Background: DJ-1, a protein involved in PD, protects neurons by acting as an oxidative stress sensor.
Results: Through adduct formation on DJ-1 cysteines, DAQs induce both structural perturbations and uncoupling of the sensor function.
Conclusion: Cys-53 is the most reactive, but Cys-106 modification induces the most severe effects.
Significance: A correlation between DJ-1 DAQ-dependent impairment and the degeneration of dopaminergic neurons observed in PD is suggested.
Keywords: Dopamine, Molecular Dynamics, NMR, Parkinson Disease, Quinones, Loss of Function, Structure-Activity Relationship
Abstract
The physiological role of DJ-1, a protein involved in familial Parkinson disease is still controversial. One of the hypotheses proposed indicates a sensor role for oxidative stress, through oxidation of a conserved cysteine residue (Cys-106). The association of DJ-1 mutations with Parkinson disease suggests a loss of function, specific to dopaminergic neurons. Under oxidative conditions, highly reactive dopamine quinones (DAQs) can be produced, which can modify cysteine residues. In cellular models, DJ-1 was found covalently modified by dopamine. We analyzed the structural modifications induced on human DJ-1 by DAQs in vitro. We described the structural perturbations induced by DAQ adduct formation on each of the three cysteine residues of DJ-1 using specific mutants. Cys-53 is the most reactive residue and forms a covalent dimer also in SH-SY5Y DJ-1-transfected cells, but modification of Cys-106 induces the most severe structural perturbations; Cys-46 is not reactive. The relevance of these covalent modifications to the several functions ascribed to DJ-1 is discussed in the context of the cell response to a dopamine-derived oxidative insult.
Introduction
Parkinson disease (PD)3 is a progressive movement disorder characterized by nigrostriatal dopaminergic degeneration and by cytoplasmic intraneuronal inclusions known as Lewy bodies (1). PD is the most common neurodegenerative disorder after Alzheimer disease (2). The mean age of onset is around 60 years, although in 5–10% of the cases, the onset is between the age of 20 and 50 (1).
The molecular etiopathogenesis of PD is not understood. Sporadic cases probably originate from a complex interaction between multiple environmental factors and genetic susceptibility. Mitochondrial dysfunction and oxidative stress were initially indicated as factors in PD pathogenesis because exposure to environmental toxins, which inhibit mitochondrial respiration and promote production of reactive oxygen species, causes loss of dopaminergic neurons in humans and animal models (3).
Although most PD cases are sporadic, several gene mutations leading to familial PD have been identified in recent decades. Monogenic forms of the disease are now reported for 5–10% of PD patients, and at least 13 loci and nine genes are associated with either autosomal dominant or recessive PD (4). Among these is PARK7, a 24-kb gene encoding DJ-1, a 189-amino acid homodimeric protein. Specific mutations of DJ-1 are responsible for a familial, autosomal recessive early onset form of PD (5). It has been suggested that DJ-1 is not involved only in inherited cases but also in the more common sporadic form of the disease; elevated levels of DJ-1 have been observed in the cerebrospinal fluid of sporadic PD patients, leading to the suggestion that DJ-1 could be a biomarker for early sporadic PD (6).
DJ-1 is ubiquitously expressed in both brain and peripheral tissue. It is predominantly a cytosolic protein, but it is also present in the nucleus of several cell types; a fraction of wild-type (WT) DJ-1 has also been shown to localize to the mitochondria (7).
The crystal structure of DJ-1 shows a single flavodoxin-like Rossmann fold domain comprising a six-stranded parallel β-sheet sandwiched by eight α-helices, a β-hairpin on one end, and a three-stranded antiparallel β-sheet on the opposite end (8). Although the well folded, compact structure of DJ-1 has been known since 2003, its physiological role is still controversial. Several functions have been ascribed to DJ-1, among which are a role in transcriptional regulation, cell signaling, and apoptosis (9, 10); a low intrinsic proteolytic activity (due to its ability to act as a latent protease zymogen) (11); and a chaperone function in inhibiting α-synuclein aggregation (12). At present, the most corroborated and investigated function of DJ-1 is its putative neuronal protective role against oxidative stress, although how exactly this function is exerted is not clear (13). Overexpression of DJ-1 has a neuronal cytoprotective effect against oxidative stress (14, 15), whereas DJ-1 deficiency leads to increased oxidative stress-induced cell death, both in culture and in animal models (16, 17). It has also been shown that the pathogenic L166P mutation impairs the neuronal cytoprotective function of DJ-1 by jeopardizing dimer formation and protein stability (18).
The existence of a tight correlation between DJ-1 and mitochondrial dysfunction was initially suggested by the fact that Drosophila flies lacking DJ-1 exhibit increased sensitivity to environmental mitochondrial toxins (17). Aberrant mitochondrial morphology has also been observed in DJ-1-deficient cell lines, cultured neurons, and mouse brains as well as in lymphoblast cells derived from PD patients carrying DJ-1 gene mutations or deletion. It has also been demonstrated that DJ-1-dependent mitochondrial defects contribute to oxidative stress-induced sensitivity to cell death (19).
In PD, dopamine neurons of the substantia nigra pars compacta have been shown to degenerate to a greater extent than other neurons. It has been recently reported that DJ-1 is covalently modified by dopamine (DA), both in rat brain mitochondrial preparations and in neuroblastoma cells (20). DJ-1 as a target of DA covalent modifications may suggest a more direct correlation between DJ-1 and the specific degeneration of dopaminergic neurons in PD. Excessive cytosolic accumulation of DA can result in self-oxidation of the catechol ring to generate reactive oxygen species and electron-deficient DA quinones (DAQs) (21). The highly reactive DAQs can react with cellular nucleophiles, such as the reduced sulfhydryl group on cysteinyl residues (22, 23), generating 5-S-cysteinyl-DA as the major species (24) and leading to inactivation of protein function.
The observed susceptibility of DJ-1 to covalent modifications by DA (20), together with its established role in oxidative stress and mitochondrial dysfunction, suggests that the investigation of the structural perturbations induced on DJ-1 by DAQs may provide valuable insights for the comprehension of the molecular mechanism of PD. Three cysteine residues are present in the amino acid sequence of DJ-1. Cys-46 and Cys-53 are located on two consecutive β-strands that form part of the dimer interface. The two Cys-53 residues belonging to the same dimer are located within 4 Å of each other at the dimer interface. The third residue, Cys-106, is located at the turn between a β-strand and an α-helix with energetically strained backbone torsion angles. Although it is positioned at the bottom of a narrow cleft, Cys-106 is still solvent-accessible (25). The solvent-accessible area of the sulfur atom is 7.72 Å2 in Cys-53 and 4.68 Å2 in Cys-106, whereas the sulfur atom in Cys-46 is completely buried.
