Background: Calmodulin is a Ca2+ binding protein and a major regulator of multiple signaling pathways.
Results: Inactivation of the Ca2+ binding sites in the N- and C-terminal lobe of CaM affects cell viability differentially.
Conclusion: Ca2+ binding to CaM is required for vertebrate cell survival.
Significance: A novel vertebrate knock-out/knock-in system for studying the function of CaM is described.
Keywords: Calcium Binding Proteins, Calcium Signaling, Calmodulin, Gene Knock-out, Signaling, DT40
Abstract
Calmodulin (CaM) was shown to be essential for survival of lower eukaryotes by gene deletion experiments. So far, no CaM gene deletion was reported in higher eukaryotes. In vertebrates, CaM is expressed from several genes, which encode an identical protein, making it difficult to generate a model system to study the effect of CaM gene deletion. Here, we present a novel genetic system based on the chicken DT40 cell line, in which the two functional CaM genes were deleted and one allele replaced with a CaM transgene that can be artificially regulated. We show that CaM is essential for survival of vertebrate cells as they die in the absence of CaM expression. Reversal of CaM repression or ectopic expression of HA-tagged CaM rescued the cells. Cells exclusively expressing HA-CaM with impaired individual calcium binding domains as well as HA-CaM lacking the ability to be phosphorylated at residues Tyr99/Tyr138 or trimethylated at Lys115 survived and grew well. CaM mutated at both Ca2+ binding sites 3 and 4 as well as at both sites 1 and 2, but to a lesser degree, showed decreased ability to support cell growth. Cells expressing CaM with all calcium binding sites impaired died with kinetics similar to that of cells expressing no CaM. This system offers a unique opportunity to analyze CaM structure-function relationships in vivo without the use of pharmacological inhibitors and to analyze the function of wild type and mutated CaM in modulating the activity of different target systems without interference of endogenous CaM.
Introduction
Calmodulin (CaM)7 is a highly conserved Ca2+ binding protein expressed in animals, plants, fungi, and protozoa (1). CaM is involved in a plethora of cellular functions, including growth factor signaling, cell cycle progression, transcriptional regulation, and programmed cell death. More than 100 CaM binding proteins have been identified, of which several are critical for the regulation of cell proliferation (2–12). However, analysis of the functional involvement of CaM in target protein activities in cells is far from complete. CaM null mutations are lethal in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Aspergillus nidulans, and Drosophila melanogaster (reviewed in Ref. 13). So far, no CaM gene deletions in vertebrates have been reported, and it is not known how CaM supports cell survival in these organisms. One potential obstacle might have been that higher eukaryotes express an identical CaM protein from multiple genes, making gene deletions a difficult task (14).
CaM regulates the activity of most of its binding partners in a Ca2+-dependent manner (reviewed in Ref. 4). However, CaM can activate some of its targets in a Ca2+-independent manner (15). Moreover, different CaM target proteins appear to require a distinct number of calcium ions in the Ca2+-CaM complex to attain maximum binding efficiency (16). Upon Ca2+ binding, CaM undergoes large conformational changes. Both pairs of EF-hands adopt an open conformation, thereby exposing hydrophobic patches to potential binding proteins (1). Six different classes of CaM binding proteins have been defined depending on the mode of CaM interaction and activation of the target protein (1, 17).
The N-terminal domain of Ca2+-free CaM (apo-CaM), consisting of EF-hand 1 and 2, has a closed conformation with both EF-hands packed together, which is believed to inhibit target protein interaction (18). The C-terminal domain of apo-CaM, however, adopts a semiopen conformation, which may allow binding between certain target proteins and partially exposed hydrophobic patches of CaM in the absence of Ca2+. Furthermore, the affinity for Ca2+ of the C-terminal EF-hands (Kd ≈ 10−6 m) is 10-fold greater than the affinity of the N-terminal EF-sites, with a Kd of ≈ 10−5 m (reviewed in Ref. 19). Binding of a single calcium ion to one of the C- or N-terminal EF-hands increases the affinity of the adjacent EF-hand domain denoted as positive cooperativity among paired EF-hands. So far, investigations on the role of Ca2+ binding to CaM in cell proliferation and cell death have been restricted to lower eukaryotes (20, 21).
Posttranslational modification of CaM such as tyrosine phosphorylation at residues 99 and 138 and lysine trimethylation at position 115 may play crucial roles in the modulation of the distinct cellular processes regulated by CaM (22, 23). So far, the insulin receptor and the EGF receptor are the only members of the protein tyrosine kinase receptor family which have been shown to phosphorylate CaM at its tyrosine residues. Tyrosine phosphorylation by non-receptor protein tyrosine kinases such as Src family kinases has been described as well (reviewed in Ref. 22). Although tyrosine phosphorylation of mammalian CaM has been shown to affect target interaction and activation in vitro, its significance for cell proliferation is not yet known. Trimethylation of CaM at Lys115 is known as a conserved modification found in many organisms (reviewed in Ref. 24). However, except from its function as a regulator of NAD kinase in plants (25), its functional significance in higher eukaryotes is unknown so far. Here, we describe the establishment of a CaM knock-out system in chicken pre-B lymphoma DT40 cells allowing conditional CaM expression and demonstrate its usefulness for studying the significance of Ca2+ binding and posttranslational modifications of CaM for cell survival.
EXPERIMENTAL PROCEDURES
Cell Lines and Plasmid Constructs
The chicken bursa of Fabricius cell line DT40 WT was provided by Dr. T. Kurosaki (Kansai Medical University, Osaka, Japan), and the DT40 clone 8-5 was described previously (13). For propagation of cells, design of CaMI gene targeting, rat CaM knock-in (pCaMI KO) and HA-CaM vectors, see supplemental material.
