and F) show that the UIM surface perturbed by either Lys48- or Lys63-Ub2 is essentially the same for both chains and similar to the one involved in the mono-Ub binding (Fig. 1, A and B). Likewise, there is a striking similarity between the UIM-binding surfaces on the two Ub units in each chain as well as between the chains (Fig. 1, G and H). Moreover, the perturbed residues in the Ub units in these Ub2s are essentially identical to those involved in mono-Ub binding to the UIM (Fig. 1, C and D). These results suggest that the interaction of UIM with the chains occurs via its binding to each individual Ub unit separately and regardless of the chain linkage.
UIM Adopts the Same Mode of Binding with Mono-Ub and Ub2
To verify the aforementioned prediction, we set out to quantify the stoichiometry and the strength of these interactions. To monitor the stoichiometry of the complexes studied here, we used transverse spin relaxation rate (15N R2) measurements, as R2 reflects the rate of overall tumbling (hence the size) of the molecular object under investigation. The molecular mass dependence of R2 was utilized as a “molecular mass ruler” (calibrated using R2 data for Ub and Ub2) to assess the mass of a molecular complex and hence the stoichiometry (supplemental Fig. S3B). Measurements for the target protein (UIM, mono-Ub, or Ub2) in the corresponding complexes were carried out under saturating conditions of its binding partner. Residues experiencing fast local dynamics or conformational exchange were removed from the analysis. For mono-Ub, the average R2 for residues in the secondary structure increased from 7.3 ± 0.4 to 11.1 ± 0.8 s−1 upon binding to UIM. The R2 value of UIM was 5.1 ± 0.1 s−1 in the free state, and increased to 11.5 ± 0.5 s−1 in the mono-Ub-bound state (see supplemental Fig. S3B). These R2 values for the UIM/mono-Ub complex correspond to a molecular mass range from 12.0 to 14.5 kDa, in good agreement with the expected molecular mass of 12 kDa for a 1:1 UIM/mono-Ub equilibrium. For the Ub2/UIM complexes, we measured an average R2 value of 21.3 ± 1.4 and 20.6 ± 1.6 s−1 for the distal Ub in Lys48- and Lys63-Ub2, respectively, compared with the value of 13.3 ± 0.9 s−1 measured for the same Ub unit in the free form of Lys63-Ub2. These R2 values for UIM-bound Ub2 correspond to a molecular mass range from 24 to 28.5 kDa, pointing to a 2:1 (UIM:Ub2) binding stoichiometry (the expected molecular mass is 24 kDa). Thus, our 15N relaxation data clearly support the model where one UIM binds to one mono-Ub molecule, whereas two UIMs can bind to each Lys48- and Lys63-Ub2.
To quantify the strength of the binding interactions, we derived the dissociation constants from the NMR titration data for each interacting partner in the complexes under investigation. For the UIM/mono-Ub binding equilibrium, the dissociation constant was derived using a 1:1 stoichiometry model, which gave an average Kd of 287 ± 36 μm (Table 1 and supplemental Fig. S4). For the UIM/Ub2 interaction, a 2:1 stoichiometry model yielded the microscopic Kd values of 325 ± 32 and 304 ± 38 μm for UIM binding to Lys48-Ub2 and Lys63-Ub2, respectively (see supplemental Fig. S5). Thus, our quantitative analysis supports the conclusion that the isolated UIM interacts with each individual Ub unit in Lys48- and Lys63-linked Ub2 chains separately and with no linkage selectivity.
TABLE 1.
Summary of the dissociation constants derived from NMR titration curves for the UIM/mono-Ub and UIM/Ub2 binding equilibria
Kd values are averaged over several residues that showed strong CSPs upon binding. Standard deviations are shown in parentheses.
| Sample | Domain analyzed | Kd |
|---|---|---|
| μm | ||
| UIM/mono-Ub | UIM | 266 (35)a |
| Mono-Ub | 309 (37)a | |
| UIM/Lys48-Ub2 | Distal Ub | 304 (32)b |
| Proximal Ub | 325 (36)b | |
| UIM | 345 (28)b | |
| UIM/Lys63-Ub2 | Distal Ub | 310 (44)b |
| Proximal Ub | 334 (30)b | |
| UIM | 267 (40)b |
a Kd obtained using a 1:1 binding model.
b Kd obtained using a 2:1 binding model.
Interaction of VHS-UIM with Mono-Ub and Ub2 Chains
In the VHS-UIM construct, as in ESCRT-0, the VHS and UIM domains are separated by a 20-amino acid linker, which is highly flexible according to our 15N relaxation data (see supplemental Fig. S7). Moreover, measurements of the 15N longitudinal (R1) as well as the transverse relaxation rate (R2) indicate that the VHS and the UIM domains tumble essentially independently. For instance, the average R2 values measured for the VHS and UIM domains in VHS-UIM are 17.0 ± 1.2 and 5.1 ± 0.6 s−1, respectively. These values are comparable with those obtained for the isolated VHS and UIM domains (23) and correspond to the molecular mass of the individual domains (i.e. ∼21.0 ± 2.5 and ∼4.0 ± 1.0 kDa, respectively). Model-free analysis of 15N relaxation data resulted in an average squared order parameter of S2 = 0.63 ± 0.05 for the UIM part (see supplemental Fig. S7B), which indicates an increased amplitude of the backbone motion compared with the VHS part (S2 = 0.88 ± 0.07). It is worth noting that despite the higher backbone flexibility on the ps-ns time scale, the UIM part retains its helical fold, as evident from the various spectroscopic data presented above.
In the 1H,15N-HSQC spectrum of VHS-UIM, the VHS and UIM parts overlap with their respective spectra as isolated units. This fact provides clear evidence that the VHS and UIM domains do not interact directly in the VHS-UIM construct (see supplemental Fig. S8). Therefore, any perturbation observed in VHS-UIM spectra upon addition of mono-Ub or (poly)Ub is caused by the interaction of VHS-UIM with the ligand.