In the present work, we analyzed the modifications induced on DJ-1 by DAQs, using different biochemical, biophysical, and computational techniques, and a neuroblastoma cellular model to gain structural information to be correlated to protein functional failure/perturbation. We investigated the reactivity of each cysteine residue toward DA oxidation products, and we further analyzed the specific effects induced by the modification of each cysteine residue on the overall protein structure. The three cysteine residues behave in very different ways, in agreement with their most likely different roles.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human wild-type DJ-1 cDNA was amplified by PCR using the pcDNA3.1/GS-DJ-1 vector, containing the full-length DJ-1 coding region as template (a generous gift of Dr. M. R. Cookson) and synthetic oligonucleotides (Sigma-Genosys) containing the NcoI and XhoI restriction sites. After digestion with the appropriate restriction enzymes, the PCR product was subcloned into the NcoI-XhoI linearized pET28 expression plasmid (Novagen) and introduced into Escherichia coli BL21(DE3) strain. The C53A, C106A, C46S, and C53A/C106A mutants were generated by site-directed mutagenesis using specific oligonucleotides. Overexpression of the proteins was achieved by growing cells in LB medium at 37 °C to an A600 of 0.6 followed by induction with 0.6 mm isopropyl β-d-thiogalactopyranoside for 4–5 h. 15N-Labeled proteins were expressed by growing cells in M9 minimal medium, supplemented with 1 g/liter [15N]ammonium chloride. After sonication and centrifugation, the soluble fraction, containing DJ-1, was subjected to a two-step (70 and 90%) ammonium sulfate precipitation. The pellet was then resuspended; dialyzed against 20 mm Tris-HCl, pH 8.0, 3 mm dithiothreitol (DTT); and purified through a 6-ml Resource Q column (Amersham Biosciences). After purification, wild-type DJ-1 and its mutants were stored at 4 °C in 20 mm Tris-HCl, pH 8.0, 10 mm DTT for no more than 2 weeks. Protein concentration was estimated using the extinction coefficient of the monomeric DJ-1 form, ϵ = 4200 m−1 cm−1.
Nitro Blue Tetrazolium/Glycinate Redox Cycling Staining
Quinone-modified proteins were detected by redox-cycling staining (26). Briefly, the protein samples, separated by SDS-PAGE, were transferred to nitrocellulose membranes at 50 V for 90 min at 4 °C. The membrane was first stained with Ponceau S (0.1% in 5% acetic acid), resulting in a red staining of each protein band present. After washing with water, protein-bound quinonoids were detected by immersing the membrane in a solution of 0.24 mm nitro blue tetrazolium, 2 m potassium glycinate (pH 10.0) for 45 min in the dark, resulting in a blue-purple stain of quinoprotein bands and no staining of other proteins. The reaction was finally blocked by immersing the membrane in a solution of 0.1 m borate buffer, pH 10.0.
Radioactivity Assays
The reactions were performed in a final volume of 20 μl, at 37 °C, in 20 mm phosphate buffer, pH 7.4, in the presence of 150 μm protein and 50 μm 14C-DA (containing 0.05 μCi of radioactivity). The protein/DA ratios were 1:1, 3:2, and 3:1 for the wild-type, single mutants and double mutant, respectively, so that the cysteine/DA ratios were 3:1 in all cases. Cold DA was added when required to obtain the desired final DA concentration. After the addition of 10 units of tyrosinase (Ty) (from mushroom; Sigma), the reaction was carried out for 45 min. The reaction products were separated by 13% SDS-PAGE and detected by autoradiography.
Molecular Dynamics Simulations and Analysis
The x-ray structure with the highest resolution of the preoxidation complex of human DJ-1 was selected for all of the calculations (PDB code 2OR3) (27). MD simulations were performed using the software GROMACS 3.3 (28–31) (see supplemental material). The models of the covalent complexes of DJ-1 with DA were obtained using the human DJ-1 crystal structure covalently bound to DA (PDB code 2OR3) (27). We selected the best geometries obtained from a docking study using the covalent bond constraint tool part of the GOLD program suite (32). Trajectory analysis was performed using GROMACS and the VMD software package (33) and plotted with QTIPLOT and GRACE.
Circular Dichroism
CD measurements were carried out on a JASCO J-715 spectropolarimeter. The CD spectra were acquired and processed using the J-700 software. A HELLMA quartz cuvette with an optical path length of 0.02 cm was used. Thermal stability was assessed following the change of the CD signal at 222 nm while increasing the temperature from 25 to 75 °C at a constant rate of 1 °C/min. All melting curves were recorded using a bandwidth of 2 nm and an integration time of 6 s/point. The CD spectra were recorded using a bandwidth of 2 nm and a time constant of 4 s at a scan speed of 20 nm/min. The signal/noise ratio was improved by accumulating four scans. Spectra were acquired on 150 μm DJ-1 solutions in 20 mm phosphate buffer at pH 7.4. CD data were also recorded on DJ-1 samples after reaction with dopamine in a 3:2 (DJ-1/DA) molar ratio in the presence of Ty at room temperature.
NMR Studies
All NMR experiments were carried out at 25 °C on a Bruker Avance DMX600 spectrometer equipped with a gradient triple resonance probe interfaced with a Red-Hat Linux work station. The spectra were collected using TopSpin 1.3, processed using the NMRPIPE software suite, and analyzed using the software CARA. The NMR samples contained ∼0.5 mm protein in H2O/D2O (90:10, v/v), 20 mm phosphate buffer (pH 7.4), 1 mm DTT. NMR data were also recorded on DJ-1 samples after reaction with dopamine in a 3:2 (DJ-1/DA) molar ratio in the presence of Ty at room temperature.
Cellular Model
Human neuroblastoma SH-SY5Y were obtained from the “Istituto Nazionale per la Ricerca sul Cancro” (Genova, Italy). DMEM/F-12 (1:1), fetal bovine serum, Lipofectamine 2000 reagent, Opti-MEM®I reduced serum medium, and 2.5% trypsin were obtained from Invitrogen.
Western Blot Materials
For Western blot, we used the following: nitrocellulose membrane (Sigma-Aldrich), DJ-1 monoclonal antibody (Stressgen), monoclonal anti-β tubulin (Sigma-Aldrich), horseradish peroxidase-conjugated anti-mouse (Sigma-Aldrich), mouse anti-V5 antibody (Invitrogen), BCA protein assay kit (Thermo Scientific), and ECL plus (GE Healthcare).
Cell Culture and Treatment
SH-SY5Y cells were grown under sterile conditions as monolayer in DMEM/F-12 medium (1:1) supplemented with 10% heat-inactivated fetal bovine serum (FBS) in a 5% CO2 humidified atmosphere at 37 °C. For the experiments on naive cells, cells were seeded in a 100-mm dish and allowed to reach confluence.
Cellular Transfection and Treatments
SH-SY5Y cells were seeded in a 6-well plate at 50% of confluence. The following day, cells were transiently transfected with 2 μg of pcDNA3.1/GS plasmid containing wild-type DJ-1 and the two mutants C53A and C106A, respectively. Transfections were performed using the Lipofectamine 2000 reagent according to the manufacturer's instructions.
Protein Extraction and Western Blotting
Briefly, cells were trypsinized, centrifuged, and resuspended in 100 μl of lysis buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton, and protease inhibitor mixture) to prepare whole-cell lysates and maintained in ice for 30 min. Subsequently, lysates were centrifuged at 13,000 × g for 30 min to remove cell debris, and the supernatant was collected. Protein concentration was determined by BCA method. Lysates were treated with 100 units of tyrosinase, 400 μm dopamine, or both for 40 min. 50 μg of total protein cell lysate were separated in a 13% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membranes were probed with mouse DJ-1 antibody (1:1000; Stressgen) for the naive neuroblastoma cells and with mouse anti-V5 antibody in the case of overexpression. Mouse anti-β tubulin (1:2000) was used as control. The incubation with primary antibody was followed by horseradish peroxidase-conjugated secondary antibody (1:2000) and developed with the ECL (GE Healthcare).