Cell Growth and Viability Experiments
Cell density was adjusted to 0.5 × 105/ml, and the cells were further grown with or without tetracycline (1 μg/ml). Every other day, the cell densities were readjusted to 0.5 × 105/ml to maintain exponential growth. For 4–6 days, cells were counted twice a day. Doubling time was estimated from linear trend lines. Percentage of viable cells was estimated using flow cytometry.
Western Blot Analysis
Extraction of total cellular proteins and Western blotting was done as described previously (26). After transfer, the membranes were treated with 0.2% (v/v) glutaraldehyde to immobilize CaM (reviewed in Ref. 24). Antibodies used for this work were as follows: monoclonal mouse anti-CaM antibody (Millipore 05-173), HA11 antibody (Covance 16B12), and rabbit polyclonal anti-phospho-Tyr99 CaM antibodies (Millipore 09-295). Quantification of protein signals was done by densitometry of scanned films using ImageJ software.
Southern and Northern Blot Analyses
A chicken CaMI-specific cDNA probe (a fragment comprising most of CaMI intron 1 and part of exon 2) was used for hybridization with genomic DNA digested with EcoR1. Northern blots containing total RNA, isolated as described in Ref. 27 were probed with a 400-bp chicken CaMI cDNA probe.
RESULTS
Deletion of CaMI Gene in DT40 Cells (CaMII−/−) and Replacement by Repressible CaM Construct
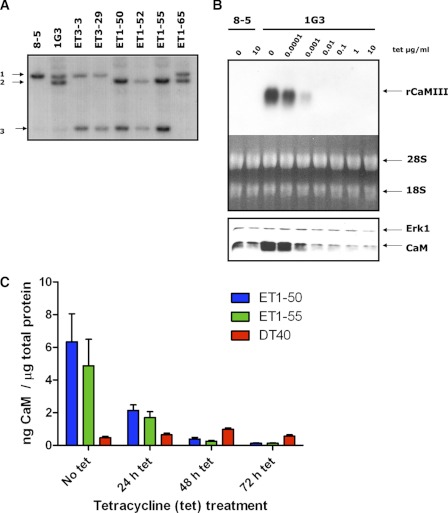
In chicken, two CaM genes (CaMI and CaMII) are expressed and both are translated into the same CaM protein, which is identical to all known vertebrate CaMs. For targeting the first allele of the chicken CaMI gene in the cell line DT40 8-5, which has both CaMII alleles deleted (13), a plasmid construct was prepared containing 2.2-kb 5′-flanking region and 1.8-kb chicken CaMI 3′-flanking region (see supplemental Materials and Methods and Fig. S1). Homologous recombination with chicken genomic DNA interrupted exon 3, which prevented the expression of functional chicken CaMI. The tetracycline transactivator was introduced into the cells by cotransfection. Two clones (1G3 and 1B3) were found to exhibit the correct genomic status (CaMII−/−, CaMI−/+ rat CaMIII, shown for 1G3 in Fig. 1A). These cells expressed much higher levels of CaM as compared with the levels of CaM in the parental cell line 8-5 in the absence of tetracycline (Fig. 1B, bottom).
FIGURE 1.
Generation and analysis of cell lines with both endogeneous CaM genes deleted and expressing repressible CaM. A, Southern blot of EcoR1-restricted genomic DNA from various cell lines. The hybridization probe was a 1.8-kb fragment of the 5′-flanking region (Nhe1/Xho1 fragment) of the chicken CaMI gene used for homologous recombination. Cell line 8-5 (CaMII −/−) has both CaMII alleles deleted but intact CaMI alleles. 1G3 (CaMII−/−, CaMI−/+rCaMII) is the parental cell for the CaMI second allele knock-out. ET1-50, ET1-52, and ET1-55 are cell lines with both CaMI alleles inactivated. Cell lines ET3-3 and ET3-29 still contain one unchanged CaMI allele as the already inactivated CaM I allele was targeted. Arrows: 1, CaMI (both alleles); 2, CaMI allele replaced by rCaMIII; 3, CaMI allele interrupted with the Zeocin casette. The ET1-65 cell line did not undergo recombination at the CaMI locus. B, top panel: Northern blot of RNA isolated from cell lines 8-5 and 1G3 after 48 h of treatment with the indicated tetracycline (tet) concentrations (μg/ml). As a loading control, total RNA was stained with ethidium bromide (bottom). Lower panel: detection of CaM protein by Western blotting. Erk1 immunodetection served as loading control. C, quantitation of CaM expression in wt and the CaM repressible cell lines ET1-50 and ET1-55 at various time points after treatment with tetracycline. Values represent mean of three experiments + S.E.
The 1G3 cell line was further used to eliminate the remaining chicken CaMI allele. The cells were transfected with the same construct as used to knock out the first CaMI allele but with a Zeocin resistance cassette and without the rat CaMIII cassette (supplemental Fig. S1). Southern blot analysis showed that three of the newly generated clones (ET1-50, ET1-52, and ET1-55) had both CaMI alleles disrupted (Fig. 1A). Repression of CaM expression was analyzed by growing the cells under various tetracycline concentrations. Northern blot analysis showed that after 48 h of tetracycline treatment, rat CaMIII transcripts were no longer detectable when at least 0.01 μg/ml tetracycline or more was applied (Fig. 1B, top; shown for clone 1G3). The level of the remaining CaM (Fig. 1B, bottom), originating from the second allele of chicken CaMI after down-regulation of rat CaMIII, was sufficient to allow cell growth.
