Background: (S)-Ureidoglycine aminohydrolase plays a key role in purine catabolism of plants and some bacteria.
Results: Substrate (S)-ureidoglycine is bound to the Mn2+ ion at the active site of homo-octameric enzyme.
Conclusion: The binding mode of (S)-ureidoglycine dictates the enantioselectivity of the reaction.
Significance: (S)-Ureidoglycine aminohydrolase liberates one of purine ring nitrogen as ammonia, which could be utilized for further nitrogen assimilation.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Structure, Nitrogen Metabolism, Protein Structure, Purine Catabolism, Ureide Pathway
Abstract
The ureide pathway has recently been identified as the metabolic route of purine catabolism in plants and some bacteria. In this pathway, uric acid, which is a major product of the early stage of purine catabolism, is degraded into glyoxylate and ammonia via stepwise reactions of seven different enzymes. Therefore, the pathway has a possible physiological role in mobilization of purine ring nitrogen for further assimilation. (S)-Ureidoglycine aminohydrolase enzyme converts (S)-ureidoglycine into (S)-ureidoglycolate and ammonia, providing the final substrate to the pathway. Here, we report a structural and functional analysis of this enzyme from Arabidopsis thaliana (AtUGlyAH). The crystal structure of AtUGlyAH in the ligand-free form shows a monomer structure in the bicupin fold of the β-barrel and an octameric functional unit as well as a Mn2+ ion binding site. The structure of AtUGlyAH in complex with (S)-ureidoglycine revealed that the Mn2+ ion acts as a molecular anchor to bind (S)-ureidoglycine, and its binding mode dictates the enantioselectivity of the reaction. Further kinetic analysis characterized the functional roles of the active site residues, including the Mn2+ ion binding site and residues in the vicinity of (S)-ureidoglycine. These analyses provide molecular insights into the structure of the enzyme and its possible catalytic mechanism.
Introduction
Nitrogen metabolism is a biological process present in a wide variety of living organisms, and it is crucial in cell physiology because it supplies an essential nutrient. Nitrogen metabolism includes metabolic routes for nitrogen uptake and assimilation and catabolism of nitrogen-rich metabolites (1). Of these, nitrogen assimilation is a universal, indispensible process common to all organisms, as identified in de novo biosynthesis of purines and pyrimidines (2, 3). In contrast, both nitrogen uptake and degradation of nitrogenous compounds manifest in distinct metabolic pathways in different organisms (4).
Recently, the unique nitrogen utilization features of the pyrimidine and purine catabolic pathways have been revealed. For example, two oxidative and reductive pyrimidine degradation pathways have been characterized in Escherichia coli (5). An additional Rut pathway was also recently identified in this bacterium (6, 7). In contrast, an oxidative purine degradation pathway is conserved in most organisms, the primary product of which is uric acid (1). Although uric acid has a high content of nitrogen, with a nitrogen-to-carbon ratio of 0.8 (supplemental Fig. S1), humans and most mammals are unable to utilize the ring nitrogen in uric acid due to a lack of the necessary catabolic pathways. However, there exists a recently elucidated uric acid degradation pathway that could remobilize the ring nitrogen atoms for further assimilation. This catabolic pathway in purine metabolism, called the ureide pathway, has been characterized in Arabidopsis thaliana, and the presence of genes for the pathway were also identified in all plants that have been sequenced to date as well as in some bacteria and fungi (8–10). These two independent catabolic pathways (the Rut pathway for pyrimidines and the ureide pathway for purines) do not show any genetic or biochemical resemblance but exhibit common features for producing ammonia as a reaction byproduct. Therefore, the possible physiological role of these degradation pathways has been suggested for nitrogen mobilization from nitrogen-rich metabolites in their respective organisms (4, 7, 9, 10).