RESULTS
To document the formation of a DA-DJ-1 adduct, the protein was treated with DA in a 1:1 molar ratio in the presence of Ty, and the reaction products were subsequently separated on an SDS-polyacrylamide gel (Fig. 1A). The gel was developed using the redox-cycling staining technique (26). The reaction of WT DJ-1 with DAQs, caused by the simultaneous presence of dopamine and tyrosinase in the reaction mixture, yielded both DJ-1 monomeric and dimeric species covalently bound to quinoid compounds (Fig. 1B).
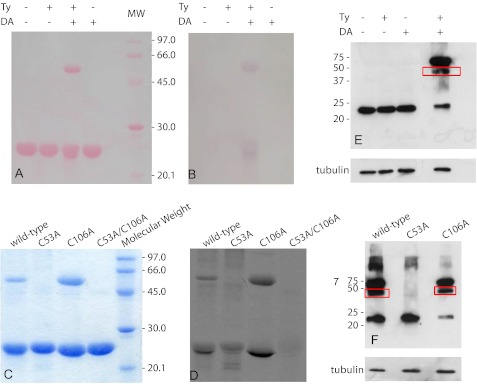
FIGURE 1.
Identification of DAQ adducts formed on DJ-1. WT DJ-1 samples reacted with DA and/or Ty were run on an SDS-polyacrylamide gel. A, Ponceau S staining of the nitrocellulose membrane on which the SDS-polyacrylamide gel was transferred. B, redox-cycling staining assay run on the nitrocellulose membrane reported in A. C, SDS-polyacrylamide gel of WT DJ-1 and mutant protein samples previously exposed to 14C-DA, in the presence of Ty, in a 3:1 cysteine/DA ratio. D, autoradiography obtained from the SDS-polyacrylamide gel reported in C. E, Western blot analysis performed using a monoclonal DJ-1 antibody on endogenous DJ-1 from cell lysates after treatment with DA and/or Ty. F, Western blot analysis performed using a V5 antibody on transfected cells containing WT DJ-1, C53A, or C106A mutants. Cells were lysed and treated with DA and/or Ty before Western blot analysis. Mouse anti-β-tubulin was used as an internal control.
The modifications induced on DJ by DAQs were characterized by electrospray ionization mass spectrometry (MS). A mixture of DJ-1 and DA (1:1 molar ratio), reacted for 30 min in the presence of Ty, was eluted from a reverse phase C4 column. The HPLC analysis of the reaction mixture showed the presence of an additional peak with a different retention time compared with the WT protein (supplemental Fig. S1). MS analysis revealed the presence of different amounts of protein modified by one (+150 Da) or two (+300 Da) DAQs, suggesting that no more than two quinones bind to each DJ-1 monomer.
To identify which residues are modified, and also to seek a possible correlation between individual DAQ adducts and structural perturbations induced, we decided to systematically mutate each cysteine residue. Mutant proteins DJ-1(C53A) and DJ-1(C106A) were cloned and purified. The single mutation of Cys-46 (C46A) produces an unstable protein, which is prone to degradation (25, 34), and the C46S, which we cloned and purified, is more unstable than the other single mutant proteins. Therefore, to obtain information on Cys-46 indirectly, a double mutant, DJ-1(C53A/C106A), was prepared. The difficulty in expressing the single C46A mutant and the low stability of the single C46S mutant strongly suggest that Cys-46 has a more relevant role in structure preservation than the other two.
A radioactivity assay was performed on WT DJ-1 and on the relevant mutants available. Proteins were exposed to 14C-DA, in the presence of Ty, in a 3:1 cysteine/DA ratio. The reaction products were then separated by SDS-PAGE (Fig. 1C) and detected by autoradiography (Fig. 1D). Distinct spots of radioactivity indicated protein targets covalently modified by 14C-DAQs.
We did not observe any significant band corresponding to DAQ conjugates for DJ-1(C53A/C106A), suggesting that Cys-46, which is the least solvent-exposed cysteine, is hardly reactive toward DAQs. On the contrary, Cys-106 and Cys-53 are both reactive toward quinones, although the reaction products are different. Cys-53 seems to be the most susceptible to the attack by the DAQs; the presence of covalent dimers in addition to the modified monomer (Fig. 1D) can be explained by the location of Cys-53 at the dimer interface, close to Cys-53′. The reaction of both Cys-53 and Cys-53′ with the same DAQ molecule would result in the formation of covalent dimeric species bridged by DAQ. Modification of Cys-106 by DAQs preferentially generates high molecular weight species (Fig. 1D). In the case of WT DJ-1, both DAQ-modified dimers and high molecular weight species are formed, indicating that modifications on both residues are simultaneously present.
The DAQ-dependent modifications of DJ-1 and its mutants were also evaluated in a cellular model. Using the procedure previously described for the detection of parkin-DAQ adducts in cell lysates (35), we investigated the effects induced by dopamine oxidation products on endogenous WT DJ-1 using a monoclonal DJ-1 antibody in a Western blot analysis (Fig. 1E). In analogy with what we described previously for the recombinant protein in vitro, we observed that the formation of dimers of endogenous WT DJ-1 occurs only in the presence of both dopamine and tyrosinase.
To confirm the behavior of DJ-1 mutants toward DAQs in a cellular model, we transiently transfected SH-SY5Y cells with a pcDNA3.1/GS plasmid containing either WT DJ-1 or one of the mutants, C53A or C106A. Western blot analysis was performed on cell lysates pretreated with both DA and Ty, using a V5 antibody to visualize only the overexpressed proteins (Fig. 1F). Although the WT and the C106A mutant show a comparable pattern of monomeric and dimeric forms, the mutation of Cys-53 precludes the formation of the dimers with a behavior that reproduces the results obtained in vitro.
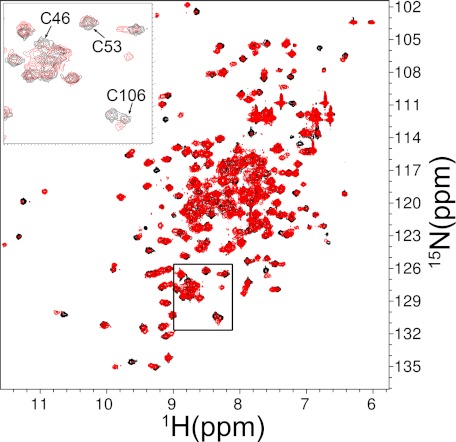
A more detailed structural analysis of the effects of DAQs on DJ-1 was carried out by NMR spectroscopy. 1H-15N HSQC spectra were initially recorded on both WT DJ-1 and the protein treated with DA oxidation products in a 1:1 molar ratio. The protein/DA molar ratio was later reduced to 3:2 to improve the quality of the NMR spectra. A significant decrease of signal intensity in the HSQC spectrum of the modified protein was detected for almost all of the residues assigned. A significant shift of some peaks, the disappearance of others, and the appearance of new ones support a modification of the chemical environment throughout the protein sequence (Fig. 2). These data suggest that the perturbation induced by DAQs is extended to most of the protein structure. The total number of peaks after the reaction is compatible with a heterogeneous sample.
FIGURE 2.