Time Course of Ectopic CaM Expression in CaM Knock-out Cell Lines and Cell Growth
Both ET1-50 and ET1-55 cell lines showed markedly elevated CaM levels due to ectopic expression as compared with DT40 wild type cells (Fig. 1C). CaM concentrations were 13.5 times higher in ET1-50 cells and 10.4 times higher in ET1-55 cells as compared with DT40 WT cells. However, in both cell lines, the CaM expression levels were 4.4 times lower than in WT cells 72 h after addition of tetracycline.
CaM Is Essential for DT40 Cell Proliferation
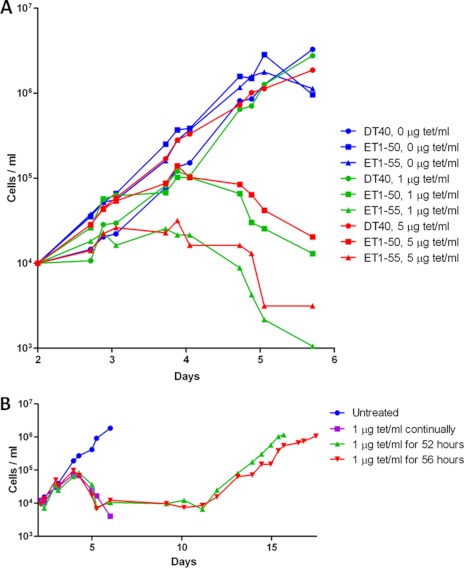
A marked reduction in cell proliferation was noticed after addition of tetracycline in the two knock-out cell lines. Growth of ET1-55 cells stopped 75 h after tetracycline addition and growth of ET1-50 cells after 92 h (Fig. 2A). The latter cell line expresses higher levels of CaM without tetracycline repression and had a slower decline in CaM epression following tetracycline repression (Fig. 1C). These experiments were done with 1 and 5 μg/ml tetracycline, which are nontoxic concentrations as shown for the DT40 WT cell line, which grew similar to untreated cells in the presence of 5 μg/ml tetracycline (Fig. 2A). These data demonstrate that CaM is essential for growth and survival of DT40 cells. Cell growth could be recovered by removing tetracycline from the medium 2 to 3 days after the start of the tetracycline treatment (Fig. 2B).
FIGURE 2.
Calmodulin is essential for vertebrate cell proliferation. A, cells were seeded at 104 cells/ml and grown under standard conditions with different tetracycline (tet) concentrations. CaM expression was repressed by tetracycline for a 48 h time period before cellular growth was measured. The figure shows one of three experiments with similar results. B, growth curves of cell cultures grown in the presence of tetracycline (1 μg/mg) during the indicated time period followed by growth without tetracycline. Experimental conditions were as described in A. Cell counting for continuous tetracycline treatment was stopped when the cells were all dead (day 9) and for untreated cells when high density was reached (day 6).
HA-CaMwt Supports Cell Growth in Absence of Endogeneous CaM
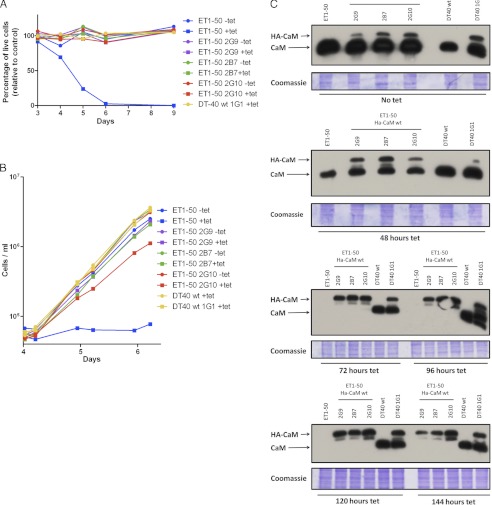
To study the significance of CaM for cell growth and survival, we ectopically expressed HA-tagged CaMwt and CaM mutants in the above described ET1-50 knock-out cells. After the tetracycline-dependent down-regulation of CaM expression, these cells relied completely on HA-CaM (Fig. 3A). This experimental strategy allows the stable expression of HA-CaM mutants independently of their ability to support growth, as CaMwt is present as long as its synthesis was not stopped by adding tetracycline. The HA tag allows differentiation between tetracycline-dependent CaM and mutant HA-CaM, as the latter runs slower on SDS gels and may further be detected independently with an anti-HA antibody. To compensate for variations in HA-CaM expression, several clones stably expressing HA-CaM at various levels derived from DT40 and ET1-50 cells were established. Three days after addition of tetracycline, no significant differences in the relative cell survival were observed in any cell line (Fig. 3A). From day 4 to day 7, the relative survival of tetracycline treated ET1-50 cells decreased dramatically, and repression of CaM expression was lethal for ET1-50 cells, as from day 6 on no living cells could be detected (Fig. 3A). In contrast, the survival of cell lines expressing HA-CaM did not decrease during the entire period measured. Furthermore, the relative survival of untreated and tetracycline-treated HA-CaM cell lines were indistinguishable. These data indicate that HA-CaM can support cell survival after repression of CaM synthesis. Moreover, our results provide evidence for the essential survival function of CaM in vertebrate cells, as down-regulation of CaM in ET1-50 led to cell death. DT40 wt cells without inducible CaM expression and wt cells stably expressing HA-CaM showed similar growth and survival characteristics, indicating that neither the HA tag nor the transfection itself had any deleterious effects. In addition to cell survival, cell proliferation of the HA-CaM cell lines was measured from day 4 on, when ET1-50 cells had a significantly reduced survival following repression of CaM (Fig. 3, B and C). The three HA-CaM cell lines had a similar doubling time as ET1-50 except that ET1-50 2G10 grew with a slightly increased doubling time in the presence of tetracycline. In contrast, ET1-50 cells did not proliferate 96 h after addition of tetracycline. This is consistent with the increased cell death following CaM repression (Fig. 3A).