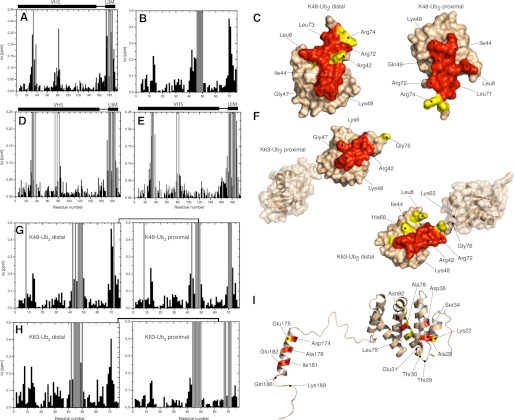
Our NMR mapping data suggest a specific interaction between VHS-UIM and mono-Ub, where both the VHS and UIM domains are involved in binding, each to a separate ubiquitin molecule. On the VHS-UIM side, the largest perturbations stretch along the α2 and α4 helices of the VHS domain, which comprise residues Thr29– Asp38 and Leu75-Asn82, respectively (Fig. 2A). The UIM domain was also affected by Ub binding, through residues located on one side of the α-helix; these include Ala178, Ile181, Glu182, Leu185, and Gln186 that experienced strong signal attenuations (hence intermediate exchange) and Asp174, Asp176, and Leu183 that showed significant CSPs. On the mono-Ub side, strong perturbations clustered around the hydrophobic residues Ile44, Val70, and Ile13 (see Fig. 2B). Surprisingly, Gly75 and Gly76 were significantly perturbed compared with the interactions of mono-Ub with the individual UIM (see above) or VHS domains (23). This suggests additional interactions, involving the C terminus of Ub and possibly with the VHS-UIM linker. The dissociation constant for the VHS-UIM/mono-Ub equilibrium was derived by fitting the CSPs data for each individual residue in the course of titration (supplemental Fig. S9). From the Ub side, the dissociation constant was 102 ± 38 μm, and a similar average value of 117 ± 33 μm was obtained for the VHS-UIM residues, thus giving a total average Kd of 112 ± 35 μm (Table 2).
FIGURE 2.
NMR mapping of the interface between VHS-UIM and mono-Ub, Lys48- or Lys63-Ub2. Shown are CSPs observed in VHS-UIM at the end point of titration with mono-Ub (A), Lys48-Ub2 (E), Lys63-Ub2 (D) and in mono-Ub (B), Lys48-Ub2 (G), and Lys63-Ub2 (H) upon saturation with VHS-UIM. Residues experiencing intermediate exchange during the course of titration are represented by gray bars. C and F, mapping of the residues affected by VHS-UIM/Ub2 binding on the three-dimensional surface of Lys48-Ub2 (C) and Lys63-Ub2 (F). I, representation of the residues affected by VHS-UIM/mono-Ub binding on the three-dimensional structure of VHS-UIM. Note that the three-dimensional structure of VHS-UIM results from homology modeling (see “Experimental Procedures”). In Ub2, residues that show significant CSPs (Δδ > 0.15) and/or intermediate exchange are colored red and residues with 0.15 ≥ Δδ > 0.1 are colored yellow. In VHS-UIM, the VHS domain residues that exhibit significant CSPs (Δδ > 0.1) and/or intermediate exchange are colored red and those with 0.1 ≥ Δδ > 0.05 are colored yellow. Residues in the UIM domain that show Δδ > 0.2 and/or intermediate exchange are colored red, residues with 0.2 ≥ Δδ > 0.15 are colored yellow.
TABLE 2.
Summary of the dissociation constants derived from NMR titration curves for the VHS-UIM/mono-Ub and VHS-UIM/Ub2 binding equilibria
Kd values are averaged over several residues that showed strong CSPs upon binding. Standard deviations are shown in parentheses.
| Sample | Domain analyzed | Kd |
|---|---|---|
| μm | ||
| VHS-UIM/mono-Ub | VHS | 81 (28)a |
| UIM | 153 (38)a | |
| Mono-Ub | 102 (38)a | |
| VHS-UIM/Lys48-Ub2 | Distal Ub | 57 (18) |
| Proximal Ub | 67 (25) | |
| VHS | 69 (32) | |
| UIM | 270 (65) | |
| VHS-UIM/Lys63-Ub2 | Distal Ub | Kd1 + 47 (8)b |
| Kd2 =7 (3)b | ||
| Proximal Ub | Kd1 + 40 (14)b | |
| Kd2 + 12(6)b | ||
| VHS-UIM | Kd1 + 35(10)b | |
| Kd2 =2 (2)b |
a Data were fit using a single-site binding equation.
b Data were fit using sequential binding model (see supplemental materials) assuming two consecutive binding events, characterized with two binding constants, Kd1 and Kd2.
VHS-UIM Construct Binds Differently to Lys48-Ub2 and Lys63-Ub2
According to the aforementioned results, the isolated UIM has the same dissociation constant and the same mode of binding when forming a complex with mono-Ub, Lys48-Ub2, or Lys63-Ub2. By contrast, we have previously reported a different behavior for the isolated VHS domain of STAM2, which binds Lys48-Ub2 and Lys63-Ub2 differently (23). In that case, up to two VHS molecules can bind to Lys63-Ub2, whereas only one VHS domain can bind to Lys48-Ub2. This raises an important question: how do the two UBDs, UIM and VHS, interact with Ub2 chains when they are in tandem?
To gain insights into the interaction of VHS-UIM with Ub2, we monitored CSPs on distal and proximal Ub of both Lys48- and Lys63-Ub2, as well as on VHS-UIM. Any change in the chemical shift (i.e. resonance frequency) reflects a change in the electronic environment of the nucleus under observation. Therefore, similarities in the directions of the signal shifts in the 1H-15N spectra upon binding would suggest similarities in the changes of the electronic environment of 1H nuclei and (separately) of 15N nuclei in the corresponding complexes, thus similar changes of the chemical environment. On the VHS-UIM side, the CSP pattern for the VHS-UIM/Ub2 complex is similar to the one seen upon VHS-UIM binding to mono-Ub (Fig. 2, D, E, and I). It is noteworthy that several residues experienced strong signal attenuation during titration, but there were no additional perturbed sites compared with VHS-UIM/mono-Ub binding. Strong signal attenuations were also detected in several residues located around Ile44 and Val70 on both Ub units of Lys48- and Lys63-Ub2 (Fig. 2, G and H). As seen in Fig. 2, C and G, the perturbation patterns for the proximal and distal Ubs of Lys48-Ub2 show no significant differences from each other or from the perturbations observed in mono-Ub (Fig. 2B) upon binding to VHS-UIM. By contrast, there is a striking difference between the perturbations in the distal and the proximal Ubs of Lys63-Ub2 (Fig. 2, F and H, and supplemental Fig. S10). It is noteworthy that the CSP profile obtained for the distal Ub in Lys63-Ub2 is in many aspects similar to the one observed upon binding to the isolated UIM (Fig. 1C). This is especially true for residues Leu8, Ile61, and around Ile30, which display significant signal shifts upon binding to UIM but not VHS (23). On the other hand, the CSP pattern observed in the proximal Ub is quite similar to the one for the mono-Ub/VHS complex (23). These results suggest that in the Lys63-Ub2/VHS-UIM complex, the distal Ub binds the UIM and the proximal Ub binds the VHS domain, whereas in the case of Lys48-Ub2 the binding preferences are not that pronounced.