1H-15N HSQC spectra. Shown is WT DJ-1 before (black) and after (red) DAQ exposure in a 3:2 protein/DA ratio. Spectra were recorded at 298 K, 0.5 mm protein concentration, pH 7.2. Inset, enlarged view of the boxed region.
The HSQC spectra of the C53A and C106A mutants showed similar signal dispersion compared with the WT protein, and the peaks were essentially in the same positions. The few exceptions were those resonances arising from residues immediately adjacent to the mutation site, which experience larger changes in chemical shifts. These data strongly indicate that mutation of either cysteine does not lead to significant modifications of the protein folding. Because no quinoprotein formation was detected in the radioactivity assay for the double mutant, no further analysis was performed on this protein.
DAQ modifications on the C53A mutant induced significant perturbations of the spectrum compared with the non-reacted protein, as revealed by the overlap of the HSQC spectra reported in Fig. 3A. In the HSQC spectrum of the modified protein, many of the newly formed peaks display a significant reduction of chemical shift dispersion, indicating that adduct formation on Cys-106 induces also a partial unfolding of the mutant protein. On the contrary, the C106A mutant appears to be much less affected by exposure to DAQs (Fig. 3B).
FIGURE 3.
1H-15N HSQC spectra of DJ-1 mutants. Spectra were recorded before (black) and after (red) DAQ exposure in a 3:2 protein/DA ratio. Spectra were recorded at 298 K, 0.5 mm protein concentration, pH 7.2. A, C53A; B, C106A.
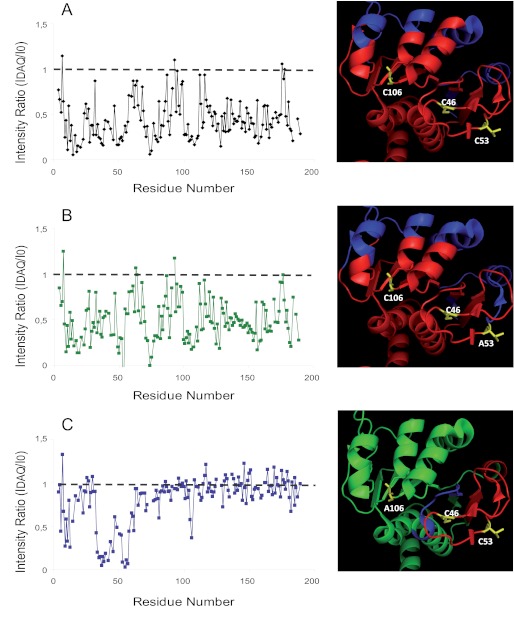
In Fig. 4, the normalized intensity ratio of the HSQC peaks before and after DAQ modification are reported for each DJ-1 residue, both for the WT and for the two mutant proteins. The patterns of signal intensity loss for WT DJ-1 and for the C53A mutant are very similar (Fig. 4, A and B), suggesting that the perturbation of the overall structure induced by DAQs on the two proteins is similar. For the C106A mutant, the few detected significant chemical shift variations and signal intensity reductions can be ascribed to residues around Cys-53 (i.e. the cysteine modified by DAQs) (Fig. 4C).
FIGURE 4.
Effect of the DAQs on the 1H-15N HSQC spectra of DJ-1. Left panels, normalized intensity ratios (R) of the HSQC peaks after and before DAQ modification as a function of residue number. A, WT; B, C53A; C, C106A. Right panels, intensity ratios as in the left panels mapped on the published crystal structure (PDB code 1P5F) of the WT protein. Red, R < 0.5; blue, 0.5 < R < 0.8; green, R > 0.8. A, WT; B, C53A; C, C106A.
The intensity ratios of the HSQC peaks after and before DAQ modifications were mapped on the published crystal structure (PDB code 1P5F) (8) of the WT protein using different colors (from green to red) according to the increasing induced structural perturbations (Fig. 4, right panels). DAQ modification on the WT protein causes a significant alteration of the overall protein, which is not relieved in the C53A mutant. On the contrary, mutation of residue 106 leads to a surprisingly localized perturbation. A possible involvement of Cys-46 was excluded by the radioactivity assays on DJ-1(C53A/C106A), making modifications of only Cys-53 responsible for the effects observed in the spectra of the C106A mutant. A possible reaction of Cys-46, modulated by the presence of Cys-53, is also ruled out by the extremely similar signal intensity loss observed for DAQ-modified WT and C53A mutant proteins (Fig. 4B). Taken together, these data suggest that the most important structural changes are induced by modification of Cys-106.
The structural effects induced on WT DJ-1 by DAQ covalent modification were also investigated using CD spectroscopy. At pH 7.4 and 25 °C, the far-UV CD spectrum of WT DJ-1 is typical of a well folded polypeptide with a substantial amount of helical content (Fig. 5A), as already reported (18). WT DJ-1 was reacted with the oxidation products of DA in a 3:2 molar ratio. The CD spectrum of the DAQ-modified WT protein was virtually unchanged (Fig. 5A), suggesting that the modifications induced by DAQs on the overall secondary structure of DJ-1, if present, are below the detection limits of this technique.
FIGURE 5.
Effect of the DAQs on the CD spectra of DJ-1. A, far-UV CD spectra of WT DJ-1 recorded at 298 K before (continuous line) and after (dashed line) DAQ exposure in a 3:2 protein/DA ratio. Protein concentration was 150 μm in 20 mm phosphate buffer, pH 7.4. B, thermal stability curves recorded before (black) and after (gray) DAQ exposure in a 3:2 protein/DA ratio. Solid line, WT DJ-1; dashed line, C53A; dotted line, C106A.
To assess whether the perturbations observed in the NMR spectra affect the structural stability of DJ-1, we performed thermal denaturation experiments by monitoring the ellipticity at 220 nm while increasing the temperature (1 °C/min) from 25 to 75 °C (Fig. 5B). The WT protein underwent unfolding with a melting temperature (Tm) of 60 °C. Although the melting temperature of the DAQ-modified protein is only slightly lower (Tm = 59 °C) than that of the WT, the transition displays a significant loss of cooperativity (Fig. 5B). For both modified and non-modified proteins, significant precipitation occurred during the unfolding process (data not shown).
The temperature melting curves of DJ-1(C53A) and DJ-1(C106A) at 220 nm are also reported in Fig. 5B. Both mutant proteins exhibit sigmoidal unfolding transitions, as observed for the WT; the observed midpoint temperatures suggest a decreased stability of the C53A mutant (Tm = 57 °C) and an increased stability of the C106A mutant (Tm = 64 °C).
Similar to the WT protein, a decrease in thermal stability was observed for both mutants treated with quinones. Although mutant C106A is only slightly affected by DAQ modification (ΔTm = −1 °C), a significant thermal destabilization is observed for the DAQ-modified C53A mutant (ΔTm = −5 °C) (Fig. 5B). The unfolding transition curve of the latter DAQ-modified mutant is less cooperative than that of the modified WT protein, although the final unfolded state is the same as for the unmodified mutant, at variance with the WT protein.
The sigmoidal thermal unfolding curve of the C46S mutant shows a midpoint temperature of 46 °C, suggesting that this mutation induces a significant decrease in structural stability (see supplemental material). Modifications of the thermal unfolding transition induced by DAQ binding to the protein are not significant when compared with the remarkable intrinsic instability of the unmodified C46S mutant (data not shown).