FIGURE 3.
HA-CaMwt supports cell growth in the absence of endogeneous CaM. A, ET1-50 cells and three ET1-50-derived cell lines expressing HA-CaMwt (2G9, 2G10, and 2B7) were grown with or without tetracycline for 3 days. Additionally, DT40 wt cells with (clone 1G1) or without HA-CaM expression were grown with tetracycline. Cell survival was estimated using flow cytometry. Values are given as % of the live cells in the DT40 population. B, growth of cell lines relying on HA-CaMwt. The same cell cultures, which were analyzed for survival measurements shown in A were used to establish growth curves starting 4 days after tetracycline (tet) treatment. One of two experiments with similar results is shown. C, tetracycline-dependent down-regulation of CaM expression in HA-CaMwt cell lines. Protein samples were obtained from cell cultures without tetracycline treatment and at the indicated time after tetracycline addition. Western blot analysis was performed using an anti-CaM antibody. Coomassie Blue-stained postblot gels were used as loading controls.
Forty eight hours after addition of tetracycline, CaM expression in ET1-50 cells and in the three cell lines expressing HA-CaM decreased to a level below the expression level in DT40 wt cells (see Figs. 3C and 1C). After 72 h, untagged CaM was no more detectable in these cell lines as in tetracycline treated ET1-50 cells when they started to die. Thus, cell survival was maintained by HA-CaM. From hereon, the CaM expression level in HA-CaM and non HA-CaM expressing ET1-50 cell lines remained at a non-detectable level. Based on these results cell survival measurements were started at day 3. HA-CaM expression in the three analyzed clones was similar (Fig. 3C), indicating that the slightly reduced proliferation rate of ET1-50 2G10 was not caused by low expression of the HA-tagged CaM. Note that a slightly weaker signal at higher electrophoretic mobility appeared for HA-CaM (Fig. 3C) most likely derived from an alternative in frame start codon in the HA sequence.
HA-CaM Mutants with Altered Calcium Binding Domains Exhibit Various Phenotypes
The importance of Ca2+ binding to CaM for its function in cell proliferation and survival was explored by analyzing cells expressing CaM with either disrupted single or multiple Ca2+ binding sites (see supplemental “Materials and Methods” and Table 1). Conversion of Asp to Ala at position 1 of each Ca2+ binding loop in the EF-hand makes it impossible for CaM to bind Ca2+ (28). Down-regulation of CaM expression was performed in two clones for each mutation with the following controls: DT40wt, HA-CaMwt as well as ET1-50 cells, which do not express HA-CaM. After 4 days of tetracycline treatment, CaM was not detectable by Western blots in any of the cell lines with mutant HA-CaM as shown before for the HA-CaMwt cell lines (Fig. 3C).
TABLE 1.
Plasmids containing HA-CaM variants
| Plasmid name | Changes in CaM protein | Disrupted function |
|---|---|---|
| HA-CaMwt gpt | ||
| HA-CaM1 gpt | D20A | Ca2+ binding to EF-hand 1 |
| HA-CaM2 gpt | D56A | Ca2+ binding to EF-hand 2 |
| HA-CaM3 gpt | D93A | Ca2+ binding to EF-hand 3 |
| HA-CaM4 gpt | D129A | Ca2+ binding to EF-hand 4 |
| HA-CaM1,2 gpt | D20A + D56A | Ca2 + binding to EF-hands 1 + 2 |
| HA-CaM3,4 gpt | D93A + D129A | Ca2 + binding to EF-hands 3 + 4 |
| HA-CaM1,2,3,4 gpt | D20A + D56A + D93A + D129A | Ca2 + binding to EF-hands 1 + 2 + 3 + 4 |
| HA-CaM(K115R) gpt | K115R | Trimethylation of Lys115 |
| HA-CaM(Y99F) gpt | Y99F | Phosphorylation of Tyr99 |
| HA-CaM(Y138F) gpt | Y138F | Phosphorylation of Tyr138 |
| HA-CaM(Y99F/Y138F) gpt | Y99F + Y138F | Phosphorylation of Tyr99 + Tyr138 |
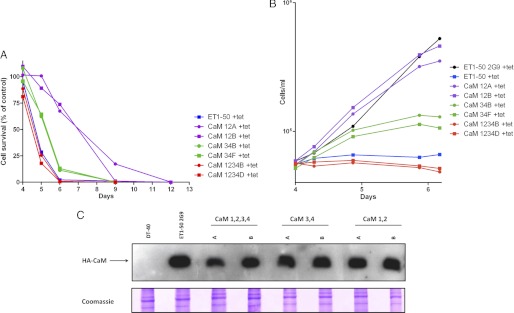
Cell lines relying on single Ca2+ binding site mutant proteins CaM1, CaM2, CaM3, and CaM4 survived and grew similarly as compared with CaMwt (Table 2). Cell lines with mutated Ca2+ binding domain 4 grew slightly slower than wild type but exhibited a similar survival profile (Table 2). Cells relying on HA-CaM1,2 grew exponentially up to 6 days (Fig. 4A), and the survival was similar to the HA-CaMwt cell line 2G9 until 5 days after tetracycline addition, but they started to die from hereon (Fig. 4A). The decrease in cell survival was markedly slower than in tetracycline-treated ET1-50 cells (Fig. 4A), and the CaM1,2 cells reached similar density as tetracycline-treated ET1-50 2G9 cells (Fig. 4B). In contrast, double mutation of sites 3 and 4 showed reduced growth already after 4 days, and the cell survival was reduced markedly at 5 days after tetracycline addition. CaM1,2,3,4 cells showed the same proliferation and survival profile as tetracycline-treated ET1-50 control cells (Fig. 4, A and B). The observed phenotypes of the cell lines CaM1,2,3,4, CaM3,4, and CaM1,2 cannot be explained by differential expression of HA-CaM as similar HA-CaM levels were observed in these cell lines after a 4-day tetracycline treatment (Fig. 4C). In addition, both cell lines with the same HA-CaM mutant exhibited very similar growth and survival characteristics (Fig. 4, A and B).