To further support these observations, the trajectories of the Ub2 signal shifts (in 1H-15N spectra) upon addition of VHS-UIM were compared with those during titration with the isolated VHS or UIM domain (supplemental Fig. S6). Indeed, the 1H-15N signals belonging to residues from the distal Ub of Lys63-Ub2 shifted in the same direction upon binding to UIM and VHS-UIM (supplemental Fig. S6). Likewise, the trajectories of the signal shifts in the proximal Ub of Lys63-Ub2 were very similar when binding to VHS-UIM and VHS. The direction of the shifts of the 1H-15N signals reflects the ratio of 1H and 15N CSPs, which is unique for each residue (N-H bond) and for the changes in its electronic environment induced by binding. Thus, similar directions of the NMR signal shifts indicate similarity in the local intermolecular contacts, further supporting the model in which VHS-UIM interacts with Lys63-Ub2 through specific pairwise contacts between the UIM and distal Ub and the VHS domain and proximal Ub. Unlike Lys63-Ub2, comparison of the signal shift trajectories for Lys48-Ub2 upon binding to VHS-UIM did not reveal any clear similarities with the shifts induced by the isolated VHS or UIM. However, one has to bear in mind that these conclusions are based largely on empirical results from numerous observations, and have not been clearly quantified by a direct comparison between alterations in the local structure and dynamics at the interface (including solvent accessibility) and the experimental or theoretical/computed chemical shifts.
We then performed 15N spin relaxation experiments to determine the stoichiometry of the VHS-UIM/Ub2 complex. In the case of VHS-UIM saturated with Lys63-Ub2, the measurements yielded a 15N R2 value of 31.5 ± 2.5 s−1 (for residues belonging to VHS) compared with a value of 17.0 ± 1.2 s−1 in the free state (see above). This increase in R2 indicates an increase in the overall size (the total molecular mass), consistent with the formation of the VHS-UIM/Lys63-Ub2 complex. This R2 value corresponds to a molecular mass range of 38–48 kDa, comparable with the sum of Ub2 and VHS-UIM masses (∼39.5 kDa), thus indicating a 1:1 stoichiometry. After diluting the sample by ∼2-fold, from 200 to 120 μm concentration of VHS-UIM, we performed 15N spin relaxation experiments. The average R2 for the well structured part of VHS was 28.4 ± 2.7 s−1. This value is somewhat lower than (although within the error bars from) the value determined for the higher concentration, which likely reflects a lesser fraction of the complex due to lower concentrations of both binding partners. This R2 corresponds to a molecular mass of 35.7–44 kDa and is in good agreement with a 1:1 stoichiometry. We were not able to extract a reliable transverse relaxation rate for the VHS-UIM/Lys48-Ub2 complex due to a significantly faster relaxation decay. This is indicative of a larger size (molecular mass) of the complex and suggests that more than one Lys48-Ub2 molecule can bind to VHS-UIM, perhaps with different populations. These results agree with the CSP mapping data shown above, which indicate a domain-specific “preferred” orientation of the VHS-UIM interactions in complex with Lys63-Ub2, and absence of clear interdomain interaction preferences in the case of Lys48-Ub2.
Lys63-Ub2 Exhibits Preferred Binding Mode with VHS-UIM
To further investigate the binding modes in the Lys48-Ub2/VHS-UIM and Lys63-Ub2/VHS-UIM complexes, we used MTSL as a paramagnetic spin label. This approach is used to derive long-distance constraints and identify low-populated states (42, 43). The paramagnetic tag causes strong signal attenuation in nuclei that are close in space (<25 Å) to the unpaired electron of MTSL via the PRE effect. MTSL was attached to a cysteine in Lys48- or Lys63-Ub2 introduced through a T12C mutation in either the distal or proximal Ub. The PRE effects were quantified from 1H,15N-HSQC spectra recorded for 15N-VHS-UIM in complex with Ub2 carrying the MTSL on the distal or proximal Ub (see “Experimental Procedures”). As shown in Fig. 3, A–D, the PREs arising from MTSL attached to Lys48- or Lys63-Ub2 result in strikingly different patterns of signal attenuations in VHS-UIM.
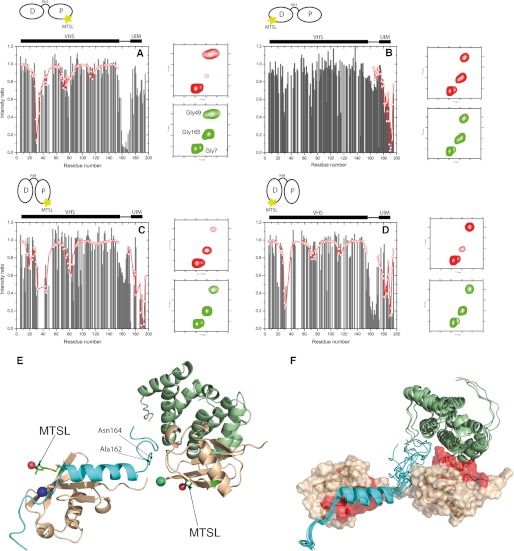
FIGURE 3.
Analysis of the PRE data for VHS-UIM in complex with MTSL-Lys63-Ub2 (A and B) and MTSL-Lys48-Ub2 (C and D), along with an expanded representative region of the 1H,15N-HSQC spectra in the oxidized (red) and reduced (green) states. The patterns of MTSL-induced signal attenuations in VHS-UIM are different, depending on whether MTSL is attached to C12 of (A) the proximal Ub of Lys63-Ub2 or (B) the distal Ub of Lys63-Ub2 or (C) the proximal Ub of Lys48-Ub2 or (D) the distal Ub of Lys48-Ub2 (E). Black bars represent experimental data, whereas open red circles connected by lines represent the back-calculated PREs for the fitted MTSL position. E, the best structure of the best cluster for the VHS-UIM/Lys63-Ub2 complex shows a good agreement between the position of MTSL derived from the measured PRE data and its position resulting from modeling (red spheres). The green sphere indicates the MTSL position derived from PREs caused by MTSL attached to the proximal Ub of Lys63-Ub2, whereas the blue sphere indicates the MTSL position derived from PREs induced by MTSL attached to the distal Ub of Lys63-Ub2. F, 10 best structures of the best cluster from the modeling of the VHS-UIM/Lys63-Ub2 complex. The red surface indicates the most perturbed residues in the distal and the proximal Ub units of Lys63-Ub2 upon binding to VHS-UIM. The structure of the VHS-UIM used in the current docking procedure was obtained by homology modeling (see “Experimental Procedures”).