The thermal stability data are in line with the structural information obtained by NMR. Mutant C106A, which is only slightly perturbed by DAQs, is also extremely resistant to thermal unfolding and largely retains the cooperative behavior of the unmodified protein. On the contrary, the C53A mutant is significantly affected by DAQs and shows remarkable thermal instability, which can be ascribed to the effect of the modification of Cys-106 by DAQ. This perturbation leads to the effects observed in the HSQC spectrum of the DAQ-modified C53A mutant and to the sizable loss in cooperativity in the thermal unfolding, without necessarily affecting the overall protein secondary structure, as observed previously in the CD spectra. Upon DAQ exposure, the presence of Cys-53 is essential for protein covalent dimerization (Fig. 3), which may be responsible for the observed loss in cooperativity during the thermal unfolding process.
Several MD simulations were performed to evaluate the possible structural effects induced by DAQ conjugation to either Cys-53 or Cys-106. The DAQ-modified covalent dimer was also analyzed.
To reduce ambiguous molecular contacts in the Cys-DAQ adducts, the initial positions of the DAQs were selected through a molecular docking study, performed with the GOLD suite package 4.0 (32). The conformation selected for MD is similar to that in the crystal structure (PDB code 2R1T), released in 2008 with no associated reference, in which a single DAQ is conjugated to DJ-1 on Cys-53.
Residue Cys-106 is located in the core of a pocket. The docking returns one energetically favorable conformation, stabilized by two hydrogen bonds between the two hydroxyl groups of dopamine and the backbone of Gly-75 and Asn-76; in addition, the side chain of Asn-76 is involved in a hydrogen bond with the DAQ amino group.
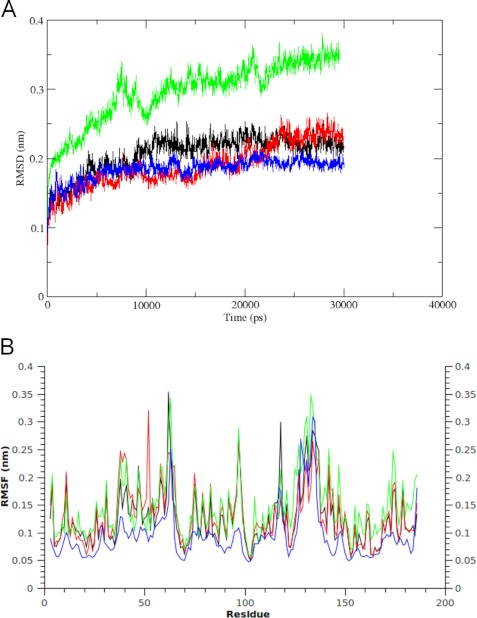
A 30-ns-long molecular dynamics simulation was used to elucidate the behavior of the DJ-1 non-covalent dimer. The backbone root mean square deviation (RMSD) of each step, in comparison with the starting geometry, does not show any significant change after 10 ns (Fig. 6A). The same elements of secondary structure are present during the entire simulation. The β-sheet forming the core of each monomer is strictly conserved, and so is the α-helix (α1) located at the dimer interface. Specifically, the accessible surface area of the three cysteines is unvaried during the simulation. The most relevant root main square fluctuations (RMSF) were observed in specific regions (i.e. 37–42(β2), 58–65(β3), and 127–139(α6)) (Fig. 6B). These regions are directly exposed to the solvent, and the degree of mobility they show does not affect the remarkable stability of the complex. These backbone mobilities are in agreement with other in silico studies, although the published MD simulations on DJ-1 mainly focus on disease-linked mutations (36–38). These results are also confirmed by published NMR relaxation measurements and B-factor values from x-ray crystallography (8, 39).
FIGURE 6.
MD simulations of DAQ-bound DJ-1. A, comparison of the RMSD profiles from the respective initial backbone structures over the simulation time. Black, WT DJ-1; red, DJ-1 with Cys-53 covalently bound to DAQ; green, DJ-1 with Cys-106 covalently bound to DAQ; blue, DAQ-bridged covalent dimer. B, residue-based RMSF during the simulation. The profiles are colored as in A.
Cysteine 53 is located in a long loop involved in protein dimerization, characterized by the presence of short β-sheet regions. The conformational perturbation induced by modification of both Cys-53 and Cys-53′ was followed through the RMSD of the trajectory compared with the starting geometry. After 22 ns, the complex lies in an equilibrium state, as shown in Fig. 6A. The RMSF profile of each residue during the simulation is similar to that of the WT protein, with only few residues showing an increased mobility (Fig. 6B). Specifically, the β2 region shows wider perturbations, compatible with its flexible nature, as already underlined in the simulation of the WT protein. The most important effect is evident on Cys-53, which is directly involved, with an RMSF of 3.2 Å. The essential dynamics analysis shows a notable agreement with what was observed in the HSQC spectra. The filtered trajectory projected along the first eigenvectors shows a residue-based RMSF profile compatible with the decrease of the signal intensity observed by NMR (Fig. 7). The superimposition of the final complex with the WT protein, based on the Rossmann fold portion, shows that the perturbation is strictly localized in the 37–68 region and that it does not affect the protein folding.
FIGURE 7.
Essential analysis of MD trajectories. A, RMSF per residue along the first eigenvector relative to the trajectory of WT DJ-1; DAQ-conjugated on Cys-53 (red) and on Cys-106 (green). B and C, residues showing the widest fluctuations in A (blue) are mapped on the structure of WT DJ-1 (gray). DAQs are highlighted (atoms in stick representation and purple surface). B, DAQ-conjugated on Cys-53; C, DAQ-conjugated on Cys-106.
The distance between the CA of the two Cys-53 residues varies between 8 and 12 Å during the simulation as a consequence of the steric hindrance between the two DAQs and of the intrinsic mobility of the loop where the cysteines are located. A similar effect is observed when only one of the two Cys-53 residues present in the dimeric structure is bound to a DAQ molecule. We suggest that the mobility of region 37–68 plays a key role in the formation of the DAQ-mediated covalent dimer. When bound to the first residue, the DA is still exposed and available to further oxidation. The second cysteine is located in the proximity of the novel DAQ species, justifying the reaction between the thiol group and the C2 carbon. Several studies have reported the different reactivity of the carbon ring, and the double conjugation at C2 and C5 is the second major product after the single conjugation on C5 (27, 40).
A 30-ns-long simulation was performed and compared with that of the WT DJ-1 protein. We observed lower RMSD values of the structure along the trajectory (Fig. 6A), which reaches a plateau after 5 ns. Specifically, a lower mobility is evident in the flexible regions β2, β3, and α6 (residues 38–70 and 126–136) as shown by the RMSF plot (Fig. 6B). The superposition of the 40–60 segments of each monomer in comparison with WT DJ-1, reported in supplemental Fig. S2, shows the conformation adopted by the DAQ in the covalent bridge and the position of the relevant cysteines in WT DJ-1. The covalent bond improves the stability of the dimeric form with no perturbations on the secondary structure and on the protein topology.
The structural effect of DAQ conjugation on Cys-106 was evaluated by a 30-ns-long MD simulation. The global folding is stable during the simulation time; the total amount of secondary structure is unchanged. The backbone RMSD during the simulation mainly shows changes in the first 12 ns, after which only the flexible segments of the protein show fluctuations (Fig. 6A). The fluctuation of the filtered trajectory along the first eigenvector highlights a major effect in comparison with Cys-53, involving several segments of the protein: 36–68, 80–95, 115–148, and 155–187 (Fig. 7). Specifically, also the β-strand forming the core of DJ-1 and representing the most stable portion of each monomer is perturbed (supplemental Figs. S2 and S3). A generalized structural perturbation is also in line with the decrease in the NMR signal intensity (Fig. 4). In these results, the Cys-106-DAQ adduct displays a local reorganization, whereas the secondary structure is substantially preserved.