TABLE 2.
Growth and survival of cells expressing mutant CaM proteins
Growth characteristics of cell lines expressing individually mutated Ca2+ binding sites 1, 2, 3, 4, CaMTyr99/138 phosphorylation sites, and Lys115 trimethylation site as well as control cell lines are shown. Data represent average values for two clones (two individual experiments for DT40 and ET1-50) ± S.E. All cell lines except DT40 and ET1-50 were treated with 1 μg/ml tetracycline. dt, average doubling time in hours between day 4 and 6; s3, s6, and s9, percentage of live cells relative to DT40 wt cells measured at days 3, 6, and 9, respectively.
| Cell line | DT40 | ET1-50 | CaMwt | CaM1 | CaM2 | CaM3 | CaM4 | CaM(Y99F) | CaM(Y138F) | CaM(Y99F/ Y138F) | CaM(K115R) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Average doubling times without tetracycline treatment | |||||||||||
| dt | 10.2 ± 0.7 | 10.7 ± 0.3 | 12.1 ± 0.0 | 11.3 ± 0.1 | 13.0 ± 0.0 | 13.1 ± 0.5 | 14.0 ± 0.2 | 10.7 ± 0.3 | 12.2 ± 1.6 | 11.0 ± 0.6 | 10.7 ± 0.8 |
| Percentage of live cells relative to DT40 cells | |||||||||||
| s3 | 100 | 89.8 ± 1.3 | 92.3 ± 2.5 | 89.7 ± 0.5 | 96.8 ± 1.1 | 94.0 ± 2.9 | 92.9 ± 0.9 | 87.2 ± 2.8 | 91.4 ± 3.0 | 94.3 ± 1.1 | 99.4 ± 0.9 |
| s6 | 100 | 88.8 ± 3.7 | 83.7 ± 8.6 | 91.9 ± 2.0 | 97.6 ± 0.3 | 96.5 ± 4.5 | 103.0 ± 3.4 | 92.8 ± 2.8 | 89.9 ± 2.5 | 91.6 ± 0.3 | 92.7 ± 0.9 |
| s9 | 100 | 85.1 ± 4.7 | 91.4 ± 1.1 | 87.9 ± 2.0 | 100.0 ± 0.5 | 99.2 ± 0.0 | 87.0 ± 5.3 | 93.4 ± 0.6 | 85.1 ± 2.9 | 91.0 ± 1.5 | 98.3 ± 0.4 |
FIGURE 4.
HA-CaM mutants with altered calcium binding domains exhibit various phenotypes. A, cell survival. ET1-50 cells expressing mutant HA-CaM were grown in the presence of tetracycline (tet). The parental ET1-50 cells and DT40 wt cells grown in the presence of tetracycline served as controls. Survival was analyzed using flow cytometry. Values are given as % of the live cells in the ET1-50 2G9 population. B, cell growth. Cells were grown with or without 1 μg/ml tetracycline for 4 days. Then, the cell densities were adjusted to 0.5 × 105/ml, and the cells were counted for the following 2 days every 10–12 h. C, HA-CaM expression in the CaM mutant cell lines CaM1,2; CaM3,4; CaM1,2,3,4 after 4 days of tetracycline treatment. DT40 and ET1-50 2G9 and HA-CaMwt served as controls. Western blot analysis was performed using an HA antibody. Coomassie Blue-stained postblot gels were used as loading controls.
As CaM has been associated with the regulation of the cell cycle (29), we examined whether a lack of CaM or replacement of CaM with various mutants unable to bind Ca2+ at either two or four Ca2+ binding sites affected the cell cycle distribution. No significant alteration of the cell cycle profile was observed in either ET1-50 cells during CaM down-regulation or representative cell lines expressing solely HA-CaMwt, HA-CaM1,2, CaM3,4, and HA-CaM1,2,3,4 (supplemental Fig. S2).
HA-CaM Mutants with Altered Tyrosine Phosphorylation Sites and Trimethylation Site Show No Alterations in Survival and Growth Characteristics
As tyrosine phosphorylation of CaM has been shown to be important for some functions of CaM (22, 30), we substituted Tyr99 and Tyr138, respectively, to Phe both as single mutations and in combination. Initial experiments showed that CaM phosphorylated at Tyr99 is present in DT40 cells (supplemental Fig. S3A). All cell lines expressing Tyr phosphorylation site mutant proteins grew with similar characteristics as compared with the parental cell line, indicating that phosphorylation of Tyr99 and/or Tyr138 is not required for growth and survival of DT40 cells (Table 2). Mass spectrometry confirmed the presence of trimethyl Lys in the CaM protein purified from DT40 cells (supplemental Fig. S3B). The mass per charge (m/z) of 2401.111 Da corresponding to the peptide HVMTNLGEK(CH3)3LTDEEVDEMIR was found in CaM from DT40 cells but not in recombinant vertebrate CaM produced in Escherichia coli. Both recombinant and DT40-derived CaM contain a peptide with the m/z of 2359.079 corresponding to the same peptide without trimethylated Lys115. Two cell lines stably expressing HA-tagged CaM(K115R) were studied following repression of CaMwt. Cell proliferation and survival of these cell lines exclusively relying on CaM(K115R) were comparable with cell lines expressing HA-CaMwt, indicating that this modfication is not essential for DT40 cell survival (Table 2). As Lys115 may be a site of ubiquitination, and methylation may decrease the susceptibility of CaM for proteasomal degradation (31), we tested the stability of HA-CaM(K115R) compared with HA-CaMwt by pulse-chase labeling and found no difference (supplemental Fig. S4).