As obvious from Fig. 3, A and B, the attenuations in VHS-UIM induced by MTSL attached to the proximal or distal Ub of Lys63-Ub2 are strikingly different, especially in the linker region around Ala160. Indeed, the linker was affected by MTSL attached to the proximal Ub, whereas MTSL on the distal Ub did not induce any significant perturbation in the same region. Interestingly, when MTSL was attached to the proximal Ub, the signal attenuation pattern in the VHS region was similar to the one seen for the VHS/mono-Ub interaction (23), where residues around Thr30 were strongly attenuated, whereas the MTSL did not significantly affect the UIM domain (Fig. 3A). Conversely, when MTSL was attached to the distal Ub of Lys63-Ub2, the main attenuations occurred on the UIM as well as the C terminus of VHS-UIM (Fig. 3B). These observations confirm the above results and point to a particular binding mode where the proximal Ub of Lys63-Ub2 predominantly binds the VHS domain, whereas the distal Ub binds the UIM domain. It should be borne in mind, however, that we cannot exclude that a small fraction of the Lys63-Ub2/VHS-UIM complexes adopts a different structural organization. Indeed, MTSL attached to the distal or the proximal Ub of Lys63-Ub2 also induced modest attenuations in VHS or UIM, respectively.
A dramatically different pattern of PRE-induced signal attenuations appeared when MTSL was attached to any of the two Ubs in Lys48-Ub2. In this case, both the VHS and UIM domains were affected by MTSL attached to the distal or the proximal Ub (Fig. 3, C and D). In particular, for MTSL attached to the proximal Ub of Lys48-Ub2, the PREs induced in the VHS part of VHS-UIM (Fig. 3C) were similar to those seen for the VHS/Lys48-Ub2 interaction (23). These observations suggest a more complicated molecular organization of the VHS-UIM/Lys48-Ub2 complex than in the VHS-UIM/Lys63-Ub2 complex.
Modeling VHS-UIM/Lys63-Ub2 Interaction
Taking into account all the aforementioned results, we asked the following questions: “how does Lys63-Ub2 accommodate VHS-UIM?” and “is the intervening linker between VHS and UIM flexible enough to allow this interaction.” To address these questions, we attempted to model the Lys63-Ub2/VHS-UIM complex using our PRE data (Fig. 3, A and B). The position of MTSL attached to the proximal Ub with respect to the VHS domain was reconstructed by taking into account the PREs in VHS from Fig. 3A. Likewise, the position of MTSL attached to the distal Ub with respect to the UIM domain was reconstructed by considering the PREs in the UIM from Fig. 3B. Due to high flexibility of the linker, the PREs in that region were not taken into account.
The modeling was performed using molecular docking with NMR constraints implemented in the Haddock 2.0 program (44, 45) (see supplemental Materials). The docking was driven by using a combination of distance restraints derived from PRE data and homology modeling complemented by ambiguous restraints from the CSP data. Based on the results presented above, our model assumed that in the VHS-UIM complex with Lys63-Ub2, the VHS domain contacts the proximal Ub, whereas the UIM domain contacts the distal Ub. The existing structure of the VHS/mono-Ub complex (23) was used as the starting structure for the VHS/proximal Ub interaction. For the UIM/Ub contact, the structure of the Vps27-UIM/Ub complex (38) was used. The resulting structures were subjected to clustering and details of the docking results are summarized in supplemental Table S1 for the 10 best structures of the best cluster; these structures are presented in Fig. 3, E and F. To evaluate the goodness of our model, the distances between the oxygen of MTSL and each amide proton in the VHS or UIM domains of the modeled complex were back-calculated. As can be seen from supplemental Fig. S11, the back-calculated distances are in good agreement with those derived from our PRE data. It is noteworthy that deviations from the experimental distances seen for the VHS and UIM domains are likely due to (i) the decrease of accuracy of PRE data for distances higher than a given threshold (25 Å), (ii) the fact that the C terminus of UIM is highly flexible (supplemental Fig. S11D), and (iii) the possibly of the presence of low-populated structures that could also affect the helix α7 of VHS (supplemental Fig. S11C). Also interesting is the fact that the linker region can approach the MTSL attached to the proximal Ub of Lys63-Ub2 at a distance as close as 7.5 Å (Ala162) and 8.8 Å (Asn164) (Fig. 3E). This observation is in fact in agreement with the moderate CSPs seen on the linker region of VHS-UIM (Fig. 2D), as well as the strong decrease in intensity in the linker due to the MTSL attached on the proximal Ub of Lys63-Ub2.
Lys63-Ub2, Unlike Lys48-Ub2, Binds VHS-UIM Cooperatively
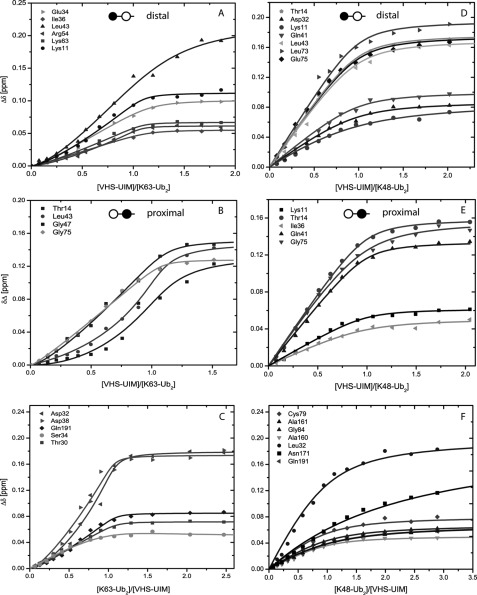
Following the aforementioned results, we wondered if the difference in structural organization between the two VHS-UIM/Ub2 complexes is reflected in the titration curves. To address this, we analyzed the NMR titration data collected from the VHS-UIM and Ub2 sides. As seen in Fig. 4, the binding isotherms for the VHS-UIM/Lys63-Ub2 complex have a sigmoidal shape, in stark contrast with the hyperbolic-shape binding isotherms for the VHS-UIM/Lys48-Ub2 complex.