The ureide pathway in A. thaliana is composed of seven enzymes and has been studied extensively via biochemical and structural investigations (9–17). The pathway begins with uric acid and produces a stereospecific (S)-allantoin through a three-step enzyme reaction occurring in peroxisome (supplemental Fig. S1) (8, 18, 19). Subsequently, (S)-allantoin is transported into the endoplasmic reticulum and is subject to further stepwise reactions of four other enzymes, resulting in the production of 1 eq of glyoxylate and 4 eq of ammonia molecules from 1 molecule of (S)-allantoin (9, 10). In contrast, bacterial enzymes catalyze the conversion of (S)-ureidoglycolate, which is the last substrate to the pathway in A. thaliana, into either oxalurate or glyoxylate depending on oxygen and/or nitrogen availability (supplemental Fig. S1).
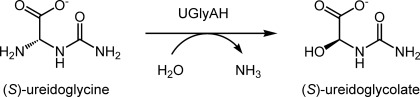
(S)-Ureidoglycolate is thus the end product of the ureide pathway common to A. thaliana as well as some bacteria and fungi. The enzyme (S)-ureidoglycine aminohydrolase (UGlyAH)2 (EC 3.5.3.-) catalyzes the conversion of (S)-ureidoglycine into (S)-ureidoglycolate in an enantioselective manner using a Mn2+ ion as a co-factor and releases ammonia (see Scheme 1) (9). In this study we carried out structural and functional analyses of UGlyAH from A. thaliana (AtUGlyAH). In particular, the structure of AtUGlyAH in the ligand-free form and as a complex with (S)-ureidoglycine substrate was determined, thus providing molecular insights into the structure of this enzyme and its mechanism.
SCHEME 1.
Conversion of (S)-ureidoglycine into (S)-ureidoglycolate by UGlyAH. The full ureide pathway is depicted in supplemental Fig. S1.
EXPERIMENTAL PROCEDURES
Construct
The UGlyAH gene from A. thaliana (GenBankTM accession number GQ303359) was modified and used as a template for PCR. The modifications were performed using the QuikChange method (Agilent) to introduce a silent mutation at the sequences for NdeI and XhoI sites residing in the internal region of the AtUGlyAH gene. The N-terminal truncated variants of AtUGlyAH, from which the signal peptide (residues 1–20) and an additional N-terminal region had been removed, were amplified using a pair of sequence-specific primers (supplemental Table S1). The resulting PCR products were introduced into the NdeI and XhoI sites of a modified pET-28a vector (Merck) containing a tobacco etch virus protease cleavage site at the junction between a His5 tag and a multiple cloning site. For the functional assay, all AtUGlyAH mutants were also prepared using the QuikChange method with mutagenic primers (supplemental Table S1).
We produced (S)-ureidoglycine, and the enzyme activity of AtUGlyAH was assayed using E. coli (S)-ureidoglycolate dehydrogenase as a coupled enzyme (supplemental Fig. S1). Therefore, expression vectors were constructed as described above for genes encoding the enzymes necessary for producing (S)-ureidoglycine from a racemic allantoin mixture and also for the gene encoding (S)-ureidoglycolate dehydrogenase. Those were as follows: allantoinase from E. coli (14), allantoate amidohydrolase from A. thaliana (gene access no. NM_11826) and E. coli strain K-12 substrain DH10b (EcAAH; gene access no. NC_010473), and (S)-ureidoglycolate dehydrogenase (gene access no. NC_010473) from E. coli strain K-12 substrain DH10b.