DISCUSSION
CaM is considered a central and essential mediator of Ca2+ signals in the regulation of numerous cellular activities of all eukaryotic cells (reviewed in Ref. 32). Extensive knowledge about the structure of CaM and its interaction with target proteins has been generated mostly by in vitro approaches. However, investigations of the role of CaM in vivo by genetic means have so far been restricted to lower eukaryotic organisms (reviewed in Ref. 13). Chicken DT40 is a well suited cell line for CaM deletion studies in vertebrate cells because chicken cells have only two genes encoding CaM, which makes CaM gene disruptions easier than e.g. in human cells, which, like all mammalian cells, express an identical CaM protein from three distinct genes (14). In addition, DT40 cells allow homologuous recombination with relatively high frequency (reviewed in Ref. 33). In this study, we describe for the first time the establishment of a vertebrate genetic system that allows the expression of ectopic wild type and mutant CaM in the absence of endogeneous CaM. We generated DT40 cell lines with both CaM genes disrupted and replaced by a repressible CaM transgene. Stable transfection of HA-tagged wild type and mutant CaM genes by random integration into the genome allowed us to test their significance for cell growth and survival. We showed that HA-tagged CaM can support cell growth in several cell lines even though CaM expression varied among different clones and was in most cell lines below the levels found in DT40 wt cells.
Our data demonstrate that cell growth and survival is supported in cell lines expressing a CaM version with a single Ca2+ binding site disrupted. A more severe phenotype of the sites 3 and 4 double mutation compared with the sites 1 and 2 double mutation was observed indicating that the C-terminal lobe is more important than the N-terminal lobe for the essential function of CaM in cell growth and survival. Interestingly, mutations in both site 1 and site 2 did not allow survival, even though it helps cells to reach higher cell densities and decelerates cell death compared with cells devoid of CaM. On the other hand, CaM with nonfunctional sites 3 and 4 could only support growth in an initial phase, and the survival performance was only slightly better than the one of a mutant with all four sites disrupted or control cells not expressing CaM after tetracycline down-regulation. Finally, we found that cells expressing CaM without the ability to bind Ca2+ show the same growth and survival characteristics as cells devoid of CaM. This indicates that although CaM is believed to have Ca2+-independent functions, these do not seem to be sufficient for allowing cellular growth and survival of DT40 cells. These results indicate that CaM targets involved in the essential functions of CaM are dependent primarily on the functionality of the C-terminal lobe, which has a higher Ca2+ affinity (reviewed in Ref. 34). Future experiments combining inactivation of Ca2+ binding sites from both N- and C-terminal lobes will further elucidate the importance of Ca2+ binding to CaM for cell growth and survival.
Different roles of Ca2+ binding to CaM in different organisms have been reported. In S. cerevisiae, disruption of all four Ca2+ binding sites of CaM did not have a significant effect on growth (20). In contrast to S. cerevisiae, CaM in A. nidulans and S. pombe requires high-affinity Ca2+ binding to support growth (21, 35). As shown by Moser et al. (21) in S. pombe, a mutant CaM protein with only an intact Ca2+ binding site 3 and the mutant protein with alterations in all four Ca2+ binding sites do not support growth, but all other mutant S. pombe CaMs with one or more intact Ca2+ binding domain allow growth at 21 °C. However, the temperature sensitivity of S. pombe strains relying on different forms of mutant CaM proteins is different (21). Based on the temperature sensitivity, Moser et al. (21) concluded that Ca2+ binding site 2 is more important than site 1, which is more important than site 4, which is more important than site 3 of S. pombe CaM. Our results with chicken cells differ markedly from the data obtained from the above mentioned species. This indicates that the basic functions of CaM in supporting cell growth and survival may be distinct in different organisms. CaM appears to control multiple processes connected to the cell cycle, through proteins such as calcineurin and CaM kinase II (reviewed in Refs. 29 and 36–39), or components of the cell cycle machinery, such as cyclin E1 (reviewed in Refs. 39–41). Down-regulation of CaM in our chicken cell system did not predominantly affect specific cell cycle phases. This may indicate that CaM depletion or replacement by mutant proteins deficient in Ca2+ binding affect multiple cell cycle phases or that the observed growth and death phenotypes are not directly related to cell cycle control mechanism.
The phosphorylation of CaM at serine/threonine and tyrosine residues by a variety of protein kinases has been described. It has been shown that these posttranslational modifications affect the regulatory properties of CaM (reviewed in Ref. 22). Substitution of the two tyrosine phosphorylation sites in CaM (Tyr99 and Tyr138) individually and in combination by Phe did however not demonstrate any impaired growth or viability of the DT40 cells in our experimental setup. Phospho-Tyr99-CaM appears to be present in the cells used in the present study suggesting that Tyr-phosphorylated CaM has other functions than supporting basic cell growth and survival, e.g. signal transduction after cell stimulation by growth factors. Another well known posttranslational and conserved modification of CaM is trimethylation at lysine 115, which has been shown to affect CaM targets differentially (25, 42) and has been suggested to prevent ubiquitination as well as degradation of CaM (31). Similar to the CaM mutations, which prevent tyrosine phosphorylation, replacing Lys115 by Arg has no obvious effect on growth and viability of DT40 cells, and it does not affect the turnover of CaM under our experimental conditions. Work to identify specific targets of posttranslationally modified CaM using proteomic approaches is in progress.