DISCUSSION
PD is characterized by the specific death of dopaminergic neurons, and oxidative stress is recognized as a factor involved in the etiopathogenesis of the disease. DJ-1 is known to have a role in oxidative stress response (10, 25), although the exact mechanism through which this function is carried out is not clear. Several studies on DJ-1 pathologic mutants suggested that, although its function is not yet fully understood, DJ-1 is implicated in PD through a loss of function mechanism (18, 41). Direct targeting of DJ-1 by products of oxidative stress could account for the observed loss of function of this protein, as has already been shown for other proteins (20). DJ-1 covalently modified by DA oxidation products has been found both in brain mitochondrial preparations and in SH-SY5Y cells (20). The structural and functional effects induced by DA oxidation products could provide the rationale to unravel the selective death of dopaminergic neurons observed in PD.
We focused our efforts on the elucidation of the perturbations induced by electrophilic attack of DAQs on DJ-1 previously observed to occur in vivo (20). We showed that WT DJ-1 reacts with DA derivatives, producing DAQ-conjugated DJ-1 as well as DAQ-modified covalent DJ-1 dimers and high molecular weight species. Two of the three cysteine residues of DJ-1 (Cys-53 and Cys-106) are reactive toward DAQs. Cys-53 is completely accessible to the solvent, whereas Cys-106 is less solvent-exposed. The third cysteine, Cys-46, does not seem to be involved in the reaction with DAQs.
The DAQ covalent modification of Cys-53 does not perturb the structure of DJ-1, as indicated by the very similar thermal stability of the C106A mutant before and after reaction with DAQs. DAQ binding to Cys-53 affects a limited number of residues, almost exclusively those in close proximity to Cys-53 (37–68 region), leading to a substantial preservation of the protein native fold. A significant portion of the protein forms a covalent dimeric species upon reaction with DAQ, through further oxidation of the Cys-53-bound DA and reaction with the free Cys-53 of the opposite monomer. The formation of a covalent dimer between Cys-53 and Cys-46 of two different DJ-1 monomers is quite unlikely, given the poor reactivity of Cys-46 and the significant perturbation of the dimeric native structure it would cause, contrary to what was observed.
The covalent dimer formation through Cys-53 in the presence of DAQs was tested also in a cellular model. In the lysate of SH-SY5Y cells transfected with WT, C106A, or C53A DJ-1 and treated with the DAQ-generating solution, dimers were detected only in the WT protein and in the C106A mutant. This result clearly indicates that the formation of a covalent bond between two DJ-1 monomers of the same dimer is an event that can happen in cells and occurs through bridging of the two Cys-53 residues by a DAQ molecule.
Our results are relevant to the pathology because two distinct studies reported different isoforms of SDS-resistant DJ-1 dimers from extracts of human frontal cortex tissues of post-mortem PD brains (42, 43). The authors did not investigate the nature of the covalent dimeric form, but bridging of the two monomers through Cys-53 by a DAQ molecule, as described here, is a strong possibility. Also, Logan et al. (38) have recently reported that the proposed chaperonic activity of pathologic mutants of DJ-1 can be restored through the formation of a disulfide bond linking the two monomers of a properly engineered V51C DJ-1 mutant. On these grounds, it is conceivable that the DAQ-mediated dimerization process may modulate the chaperonic activity of the WT protein.
DAQ conjugation to Cys-106 leads to the most significant structural destabilization, resulting in a substantial conversion of the protein into DAQ-modified high molecular weight species. The structural perturbation induced by DAQs on the C53A mutant and on the WT protein are quite similar; specifically, the core of the Rossmann fold of DJ-1 is not changed by the modification, and the most affected region comprises the residues close to Cys-106. A partial thermal destabilization also characterizes the DAQ-modified C53A mutant compared with the non-conjugated protein. It is not surprising that the DAQ can be hosted in this relatively internal region of the protein at the dimer interface because the region around Cys-106 shares a strong homology with the cysteine protease family, and consequently the presence of another chemical entity should be well accepted in the region that normally binds the substrate in the homologous proteases.
It has been suggested that aggregation of DJ-1 may contribute to the pathogenesis of several neurodegenerative diseases, including PD. Insoluble aggregates of DJ-1 have been observed in brains of patients with neurodegenerative diseases (44), and a dramatic increase of insoluble DJ-1 has been observed in brains of sporadic PD patients (45). The high molecular weight DAQ-modified oligomers, which we observed upon reaction of dopamine quinones with DJ-1, and specifically with Cys-106, might be the precursors of the aggregates observed in vivo in PD patients. The formation of these oligomers, which eventually become insoluble and precipitate, could rationalize the loss of DJ-1 function, associated with the key residue Cys-106, implicated in PD.
A DJ-1 aggregation hypothesis has been proposed through x-ray crystallography (46), PDB code 3BWE. In this structure, DJ-1 dimers are linearly stacked through phosphate-mediated interactions to form protofilaments, which are then bundled into a filamentous assembly. The overall secondary structure of each DJ-1 dimer in these oligomers/aggregates is preserved quite well, similar to what we observed in vitro, for the DAQ bound to Cys-106 (C53A DJ-1 mutant).
Cys-106 has proven to be tightly implicated in oxidative stress control and mitochondria association (25). The oxidation of the highly conserved Cys-106 to cysteine-sulfinic acid has been proposed as a key signaling mechanism to control DJ-1 mitochondria localization in response to oxidative stress. Specifically, the abilities of DJ-1 mutants to oxidize, translocate to mitochondria in response to oxidation, and protect against toxicity are correlated (25, 47). The formation of cysteine-sulfinic acid would also justify the pI shift from 6.2 to 5.8 observed upon exposure of DJ-1 to oxidative insults. Mutation of Cys-106 (C106A) prevents the formation of oxidized DJ-1 isoforms in intact cells and, as a consequence, impairs the protein's neuroprotective function (25). Strong support for such a mechanism is provided by an abundance of acidic DJ-1 forms in post-mortem brain samples of sporadic PD patients (42). The covalent adduct formed with residue Cys-106 upon reaction with DAQ would most likely compromise DJ-1 function, both inhibiting Cys-106 redox activity and affecting protein structure.
The covalent dimers, which we observed in the C106A mutant upon reaction with DAQs, would preserve the oxidative stress control activity exerted by Cys-106 at least in part. Nevertheless, because they perturb protein unfolding, they would most likely compromise the mitochondria association process.