Protein Expression, Purification, and Crystallization
For structural studies, seleno-l-methionine-substituted, N-terminal His-tagged AtUGlyAH was expressed in E. coli B834 (DE3) methionine auxotroph cells (Merck) that were grown at 37 °C in minimal medium supplemented with seleno-l-methionine. When the optical density of the culture medium reached 0.7 at 600 nm, the recombinant protein was expressed for 18 h at 20 °C with the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside. The cells were harvested and sonicated in buffer A (50 mm NaH2PO4, pH 7.4, and 200 mm NaCl), and the supernatant was obtained by centrifugation at 30,000 × g for 1 h at 4 °C. The enzyme was purified using an immobilized metal affinity column (GE Healthcare) that had been equilibrated with buffer A and was eluted with buffer B (buffer A containing 500 mm imidazole). After the pooled elution fractions were dialyzed against buffer C (50 mm NaH2PO4, pH 7.4, and 100 mm NaCl), the N-terminal His tag was removed by treatment of tobacco etch virus protease for 12 h at 4 °C. AtUGlyAH in the absence of a His tag was further purified by immobilized metal affinity chromatography followed by gel filtration chromatography using Superdex-200 (GE Healthcare) with buffer D (50 mm HEPES, pH 7.4, and 100 mm NaCl). The protein was concentrated to ∼10 mg/ml, and 1 mm MnCl2 and 5 mm DTT were added before crystallization.
Crystallization using seleno-l-methionine-AtUGlyAH was initially carried out by the sitting-drop vapor-diffusion method at 295 K. Of the various constructs, AtUGlyAH (Δ35), from which the N-terminal 35 residues had been removed, produced crystals suitable for further structural determination under two different conditions: crystal A grown under 0.1 m phosphate citrate, pH 4.2, 10% (w/v) PEG 3000, 0.2 m NaCl and crystal B grown under 0.1 m HEPES, pH 7.5, 7% (w/v) PEG 8000, 8% (v/v) ethylene glycol. These conditions were further optimized using the hanging-drop vapor-diffusion method at 295 K.
Data Collection and Structure Determination
X-ray diffraction data were collected on beamline 4A and 6C at the Pohang Accelerator Laboratory, Korea. Both crystal A and B were cryoprotected by soaking each in crystallization solution containing additional 20–25% (v/v) glycerol and then flash-frozen in a liquid nitrogen stream at 100 K. Initially, single-wavelength anomalous diffraction data were collected to 3.0 Å resolution on crystal A at a wavelength of 0.97969 Å, corresponding to an absorption peak of selenium, and 2.5 Å resolution on crystal B. Subsequently, data for the enzyme-substrate binary complex using crystal B were collected to 2.6 Å resolution. Formation of the binary complex was achieved by a soaking experiment in which crystal B was soaked for 60 s in a solution containing crystallization solution, 20% glycerol, and 3 mm (S)-ureidoglycine. To prepare (S)-ureidoglycine, 15 mm allantoate (Sigma) was incubated with allantoate amidohydrolase from A. thaliana (2.5 μg/μl; 49.2 μm) and 1 mm MnCl2 for 2 min (supplemental Fig. S2A). The reaction product was then diluted with crystallization solution containing 20% glycerol. The collected data were processed using HKL2000 (20). Crystal A belongs to a P3221 space group with a tetramer in an asymmetric unit, whereas crystal B belongs to a P21 space group with 16 monomers in an asymmetric unit (supplemental Table S2).
Initially, the structure of crystal A was determined using the SOLVE (21) and RESOLVE (22, 23) programs for phasing and density modification, respectively. Even with 3.0 Å resolution data, the initial electron density map was sufficient to trace most of the residues in each monomer. Molecular model building and refinement was performed using COOT (24) and CNS (25), respectively, and the PHENIX program (26) was used for molecular replacement. Of the four monomers in the crystal A structure, one was manually modeled and then used as a search model of molecular replacement for locating three other subunits. Further refinement of the tetrameric structure in crystal A was carried out, and Rwork and Rfree values were 23.5 and 30.6%, respectively. Subsequently, higher resolution data were available using a crystal B, and we conducted molecular replacement using the refined monomer in crystal A as a search structure. In the 2.5 Å resolution structure of crystal B, a total of 12 monomers of the 16 in the asymmetric unit were initially found. At this stage, one particular monomer was manually modeled and again used as a search model for further molecular replacement. Eventually, all 16 monomers were found and automatically built in PHENIX. These 16 monomers were then manually inspected and refined to Rwork and Rfree values of 22.4 and 28.6%, respectively. For the AtUGlyAH in complex with (S)-ureidoglycine, the refined structure of the ligand-free form was used as a starting model for refinement. During refinement, we noticed an electron density corresponding to (S)-ureidoglycine in the metal binding site, and its model was built and included in the structure for further refinement. The stereochemistry of the refined models was evaluated using a program MolProbity (27). Details of data collection and refinement are in supplemental Table S2. Structural analyses were carried out using programs in the CCP4 suite (28). The figures presented in this study were prepared using PyMol (29).