The novel CaM knock-out system in DT40 cells offers a variety of novel possibilities and advantages over existing approaches to study the role of CaM in vivo. 1) CaM expression can be regulated from a level of 13.5-fold over the wild type to close to zero to analyze the dependence of cellular processes on the level of CaM by the addition of various concentrations of tetracycline and by varying the time period of tetracycline treatment; 2) CaM can be replaced by a mutant CaM protein to study structure-function relationships and interaction with binding partners, which can be endogenously or ectopically expressed in DT40 cells in the absence of endogenous CaM. Recent work on the interaction between CaM and the EGF receptor has shown the usfulness of our experimental system (43); 3) a major advantage of the system is that CaM inhibitors, which may have unspecific side effects can be avoided; 4) this genetic system does not rely on only partial downreguation of CaM, achieved by siRNA, which in itself is an especially challenging task for CaM, as an identical protein is encoded by multiple genes in vertebrates.
Supplementary Material
Acknowledgment
Casper Hølmkjær and Anne Møller are acknowledged for excellent technical work. Dr. Pernille Winding Gojkovic is acknowledged for advice on DT40 cell work.
This work was supported by grants from the Danish Research Council, the Lundbeeck Foundation, AP Møller Fonden, Dagmar Marshall Fonden, Willumsen Fonden, Danielsen Fonden, and Wedell Wedellsborg Fonden, Frænkels Mindefond, Hansen Fonden (to M. W. B.).

This article contains supplemental “Materials and Methods,” Figs. S1–S4, and additional references.
- CaM
- calmodulin.
REFERENCES
- 1. Chin D., Means A. R. (2000) Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 10, 322–328 [DOI] [PubMed] [Google Scholar]
- 2. Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 3. O'Day D. H. (2003) CaMBOT: Profiling and characterizing calmodulin-binding proteins. Cell. Signal. 15, 347–354 [DOI] [PubMed] [Google Scholar]
- 4. Hook S. S., Means A. R. (2001) Ca2+/CaM-dependent kinases: From activation to function. Ann. Rev. Pharmacol. Toxicol. 41, 471–505 [DOI] [PubMed] [Google Scholar]
- 5. Deb T. B., Coticchia C. M., Dickson R. B. (2004) Calmodulin-mediated activation of Akt regulates survival of c-Myc-overexpressing mouse mammary carcinoma cells. J. Biol. Chem. 279, 38903–38911 [DOI] [PubMed] [Google Scholar]
- 6. Joyal J. L., Burks D. J., Pons S., Matter W. F., Vlahos C. J., White M. F., Sacks D. B. (1997) Calmodulin activates phosphatidylinositol 3-kinase. J. Biol. Chem. 272, 28183–28186 [DOI] [PubMed] [Google Scholar]
- 7. Klee C. B., Ren H., Wang X. (1998) Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273, 13367–13370 [DOI] [PubMed] [Google Scholar]
- 8. Fischer R., Julsgart J., Berchtold M. W. (1998) High affinity calmodulin target sequence in the signaling molecule PI 3-kinase. FEBS Lett. 425, 175–177 [DOI] [PubMed] [Google Scholar]
- 9. Briggs M. W., Sacks D. B. (2003) IQGAP proteins are integral components of cytoskeletal regulation. EMBO reports 4, 571–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farnsworth C. L., Freshney N. W., Rosen L. B., Ghosh A., Greenberg M. E., Feig L. A. (1995) Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature 376, 524–527 [DOI] [PubMed] [Google Scholar]
- 11. San José E., Benguría A., Geller P., Villalobo A. (1992) Calmodulin inhibits the epidermal growth factor receptor tyrosine kinase. J. Biol. Chem. 267, 15237–15245 [PubMed] [Google Scholar]
- 12. Deiss L. P., Feinstein E., Berissi H., Cohen O., Kimchi A. (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 9, 15–30 [DOI] [PubMed] [Google Scholar]
- 13. Schmalzigaug R., Ye Q., Berchtold M. W. (2001) Calmodulin protects cells from death under normal growth conditions and mitogenic starvation but plays a mediating role in cell death upon B-cell receptor stimulation. Immunology 103, 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berchtold M. W., Egli R., Rhyner J. A., Hameister H., Strehler E. E. (1993) Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics 16, 461–465 [DOI] [PubMed] [Google Scholar]
- 15. Jurado L. A., Chockalingam P. S., Jarrett H. W. (1999) Apocalmodulin. Physiol. Rev. 79, 661–682 [DOI] [PubMed] [Google Scholar]
- 16. Dagher R., Peng S., Gioria S., Fève M., Zeniou M., Zimmermann M., Pigault C., Haiech J., Kilhoffer M. C. (2011) A general strategy to characterize calmodulin-calcium complexes involved in CaM target recognition: DAPK and EGFR calmodulin binding domains interact with different calmodulin-calcium complexes. Biochim. Biophys. Acta 1813, 1059–1067 [DOI] [PubMed] [Google Scholar]
- 17. Hoeflich K. P., Ikura M. (2002) Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 108, 739–742 [DOI] [PubMed] [Google Scholar]