FIGURE 4.
Representative titration curves for 15N-labeled distal (A) and proximal (B) Ubs in Lys63-Ub2 and distal (D) and proximal (E) Ubs in Lys48-Ub2 as a function of the molar ratio of VHS-UIM, and for 15N-VHS-UIM as a function of the molar ratio of Lys63-Ub2 (C) and Lys48-Ub2 (F).
For the VHS-UIM/Lys48-Ub2 equilibrium, looking at the VHS-UIM side (see Fig. 4F), we derived a Kd of 69 ± 32 μm for residues belonging to the VHS domain and 270 ± 65 μm for the UIM domain residues (Table 2). These Kd values are comparable with those derived for the isolated VHS (64 μm, see Lange et al. (23)) and UIM (Table 1) in complex with Lys48-Ub2. From the Lys48-Ub2 side, we determined the Kd of 57 ± 22 and 67 ± 20 μm for the distal and the proximal Ubs, respectively (see Fig. 4, D and E). These values are in excellent agreement with those determined from residues in the VHS domain and suggest that of the two UBDs in VHS-UIM, Lys48-Ub2 predominantly binds to the VHS domain. Recall that Lys48-Ub2 can bind two isolated UIM domains but it can accommodate only one VHS molecule (23). Together with the observation that the titration curves in Fig. 4, D and E (also Fig. 4F), saturate at the [VHS-UIM]/[Ub2] molar ratio close to 1 (pointing to a 1:1 stoichiometry of binding), these results further support the conclusion that the VHS domain is the predominant Lys48-Ub2-binding domain in VHS-UIM.
The sigmoidal shape of the titration curves in Fig. 4, A–C, suggests that Lys63-Ub2 binding to VHS-UIM is cooperative. Note that specific perturbations observed in both VHS and UIM indicate that both UBDs in VHS-UIM are involved in Lys63-Ub2 binding, consistent with the cooperative character of this interaction. To further examine the binding process, we considered a stepwise sequential model involving two binding events (see supplemental Materials) or a multivalent binding model (46) characterized by the binding of a single Ub to a single UBD (Kd,mono) and a multivalent binding of VHS-UIM to Lys63-Ub2 (KD,mv). The various Kd values obtained from each side of the complex are summarized in Table 2 for the sequential model. The average dissociation constants for the VHS-UIM/Lys63-Ub2 complex are Kd1 = 41 ± 11 and Kd2 = 7 ± 4 μm (for the first and second binding events, respectively). When using the multivalent binding model, the residuals of the fit were twice as high compared with the sequential model, using a three-parameter fit for both models. Moreover, the multivalent binding model could not reproduce the sigmoidal shape of our titration data, indicating that the multivalent binding model is not suitable in our case. In case of a two-step binding model, we can hypothesize that binding of either VHS to the proximal Ub or UIM to the distal Ub of Lys63-Ub2 increases the probability that the other UBD of the same VHS-UIM molecule binds to the second Ub unit of the same Ub2 chain. For such a molecular model, the effective concentration or probability density for the end to end length of the linker vector can be modeled as a polymer chain (47, 48) (see supplemental Materials). By considering a 20-amino acid linker and a 23-Å end to end linker length in the bound state, we estimated an effective concentration of ∼10 mm, comparable with the multivalent binding of RAP80-tUIM to Lys63-Ub2 (46). In addition to stepwise binding and high local effective concentration, we are interested in determining the molecular origins of this cooperative effect. To gain insights into this issue, we performed heteronuclear 15N{1H}-NOE experiments on VHS-UIM upon saturation by Lys63-Ub2 (supplemental Fig. S12). Compared with the free state of VHS-UIM, we observed that NOE values increase in the linker and UIM regions upon binding to Lys63-Ub2, suggesting that the binding of UIM to the distal unit of Lys63-Ub2 may induce stabilization of its helical structure and a higher rigidity. This conclusion is further supported by the fact that the UIM resonances in the 1H,15N-HSQC spectra became more widely dispersed upon addition of Lys63-Ub2.
DISCUSSION
The human STAM2 protein is involved in lysosomal degradation and associates with Hrs to form the ESCRT-0 component of the ESCRT machinery. STAM2 possesses two UBDs: the UIM and VHS domains. Deletion of one of these domains or mutation alters the transport of ubiquitinated cargoes (19, 21, 49, 50). Additionally, ESCRT-0 binds polyubiquitin chains with high avidity and seems to have a preference for Lys63-linked chains (21). For instance, Lys63-linked ubiquitination is required for MVB sorting of Gap1, CPS, and probably other cargoes (12). Because it has been reported that the binding of ESCRT-0 showed a modest increase in affinity for Lys63-Ub2 over Lys48-Ub2 and “linear,” head-to-tail linked chains (NC-Ub2) (21), it is likely that other factors are responsible for the specificity of Lys63-linked chains toward cargo sorting. We previously characterized the interaction of an isolated VHS domain with mono-Ub, Lys48- and Lys63-Ub2 and reported that Lys63- and Lys48-Ub2 chains bind VHS via different binding modes (23).
In the present study, we first examined whether the UIM domain of STAM2 also exhibits such structural features when binding to Lys48- and Lys63-Ub2 chains. A UIM was originally identified in the S5a subunit of the 26 S proteasome and is often found in proteins involved in the proteasomal and lysosomal degradation (51). UIMs fold into a single α-helix and are embedded as independent domains into various modular proteins. Our NMR mapping revealed that the Ub-binding surface is located on one face of the helical region of STAM2 UIM (supplemental Fig. S13B) and involves a set of conserved hydrophobic residues (supplemental Fig. S13A). These findings are in good agreement with the Vps27-UIM1/mono-Ub and STAM-UIM/mono-Ub structures (38, 41). Using NMR titration experiments, we determined the STAM2-UIM/mono-Ub dissociation constant of 287 ± 33 μm, which implies that the UIM domain of STAM2 binds to mono-Ub weaker than VHS (23). Furthermore, our study revealed that Lys48- and Lys63-Ub2 show comparable affinities for UIM (Table 1) and can accommodate up to two STAM2-UIM molecules per chain. A similar structural organization has already been seen in the case of the UIM2 domain of S5a (52). According to our results, the UIM domain of STAM2 does not discriminate between mono-Ub, Lys48- and Lys63-Ub2.