Most recently, the cytoprotective effects of DJ-1 have been suggested to be mediated by the suppression of ASK1 (apoptosis signal-regulating kinase 1) pathways. Specifically, it has been proposed that DJ-1 binds ASK1 in a Cys-106-dependent manner (34, 48) because mutation of this residue inhibits not only the protein's cytoprotective activity but also its actual binding to ASK1. Although the engineered oxidation-mimicking mutants C106D and C106E did not bind to ASK1, the substitution of Cys-106 with two acidic residues as in the C106DD and C106EE mutants, resulted in binding to ASK1 and in partial cytoprotection. Therefore, the full cytoprotective activity of DJ-1 seems to require a more complete, mixed disulfide-mediated incorporation into the ASK1 signalosome, for which Cys-106 is necessary. It has been suggested that the C106DD mutation only “opens” the conformation of DJ-1 to reveal ASK1 binding site(s), resulting in the observed ASK1 binding and partial cytoprotection (34). Also this functional hypothesis requires Cys-106, which would be severely compromised by DAQ binding. The limited structural alteration that we reported when DAQs bind to Cys-53 would also partially affect DJ-1 activity by modulating it but most likely would not fully compromise its function.
One of the functions accredited to DJ-1 and initially widely endorsed was its ability to act as a chaperone by preventing the aggregation of some proteins, including α-synuclein. Initially, Cys-53 was identified as the key residue for the redox-sensitive chaperone activity of DJ-1 (12). However, this is in conflict with more recent data, which show that oxidation of Cys-106 to sulfinic acid is critical to prevent α-synuclein aggregation (49). The sulfinic Cys-106 form seems to be the only one able to trigger the chaperone activity of DJ-1 because further oxidation leads to partial loss of protein structure and consequent abolition of its chaperone activity (49). Protein inactivation induced by strong oxidation of Cys-106 is caused by partial protein unfolding and the formation of high molecular weight species. These effects strongly resemble the ones induced by DAQ attack on Cys-106 and characterized in this work.
Independent of what the real physiological role of DJ-1 is and of what molecular pathway correlates the protein to PD, the data presented here highlight the delicate role played by residue Cys-106 in DJ-1 under stress conditions induced by DA oxidation products. We described the structural perturbations induced by DAQ adduct formation on each of the three cysteine residues in the sequence of DJ-1. Cys-53 is the most reactive residue, but modification of Cys-106 induces the most severe structural perturbations. Any of the multiple Cys-106-dependent functions that have been ascribed to this protein would most likely be lost if DAQs bind to Cys-106, whereas modifications at Cys-53 might reduce but not eliminate the ability of DJ-1 to control oxidative stress. On the other hand, DAQ binding to Cys-53 leads to the formation of DAQ-modified covalent dimers, which resemble the SDS-resistant DJ-1 oligomers detected in human frontal cortex tissues of post-mortem PD brains (42, 43).
Our findings suggest that oxidative stress, and specifically highly reactive dopamine-derived quinones, are responsible for DJ-1 function impairment, which can lead to loss of cellular control over oxidative stress and to incorrect protein compartmentalization. DJ-1 as a target of dopamine covalent modifications suggests a direct correlation between DJ-1 impairment and the specific degeneration of dopaminergic neurons observed in Parkinson disease.
Supplementary Material
Acknowledgments
We thank Dr. Patrizia Polverino de Laureto for the collection of DAQ-modified WT DJ-1 mass spectra, Dr. Mark R. Cookson for providing the cDNA of WT DJ-1 and of Cys to Ala DJ-1 mutants, Dr. David Eliezer for providing the DJ-1 NMR backbone assignment, and the Molecular Modeling section of the Department of Pharmaceutical Sciences (Padova, Italy) for computational support.
This work was supported by University of Padova Progetto di Ricerca di Ateneo 2008 Grant CPDA 087059 and Progetto di Ricerca di Ateneo 2010 Grant CPDA 103503 and by Ministero Istruzione Università Ricerca, Progetti di Ricerca di Interesse Nazionale, 2008 Grant 2008 5SYP79.

This article contains supplemental Experimental Procedures and Figs. S1–S3.
- PD
- Parkinson disease
- DA
- dopamine
- DAQ
- dopamine quinone
- Ty
- tyrosinase
- PDB
- Protein Data Bank
- MD
- molecular dynamics
- RMSD
- root mean square deviation
- RMSF
- root mean square fluctuation(s).
REFERENCES
- 1. Samii A., Nutt J. G., Ransom B. R. (2004) Parkinson's disease. Lancet 363, 1783–1793 [DOI] [PubMed] [Google Scholar]
- 2. de Lau L. M., Breteler M. M. (2006) Epidemiology of Parkinson's disease. Lancet Neurol. 5, 525–535 [DOI] [PubMed] [Google Scholar]
- 3. Greenamyre J. T., Hastings T. G. (2004) Biomedicine. Parkinson's. Divergent causes, convergent mechanisms. Science 304, 1120–1122 [DOI] [PubMed] [Google Scholar]
- 4. Lesage S., Brice A. (2009) Parkinson's disease. From monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 18, R48–R59 [DOI] [PubMed] [Google Scholar]
- 5. Bonifati V., Rizzu P., van Baren M. J., Schaap O., Breedveld G. J., Krieger E., Dekker M. C., Squitieri F., Ibanez P., Joosse M., van Dongen J. W., Vanacore N., van Swieten J. C., Brice A., Meco G., van Duijn C. M., Oostra B. A., Heutink P. (2003) Mutations in the DJ-1 gene associated with autosomal recessive early onset parkinsonism. Science 299, 256–259 [DOI] [PubMed] [Google Scholar]
- 6. Waragai M., Wei J., Fujita M., Nakai M., Ho G. J., Masliah E., Akatsu H., Yamada T., Hashimoto M. (2006) Increased level of DJ-1 in the cerebrospinal fluids of sporadic Parkinson's disease. Biochem. Biophys. Res. Commun. 345, 967–972 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L., Shimoji M., Thomas B., Moore D. J., Yu S. W., Marupudi N. I., Torp R., Torgner I. A., Ottersen O. P., Dawson T. M., Dawson V. L. (2005) Mitochondrial localization of the Parkinson's disease-related protein DJ-1. Implications for pathogenesis. Hum. Mol. Genet. 14, 2063–2073 [DOI] [PubMed] [Google Scholar]
- 8. Wilson M. A., Collins J. L., Hod Y., Ringe D., Petsko G. A. (2003) The 1.1 Å resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 100, 9256–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu J., Zhong N., Wang H., Elias J. E., Kim C. Y., Woldman I., Pifl C., Gygi S. P., Geula C., Yankner B. A. (2005) The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum. Mol. Genet. 14, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 10. Junn E., Taniguchi H., Jeong B. S., Zhao X., Ichijo H., Mouradian M. M. (2005) Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. U.S.A. 102, 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J., Li L., Chin L. S. (2010) Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum. Mol. Genet. 19, 2395–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shendelman S., Jonason A., Martinat C., Leete T., Abeliovich A. (2004) DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. PLoS Biol. 2, e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dodson M. W., Guo M. (2007) Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson's disease. Curr. Opin. Neurobiol. 17, 331–337 [DOI] [PubMed] [Google Scholar]