Enzyme Assay
For functional analysis, all enzymes used in this study were prepared as described above, with the exception of allantoinase. For expression, E. coli BL21 (DE3) (Merck) harboring the plasmid of interest was cultured in Luria-Bertani medium. Subsequently, the N-terminal His-tagged protein was purified by immobilized metal affinity chromatography and dialyzed against buffer (50 mm Tris-HCl, pH 7.4, and 200 mm NaCl). For allantoinase, functionally active enzyme was expressed and purified as described previously (14).
Enzyme assays were performed at 30 °C using a UV-visible spectrophotometer (Jasco V-560) equipped with a cuvette holder and connected to a temperature-controlling water circulator. We employed two different assay protocols; one for monitoring the stepwise release of ammonia during the formation of (S)-ureidoglycolate and the other for measuring the initial velocity of the formation of (S)-ureidoglycolate in a coupled reaction with an NAD-dependent (S)-ureidoglycolate dehydrogenase reaction.
In the first method the release of ammonia was monitored in a coupled assay with an NADPH-dependent glutamate dehydrogenase in the presence of α-ketoglutarate by measuring the decrease in absorbance at 340 nm due to the oxidation of NADPH (9, 30). This method was performed for various control experiments and used to verify the substrate specificity of each purified enzyme (supplemental Fig. S2, A and B). The reaction mixture (2 ml) contained 0.2 m Tris-HCl buffer, pH 8.5, 0.3 mm NADPH, 2.5 mm α-ketoglutarate, 10 units of glutamate dehydrogenase from Proteus sp. (Sigma), 100 μm MnCl2, and 800 μg of EcAAH (8.3 μm). The reaction was then initiated by the addition of 75 μm allantoate (Sigma) or generated from allantoin (see below) followed by the addition of AtUGlyAH (0.0581–0.232 μm). The reaction progress curve proved that the enzymes used in this assay were functionally active and exhibited high substrate specificity.
Measurements of the steady-state kinetic parameters of AtUGlyAH WT and mutant enzymes were performed using the second method, mainly because the first protocol was impractical at high substrate concentrations. Specifically, in our protocol, formation of (S)-ureidoglycolate by AtUGlyAH was monitored in a coupled assay with (S)-ureidoglycolate dehydrogenase in which the (S)-ureidoglycolate produced is subsequently converted into oxalurate, with concurrent reduction of NAD+ into NADH accompanying an increase in absorbance at 340 nm. We verified the following requirements for reliable kinetic measurements; under the experimental conditions outlined below, the increase in absorbance at 340 nm was dependent only on the concentrations of (S)-ureidoglycine and AtUGlyAH but not of other enzymes (supplemental Fig. S2C), and allantoin was completely converted into (S)-ureidoglycine (supplemental Fig. S2B). Initially, a 50 mm allantoin racemic mixture (i.e. 25 mm (S)-allantoin) was dissolved in water at 37 °C for 1 h and cooled to 30 °C and then diluted with enzyme reaction buffer (50 mm HEPES, pH 7.5) to a final concentration of 20 mm (S)-allantoin. The reaction was initiated by the addition of allantoinase (0.525 mg/ml; 10 μm) at 30 °C for 1 h to produce 20 mm allantoate. This allantoate solution was stored at −70 °C and used without further purification. The assay mixture solution (2 ml) contained 50 mm HEPES buffer, pH 7.5, 100 μm MnCl2, (S)-ureidoglycolate