Our data suggest that the UIM adopts the same mode of binding to mono-Ub, Lys48- and Lys63-Ub2 chains. Several lines of evidence support this conclusion. First, the trajectories of the NMR signals in the course of titration with UIM are similar for mono-Ub and the distal and proximal Ubs of Lys48-Ub2 and Lys63-Ub2 chains (see supplemental Fig. S6). Second, the perturbation maps (i.e. residues showing strong CSPs and/or signal attenuations as a result of UIM binding) are similar for mono-Ub, Lys48- and Lys63-Ub2 chains. Based on the present results on UIM binding and previous work on VHS (23), we can rule out the possibility that the individual UIM or VHS domains are solely responsible for the better affinity of STAM2 for Lys63-Ub2.
To determine the impact of having two UBDs in tandem (as in STAM2) on binding to ubiquitin and polyubiquitin chains, we characterized the interaction of the VHS-UIM construct from STAM2 with mono-Ub, Lys48- and Lys63-Ub2 chains. Our titration experiments suggest that both UBDs in VHS-UIM bind mono-Ub independently and with affinities similar to those observed for the isolated VHS and UIM domains (see Lange et al. (23) and Table 2). Note that VHS binds Ub stronger than UIM does. Switching from mono-Ub to Ub2, the VHS-UIM/Lys48-Ub2 interaction gives a somewhat more complicated picture of the equilibrium, where various intermolecular arrangements (and perhaps with different populations) can exist. The VHS-UIM construct binds Lys48-Ub2 with the same affinity as mono-Ub (average Kd = 115 ± 41 μm), and thus does not bind avidly to Lys48-Ub2 chains. As shown above, the isolated VHS and UIM exhibit different stoichiometry in binding to Lys48-Ub2. Then one might hypothesize that, due to the steric hindrance introduced by the VHS domain, it is unlikely that the UIM domain of VHS-UIM could bind another Ub unit of the same Lys48-Ub2 chain. An attempt to model the VHS-UIM/Lys48-Ub2 complex with a 1:1 stoichiometry supports this hypothesis. Indeed, the resulting structure is in disagreement with our CSP data and would induce a severe bending of the VHS-UIM linker (supplemental Fig. S14). In addition, it should be borne in mind that Lys48-Ub2 is in dynamic equilibrium between a closed conformation, in which the functionally important hydrophobic patch residues of each Ub unit are sequestered at the Ub/Ub interface, and one or more open conformations (24, 37). VHS-UIM binding would require opening of the interface to allow contacts between the hydrophobic residues of the two Ub units and VHS-UIM. In such a situation, the molecular recognition process would likely proceed through a conformational selection mechanism, namely selection of the right (open) conformation of Lys48-Ub2. At neutral pH the closed state is predominantly populated (∼85%), and the time of interconversion between the closed and open conformations, estimated by NMR to be ∼10 ns, is comparable with the overall tumbling time (37). Analysis of our relaxation data indicates that UIM and VHS tumble essentially independently from each other with respective rotational correlation times of 3.8 ± 0.5 and 15.0 ± 0.4 ns, respectively. Thus, slower tumbling of the VHS domain could be a limiting factor in the recognition and binding to the open state of Lys48-Ub2, possibly explaining the less efficient sorting of Lys48-polyubiquitinated cargoes by ESCRT-0.
A strikingly different picture arises in the case of VHS-UIM interaction with Lys63-Ub2, where the VHS and the UIM domains preferentially bind to the proximal and the distal Ub of Lys63-Ub2, respectively. Lys63-Ub2 adopts an extended conformation (27, 53) with the hydrophobic patches exposed to the solvent; this allows the interaction with the VHS and UIM domains without the need to compete with the Ub/Ub interaction as in Lys48-Ub2. Interestingly, the titration curves corresponding to the interaction of VHS-UIM with Lys63-Ub2 have a sigmoidal shape. This is likely to result from two binding events and a cooperative effect, thus demonstrating a clear difference in the binding of mono-Ub, Lys48-Ub2, and Lys63-Ub2 to VHS-UIM. We derived two dissociation constants (average Kd1 = 41 ± 11 μm Kd2 = 7 ± 4 μm) that reflect an increase in the affinity of VHS-UIM for Lys63-Ub2 compared with Lys48-Ub2. Assuming that VHS binds first and UIM second, it appears that the binding of VHS to Lys63-Ub2 is only slightly stronger than to Lys48-Ub2 or mono-Ub, whereas the UIM binds ∼40 times stronger. This agrees with the notion that the presence of multiple UBDs (in tandem) can greatly enhance the affinity for binding partners and/or induce a preference for a given polyubiquitin chain (54). From a mechanistic point of view, the binding can be envisioned to occur stepwise by first binding one of the UBDs to a Ub unit. Upon binding, the first UBD becomes part of the complex, and the binding of the second UBD becomes intramolecular. One can assume that one of the roles of the flexible linker is to keep the UBD in proximity, thus increasing its effective local concentration to ∼10 mm. This value is of the same order of magnitude as the one calculated for the RAP80-tUIM/Lys63-Ub2 (∼12 mm) or S5A/Lys48-Ub2 (∼3 mm) complexes (46) and reflects the dependence of the effective local concentration on the length of the inter-UBD linker. In addition to stepwise binding and increasing the local effective concentration, one has to take into account the cooperative nature of the VHS-UIM/Lys63-Ub2 interaction. A possible mechanism to explain the cooperativity is that the binding of VHS to the first Ub unit would position UIM favorably for interaction with a nearby Ub, leading to the cooperative stabilization of the UIMs helix. The difference in tumbling between the VHS and UIM in VHS-UIM could have important consequences regarding molecular events, as the initial binding of VHS to 1 Ub unit can be followed by a rapid reorientation (on the nanosecond time scale), binding, and subsequent stabilization of the UIM helix. Binding of the first UBD to a Ub unit will result in a loss of its translational and rotational degrees of freedom (55), hence reducing the conformational entropy. However, the flexibility of the linker as well as the cooperative stabilization of the UIM helix can overcome the entropic penalty. One has to keep in mind that flexibility has an important influence on the thermodynamics of binding and may both favor and disfavor association (56). In the light of our results, the differences between the binding of VHS-UIM to mono-Ub, Lys48-Ub2, and Lys63-Ub2 could be one explanation for the better sorting efficiency of Lys63- compared with Lys48-polyubiquitinated targets in the context of the full ESCRT-0.