- 18. Swindells M. B., Ikura M. (1996) Pre-formation of the semi-open conformation by the apocalmodulin C-terminal domain and implications binding IQ motifs. Nat. Struct. Biol. 3, 501–504 [DOI] [PubMed] [Google Scholar]
- 19. Maier L. S., Bers D. M. (2002) Calcium, calmodulin, and calcium-calmodulin kinase II: Heartbeat to heartbeat and beyond. J. Mol. Cell. Cardiol. 34, 919–939 [DOI] [PubMed] [Google Scholar]
- 20. Geiser J. R., van Tuinen D., Brockerhoff S. E., Neff M. M., Davis T. N. (1991) Can calmodulin function without binding calcium? Cell 65, 949–959 [DOI] [PubMed] [Google Scholar]
- 21. Moser M. J., Lee S. Y., Klevit R. E., Davis T. N. (1995) Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. J. Biol. Chem. 270, 20643–20652 [DOI] [PubMed] [Google Scholar]
- 22. Benaim G., Villalobo A. (2002) Phosphorylation of calmodulin. Functional implications. Eur. J. Biochem. 269, 3619–3631 [DOI] [PubMed] [Google Scholar]
- 23. Magnani R., Dirk L. M., Trievel R. C., Houtz R. L. (2010) Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1, 43. [DOI] [PubMed] [Google Scholar]
- 24. Klee C. B., Vanaman T. C. (1982) Calmodulin. Adv. Protein Chem. 35, 213–321 [DOI] [PubMed] [Google Scholar]
- 25. Harding S. A., Oh S. H., Roberts D. M. (1997) Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16, 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. la Cour J. M., Mollerup J., Winding P., Tarabykina S., Sehested M., Berchtold M. W. (2003) Up-regulation of ALG-2 in hepatomas and lung cancer tissue. Am. J. Pathol. 163, 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tarabykina S., Møller A. L., Durussel I., Cox J., Berchtold M. W. (2000) Two forms of the apoptosis-linked protein ALG-2 with different Ca2+ affinities and target recognition. J. Biol. Chem. 275, 10514–10518 [DOI] [PubMed] [Google Scholar]
- 28. Xia Y., Tsai A. L., Berka V., Zweier J. L. (1998) Superoxide generation from endothelial nitric-oxide synthase. A Ca2+/calmodulin-dependent and tetrahydrobiopterin regulatory process. J. Biol. Chem. 273, 25804–25808 [DOI] [PubMed] [Google Scholar]
- 29. Kahl C. R., Means A. R. (2003) Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr. Rev. 24, 719–736 [DOI] [PubMed] [Google Scholar]
- 30. Salas V., Sánchez-Torres J., Cusidó-Hita D. M., García-Marchan Y., Sojo F., Benaim G., Villalobo A. (2005) Characterization of tyrosine phosphorylation-defective calmodulin mutants. Protein Expr. Purif. 41, 384–392 [DOI] [PubMed] [Google Scholar]
- 31. Gregori L., Marriott D., West C. M., Chau V. (1985) Specific recognition of calmodulin from Dictyostelium discoideum by the ATP, ubiquitin-dependent degradative pathway. J. Biol. Chem. 260, 5232–5235 [PubMed] [Google Scholar]
- 32. Berridge M. J., Bootman M. D., Roderick H. L. (2003) Calcium signaling: Dynamics, homeostasis, and remodeling. Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 33. Winding P., Berchtold M. W. (2001) The chicken B cell line DT40: A novel tool for gene disruption experiments. J. Immunol. Methods 249, 1–16 [DOI] [PubMed] [Google Scholar]
- 34. Meyers M. B., Zamparelli C., Verzili D., Dicker A. P., Blanck T. J., Chiancone E. (1995) Calcium-dependent translocation of sorcin to membranes: Functional relevance in contractile tissue. FEBS Lett. 357, 230–234 [DOI] [PubMed] [Google Scholar]
- 35. Joseph J. D., Means A. R. (2002) Calcium binding is required for calmodulin function in Aspergillus nidulans. Eukaryot. Cell 1, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu K. P., Means A. R. (1993) Regulation of the cell cycle by calcium and calmodulin. Endocr. Rev. 14, 40–58 [DOI] [PubMed] [Google Scholar]
- 37. Means A. R. (1994) Calcium, calmodulin, and cell cycle regulation. FEBS Lett. 347, 1–4 [DOI] [PubMed] [Google Scholar]
- 38. Takuwa N., Zhou W., Takuwa Y. (1995) Calcium, calmodulin, and cell cycle progression. Cell. Signal. 7, 93–104 [DOI] [PubMed] [Google Scholar]
- 39. Machaca K. (2010) Ca2+ signaling, genes, and the cell cycle. Cell Calcium 48, 243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi J., Husain M. (2006) Calmodulin-mediated cell cycle regulation: New mechanisms for old observations. Cell Cycle 5, 2183–2186 [DOI] [PubMed] [Google Scholar]
- 41. Takuwa N., Zhou W., Kumada M., Takuwa Y. (1993) Ca2+-dependent stimulation of retinoblastoma gene product phosphorylation and p34cdc2 kinase activation in serum-stimulated human fibroblasts. J. Biol. Chem. 268, 138–145 [PubMed] [Google Scholar]
- 42. Roberts D. M., Irving-Bell R. J. (1996) Effect of weather conditions on the flight activity of Nigerian blackflies (Diptera: Simuliidae). Med. Vet. Entomol. 10, 137–144 [DOI] [PubMed] [Google Scholar]
- 43. Li H., Panina S., Kaur A., Ruano M. J., Sánchez-González P., la Cour J. M., Stephan A., Olesen U. H., Berchtold M. W., Villalobo A. (2012) Regulation of the ligand-dependent activation of the epidermal growth factor receptor by calmodulin. J. Biol. Chem. 287, 3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.