- 14. Taira T., Saito Y., Niki T., Iguchi-Ariga S. M., Takahashi K., Ariga H. (2004) DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinat C., Shendelman S., Jonason A., Leete T., Beal M. F., Yang L., Floss T., Abeliovich A. (2004) Sensitivity to oxidative stress in DJ-1-deficient dopamine neurons. An ES-derived cell model of primary Parkinsonism. PLoS Biol. 2, e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim R. H., Smith P. D., Aleyasin H., Hayley S., Mount M. P., Pownall S., Wakeham A., You-Ten A. J., Kalia S. K., Horne P., Westaway D., Lozano A. M., Anisman H., Park D. S., Mak T. W. (2005) Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 102, 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meulener M., Whitworth A. J., Armstrong-Gold C. E., Rizzu P., Heutink P., Wes P. D., Pallanck L. J., Bonini N. M. (2005) Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson's disease. Curr. Biol. 15, 1572–1577 [DOI] [PubMed] [Google Scholar]
- 18. Olzmann J. A., Brown K., Wilkinson K. D., Rees H. D., Huai Q., Ke H., Levey A. I., Li L., Chin L. S. (2004) Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 279, 8506–8515 [DOI] [PubMed] [Google Scholar]
- 19. Irrcher I., Aleyasin H., Seifert E. L., Hewitt S. J., Chhabra S., Phillips M., Lutz A. K., Rousseaux M. W., Bevilacqua L., Jahani-Asl A., Callaghan S., MacLaurin J. G., Winklhofer K. F., Rizzu P., Rippstein P., Kim R. H., Chen C. X., Fon E. A., Slack R. S., Harper M. E., McBride H. M., Mak T. W., Park D. S. (2010) Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 19, 3734–3746 [DOI] [PubMed] [Google Scholar]
- 20. Van Laar V. S., Mishizen A. J., Cascio M., Hastings T. G. (2009) Proteomic identification of dopamine-conjugated proteins from isolated rat brain mitochondria and SH-SY5Y cells. Neurobiol. Dis. 34, 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bisaglia M., Mammi S., Bubacco L. (2007) Kinetic and structural analysis of the early oxidation products of dopamine. Analysis of the interactions with α-synuclein. J. Biol. Chem. 282, 15597–15605 [DOI] [PubMed] [Google Scholar]
- 22. Hastings T. G., Zigmond M. J. (1994) Identification of catechol-protein conjugates in neostriatal slices incubated with [3H]dopamine. Impact of ascorbic acid and glutathione. J. Neurochem. 63, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 23. Hastings T. G., Lewis D. A., Zigmond M. J. (1996) Role of oxidation in the neurotoxic effects of intrastriatal dopamine injections. Proc. Natl. Acad. Sci. U.S.A. 93, 1956–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akagawa M., Ishii Y., Ishii T., Shibata T., Yotsu-Yamashita M., Suyama K., Uchida K. (2006) Metal-catalyzed oxidation of protein-bound dopamine. Biochemistry 45, 15120–15128 [DOI] [PubMed] [Google Scholar]
- 25. Canet-Avilés R. M., Wilson M. A., Miller D. W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M. J., Ringe D., Petsko G. A., Cookson M. R. (2004) The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U.S.A. 101, 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paz M. A., Flückiger R., Boak A., Kagan H. M., Gallop P. M. (1991) Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 266, 689–692 [PubMed] [Google Scholar]
- 27. Witt A. C., Lakshminarasimhan M., Remington B. C., Hasim S., Pozharski E., Wilson M. A. (2008) Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry 47, 7430–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berendsen H. J. C., van der Spoel D., van Drunen R. (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 [Google Scholar]
- 29. Lindahl E., Hess B., van der Spoel D. (2001) GROMACS 3.0. A package for molecular simulation and trajectory analysis. J. Mol. Model. 7, 306–317 [Google Scholar]
- 30. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 31. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) GROMACS. Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 32. Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. (1997) Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 [DOI] [PubMed] [Google Scholar]
- 33. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 34. Waak J., Weber S. S., Görner K., Schall C., Ichijo H., Stehle T., Kahle P. J. (2009) Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J. Biol. Chem. 284, 14245–14257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. LaVoie M. J., Ostaszewski B. L., Weihofen A., Schlossmacher M. G., Selkoe D. J. (2005) Dopamine covalently modifies and functionally inactivates parkin. Nat. Med. 11, 1214–1221 [DOI] [PubMed] [Google Scholar]
- 36. Anderson P. C., Daggett V. (2008) Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson's disease. Biochemistry 47, 9380–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herrera F. E., Zucchelli S., Jezierska A., Lavina Z. S., Gustincich S., Carloni P. (2007) On the oligomeric state of DJ-1 protein and its mutants associated with Parkinson disease. A combined computational and in vitro study. J. Biol. Chem. 282, 24905–24914 [DOI] [PubMed] [Google Scholar]
- 38. Logan T., Clark L., Ray S. S. (2010) Engineered disulfide bonds restore chaperone-like function of DJ-1 mutants linked to familial Parkinson's disease. Biochemistry 49, 5624–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malgieri G., Eliezer D. (2008) Structural effects of Parkinson's disease linked DJ-1 mutations. Protein Sci. 17, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen X. M., Xia B., Wrona M. Z., Dryhurst G. (1996) Synthesis, redox properties, in vivo formation, and neurobehavioral effects of N-acetylcysteinyl conjugates of dopamine. Possible metabolites of relevance to Parkinson's disease. Chem. Res. Toxicol. 9, 1117–1126 [DOI] [PubMed] [Google Scholar]
- 41. da Costa C. A. (2007) DJ-1. A newcomer in Parkinson's disease pathology. Curr. Mol. Med. 7, 650–657 [DOI] [PubMed] [Google Scholar]
- 42. Choi J., Sullards M. C., Olzmann J. A., Rees H. D., Weintraub S. T., Bostwick D. E., Gearing M., Levey A. I., Chin L. S., Li L. (2006) Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 281, 10816–10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neumann M., Müller V., Görner K., Kretzschmar H. A., Haass C., Kahle P. J. (2004) Pathological properties of the Parkinson's disease-associated protein DJ-1 in α-synucleinopathies and tauopathies. Relevance for multiple system atrophy and Pick's disease. Acta Neuropathol. 107, 489–496 [DOI] [PubMed] [Google Scholar]
- 44. Jin J., Meredith G. E., Chen L., Zhou Y., Xu J., Shie F. S., Lockhart P., Zhang J. (2005) Quantitative proteomic analysis of mitochondrial proteins. Relevance to Lewy body formation and Parkinson's disease. Brain Res. Mol. Brain Res. 134, 119–138 [DOI] [PubMed] [Google Scholar]
- 45. Moore D. J., Zhang L., Troncoso J., Lee M. K., Hattori N., Mizuno Y., Dawson T. M., Dawson V. L. (2005) Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum. Mol. Genet. 14, 71–84 [DOI] [PubMed] [Google Scholar]
- 46. Cha S. S., Jung H. I., Jeon H., An Y. J., Kim I. K., Yun S., Ahn H. J., Chung K. C., Lee S. H., Suh P. G., Kang S. O. (2008) Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner. J. Biol. Chem. 283, 34069–34075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blackinton J., Lakshminarasimhan M., Thomas K. J., Ahmad R., Greggio E., Raza A. S., Cookson M. R., Wilson M. A. (2009) Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem. 284, 6476–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mo J. S., Jung J., Yoon J. H., Hong J. A., Kim M. Y., Ann E. J., Seo M. S., Choi Y. H., Park H. S. (2010) DJ-1 modulates the p38 mitogen-activated protein kinase pathway through physical interaction with apoptosis signal-regulating kinase 1. J. Cell. Biochem. 110, 229–237 [DOI] [PubMed] [Google Scholar]
- 49. Zhou W., Zhu M., Wilson M. A., Petsko G. A., Fink A. L. (2006) The oxidation state of DJ-1 regulates its chaperone activity toward α-synuclein. J. Mol. Biol. 356, 1036–1048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.