A polypeptide chain linking two successive UBDs as well as the nature of the UBDs could favor a specific ubiquitin chain linkage. Among the few existing structures reporting the interaction of multiple UBDs with polyubiquitin chains, S5a, which is involved in proteasomal degradation, contains two UIMs and binds preferentially to Lys48-Ub2. Moreover, the UIMs of S5a bind the two Ub units simultaneously and with UIM1 showing a 3:1 preference for the distal Ub of Lys48-Ub2 possibly reflecting specific interactions with the Ub-Ub linker (57). Rap80 is another protein that harbors two UIM domains linked in tandem (tUIM). The tUIM fragment specifically recognizes Lys63-Ub2 chains where the length of the linker is critical for binding with high affinity. By engineering different linker lengths, Sims and Cohen (54) demonstrated that the linker length and composition modulated the affinity of RAP80-tUIM for Lys63-Ub2 in a periodic fashion. Changes in the length of the helical linker not only modify the distance between the two UIMs, but also the relative orientation of their Ub-binding surfaces. Thus, the Rap80 linker defines selectivity through domain positioning rather than specific contact with the Ub-Ub linkage. Another example is given by NEMO (NF-κ essential modulator) that forms a heterodimer, which preferentially binds to NC-Ub2 via its UBAN domain (ubiquitin binding in ABIN and NEMO) (58, 59). The NEMO dimer accommodates two Ub2s, whereas both NEMO protomers contribute to the binding to each Ub2. In contrast to Rap80, NEMO specifically recognizes the linker region of head to tail chains and can distinguish between NC- and Lys63-linked chains. These examples demonstrate that an array of homologous UBDs can define chain linkage selectivity. Therefore, not only the combination of UBDs but also the length of the intervening linker and the arrangement of the UBDs could form the basis for linkage-specific polyubiquitin recognition. Additionally, the combination of heterologous UBDs of different sizes, like in STAM2, could influence the discrimination between different polyubiquitin chain linkages. Other factors like oligomerization, compartmentalization, and concentration of UBD-containing proteins could contribute to and possibly enhance/complicate the discrimination between different Ub chains.
In the context of the full ESCRT machinery, STAM2 and Hrs form a heterodimer/tetramer (16, 60) through their GAT domain to form the ESCRT-0 component. The fact that Lys63-linked polyubiquitin chains now emerge as a specific signal for protein sorting into the MVB pathway (12, 61, 62) could be linked to different factors, one of them being the higher affinity and cooperativity of the STAM2 VHS-UIM domains for Lys63-Ub2 chains. From the general view of the ESCRT-0 complex, two different models have been proposed. In one of them, cargoes are passed sequentially from ESCRT-0 to ESCRT-I, -II, and -III. If one considers that ESCRT-0 originally possesses four UBDs being part of STAM (VHS and UIM) and Hrs (VHS and DUIM), one can wonder how cargoes carrying Lys63-Ub2 chains can be transferred to the ESCRT-I complex, with a UEV domain of Tsg101 that binds to mono-Ub with a Kd as high as 510 μm (63). In contrast, our data tend to support a second model where UBDs within the different ESCRT complexes cooperate to increase the binding affinity for ubiquitinated cargoes and work as a “supercomplex” (64), with ESCRT-0 being the main complex to cluster ubiquitinated cargoes (65).
Supplementary Material
Acknowledgments
The eNMR project (European FP7 e-Infrastructure grant, contract no. 213010), supported by the national GRID Initiatives of Italy, Germany, and the Dutch BiG Grid project (Netherlands Organization for Scientific Research) are acknowledged for the use of web portals, computing, and storage facilities.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065334 (to D. F.) and National Science Foundation Postdoctoral Fellowship DBI-0905967 (to C. A. C.).

This article contains supplemental Figs. S1–S14, Table S1, and Materials.
- MVB
- multivesicular body
- ESCRT
- endosomal sorting complexes required for transport
- Ub
- monoubiquitin
- UIM
- ubiquitin-interacting motif
- Ub2
- diubiquitin
- UBD
- ubiquitin-binding domain
- Hrs
- hepatocyte growth factor-regulated substrate
- STAM
- signal transducing adaptor molecule
- Lys63-Ub2
- Lys63-linked diubiquitin
- Lys48-Ub2
- Lys48-linked diubiquitin
- VHS
- Vps27/Hrs/STAM
- HSQC
- heteronuclear single quantum coherence
- CSP
- chemical shift perturbation
- MTSL
- 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)methanesulfonate
- PRE
- paramagnetic relaxation enhancement
- tUIM
- tandem UIM.
REFERENCES
- 1. Platta H. W., Stenmark H. (2011) Cell structure and dynamics. Curr. Opin. Cell Biol. 23, 393–403 [DOI] [PubMed] [Google Scholar]
- 2. Sorkin A., von Zastrow M. (2009) Endocytosis and signaling. Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scita G., Di Fiore P. P. (2010) The endocytic matrix. Nature 463, 464–473 [DOI] [PubMed] [Google Scholar]
- 4. Hicke L., Dunn R. (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 5. Haglund K., Di Fiore P. P., Dikic I. (2003) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 6. Pickart C. M., Fushman D. (2004) Polyubiquitin chains. Polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 7. Clague M. J., Urbé S. (2010) Ubiquitin, same molecule, different degradation pathways. Cell 143, 682–685 [DOI] [PubMed] [Google Scholar]
- 8. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 9. Acconcia F., Sigismund S., Polo S. (2009) Ubiquitin in trafficking. The network at work. Exp. Cell Res. 315, 1610–1618 [DOI] [PubMed] [Google Scholar]
- 10. Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., Lehner P. J. (2006) Lysine 63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 12. Lauwers E., Jacob C., André B. (2009) Lys63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raiborg C., Malerød L., Pedersen N. M., Stenmark H. (2008) Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 314, 801–813 [DOI] [PubMed] [Google Scholar]
- 14. Henne W. M., Buchkovich N. J., Emr S. D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- 15. Mizuno E., Kawahata K., Okamoto A., Kitamura N., Komada M. (2004) Association with Hrs is required for the early endosomal localization, stability, and function of STAM. J. Biochem. 135, 385–396 [DOI] [PubMed] [Google Scholar]
- 16. Hurley J. H. (2010) The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45, 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hurley J. H. (2008) ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20, 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dikic I., Wakatsuki S., Walters K. J. (2009) Ubiquitin-binding domains. From structures to functions. Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizuno E., Kawahata K., Kato M., Kitamura N., Komada M. (2003) STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol. Biol. Cell 14, 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fisher R. D., Wang B., Alam S. L., Higginson D. S., Robinson H., Sundquist W. I., Hill C. P. (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J. Biol. Chem. 278, 28976–28984 [DOI] [PubMed] [Google Scholar]
- 21. Ren X., Hurley J. H. (2010) VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wollert T., Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lange A., Hoeller D., Wienk H., Marcillat O., Lancelin J. M., Walker O. (2011) NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin. Biochemistry 50, 48–62 [DOI] [PubMed] [Google Scholar]
- 24. Varadan R., Walker O., Pickart C., Fushman D. (2002) Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 324, 637–647 [DOI] [PubMed] [Google Scholar]
- 25. Pickart C. M., Raasi S. (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 26. Zhang D., Raasi S., Fushman D. (2008) Affinity makes the difference. Nonselective interaction of the UBA domain of ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., Fushman D. (2004) Solution conformation of Lys63-linked diubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 279, 7055–7063 [DOI] [PubMed] [Google Scholar]
- 28. Varadan R., Assfalg M., Fushman D. (2005) Using NMR spectroscopy to monitor ubiquitin chain conformation and interactions with ubiquitin-binding domains. Methods Enzymol. 399, 177–192 [DOI] [PubMed] [Google Scholar]
- 29. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe, a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 30. Keller R. (2004) The Computer-aided Resonance Assignment Tutorial, CARA NMR [Google Scholar]
- 31. Pervushin K., Riek R., Wider G., Wüthrich K. (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fushman D., Cahill S., Cowburn D. (1997) The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution. Analysis of 15N relaxation with monomer/dimer equilibration. J. Mol. Biol. 266, 173–194 [DOI] [PubMed] [Google Scholar]
- 33. Hall J. B., Fushman D. (2003) Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy. Differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J. Biomol. NMR 27, 261–275 [DOI] [PubMed] [Google Scholar]
- 34. Kay L. E., Torchia D. A., Bax A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy. Application to staphylococcal nuclease. Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 35. Grzesiek S., Bax A. (1993) The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 36. Goddard T. D., Kneller D. G. (2001) SPARKY 3, University of California, San Francisco, CA [Google Scholar]
- 37. Ryabov Y., Fushman D. (2006) Interdomain mobility in diubiquitin revealed by NMR. Proteins 63, 787–796 [DOI] [PubMed] [Google Scholar]
- 38. Swanson K. A., Kang R. S., Stamenova S. D., Hicke L., Radhakrishnan I. (2003) Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor down-regulation. EMBO J. 22, 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 40. Wishart D. S., Sykes B. D. (1994) The 13C chemical-shift index. A simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 41. Lim J., Son W. S., Park J. K., Kim E. E., Lee B. J., Ahn H. C. (2011) Solution structure of UIM and interaction of tandem ubiquitin binding domains in STAM1 with ubiquitin. Biochem. Biophys. Res. Commun. 405, 24–30 [DOI] [PubMed] [Google Scholar]
- 42. Clore G. M., Iwahara J. (2009) Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 109, 4108–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ubbink M. (2009) The courtship of proteins. Understanding the encounter complex. FEBS Lett. 583, 1060–1066 [DOI] [PubMed] [Google Scholar]
- 44. Dominguez C., Boelens R., Bonvin A. (2003) HADDOCK. A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 45. de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) HADDOCK versus HADDOCK. New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 46. Markin C. J., Xiao W., Spyracopoulos L. (2010) Mechanism for recognition of polyubiquitin chains. Balancing affinity through interplay between multivalent binding and dynamics. J. Am. Chem. Soc. 132, 11247–11258 [DOI] [PubMed] [Google Scholar]
- 47. Zhou H. X. (2001) Loops in proteins can be modeled as worm-like chains. J. Phys. Chem. B 105, 6763–6766 [Google Scholar]
- 48. Zhou H. X., Gilson M. K. (2009) Theory of free energy and entropy in noncovalent binding. Chem. Rev. 109, 4092–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534–539 [DOI] [PubMed] [Google Scholar]
- 50. Shih S. C., Katzmann D. J., Schnell J. D., Sutanto M., Emr S. D., Hicke L. (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389–393 [DOI] [PubMed] [Google Scholar]
- 51. Hofmann K., Falquet L. (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26, 347–350 [DOI] [PubMed] [Google Scholar]
- 52. Haririnia A., D'Onofrio M., Fushman D. (2007) Mapping the interactions between Lys48- and Lys63-linked diubiquitins and a ubiquitin-interacting motif of S5a. J. Mol. Biol. 368, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Datta A. B., Hura G. L., Wolberger C. (2009) The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 392, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sims J. J., Cohen R. E. (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of Rap80. Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Finkelstein A. V., Janin J. (1989) The price of lost freedom. Entropy of bimolecular complex formation. Protein Eng. 3, 1–3 [DOI] [PubMed] [Google Scholar]
- 56. Grünberg R., Nilges M., Leckner J. (2006) Flexibility and conformational entropy in protein-protein binding. Structure 14, 683–693 [DOI] [PubMed] [Google Scholar]
- 57. Zhang N., Wang Q., Ehlinger A., Randles L., Lary J. W., Kang Y., Haririnia A., Storaska A. J., Cole J. L., Fushman D., Walters K. J. (2009) Structure of the S5a:K48-linked diubiquitin complex and its interactions with Rpn13. Mol. Cell 35, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 59. Lo Y. C., Lin S. C., Rospigliosi C. C., Conze D. B., Wu C. J., Ashwell J. D., Eliezer D., Wu H. (2009) Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mayers J. R., Fyfe I., Schuh A. L., Chapman E. R., Edwardson J. M., Audhya A. (2011) ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J. Biol. Chem. 286, 9636–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jencks W. P. (1981) On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. U.S.A. 78, 4046–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barriere H., Nemes C., Du K., Lukacs G. L. (2007) Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol. Biol. Cell 18, 3952–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., Sundquist W. I. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 64. Shields S. B., Oestreich A. J., Winistorfer S., Nguyen D., Payne J. A., Katzmann D. J., Piper R. (2009) ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Slagsvold T., Pattni K., Malerød L., Stenmark H. (2006) Endosomal and nonendosomal functions of ESCRT proteins. Trends Cell Biol. 16, 317–326 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.