dehydrogenase (0.2 mg/ml; 5.0 μm), 3 mm NAD+, and allantoate at the concentrations indicated (0.2–16 mm). After incubation for 60 s, further conversion of allantoate into (S)-ureidoglycine was catalyzed by the addition of EcAAH (0.4 mg/ml; 8.3 μm), and (S)-ureidoglycolate formation was subsequently initiated by the addition of AtUGlyAH (3.9 nm-0.15 μm) (supplemental Fig. S2C). Because(S)-ureidoglycine is chemically labile, the time period required for complete conversion of allantoate to (S)-ureidoglycine was determined experimentally by measuring the initial velocity of AtUGlyAH as a function of incubation time. Typically, 60 s of incubation was the optimal time for complete formation of (S)-ureidoglycine under our experimental conditions. Shorter or longer incubations caused a lower velocity, which indicates that either the conversion was incomplete, or chemically labile (S)-ureidoglycine underwent spontaneous degradation, respectively. The initial velocity was determined by measuring the linear increase in absorbance at 340 nm for the first 20 or 30 s and was calculated as the NADH concentration produced per min, with a molar extinction coefficient of 6220 m−1cm−1 at 340 nm. The Km and Vmax values were obtained using SigmaPlot, and kcat values were computed by dividing Vmax by the AtUGlyAH concentration used.
Metal Analysis
For metal analysis, the WT enzyme and E235Q and E235A mutants were expressed and purified as described in “Enzyme Assay” above. Specifically, the N-terminal His-tagged protein was purified by immobilized metal affinity chromatography, and the His tag was subsequently removed by treatment of tobacco etch virus protease. The His-tag free enzyme was then incubated in the presence of 5 mm EDTA for 1 h at 4 °C and was purified by gel filtration chromatography. After the addition of 1 mm MnCl2, the resulting enzyme solution was again subjected to gel filtration chromatography with buffer (50 mm HEPES, pH 7.4, and 100 mm NaCl). Protein concentrations were about 0.11∼0.14 mm, and the metal content of the enzyme was determined by inductively coupled plasma-atomic emission spectrophotometer.
RESULTS
Molecular Architecture of Octameric AtUGlyAH in Ligand-free Form
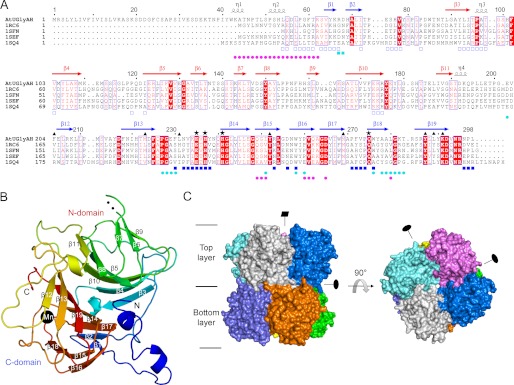
The crystal structure of AtUGlyAH was determined using AtUGlyAH Δ35, which was missing 35 residues from the N terminus including a cleavable signal peptide (Fig. 1A and supplemental Table S2). Two octamers were characterized with no non-crystallographic symmetry in an asymmetric unit of the P21 space group. The apparent molecular mass of AtUGlyAH (30.2 kDa monomer) was estimated by size-exclusion chromatography to be more than 200 kDa, suggesting that the functional unit of AtUGlyAH is likely an octamer.
FIGURE 1.
Sequence alignment and overall structure of AtUGlyAH. A, the amino acid sequences of AtUGlyAH, including the signal peptide (residues 1–20) (9), are compared with those of the E. coli ortholog and structural homologs from other bacteria, all with known structures in the absence of their functional roles. Structures of these proteins were previously determined by Structural Genomics Centers: an E. coli ortholog (PDB ID 1RC6; resolution, 2.6 Å; Z-score, 29.6; r.m.s.d., 2.1 Å; sequence identities, 28%) and structural homologs from Deinococcus radiodurans R1 (PDB ID 1SFN; 2.46 Å; 34.8; 1.4 Å; 40%), Enterococcus faecalis V583 (PDB ID 1SEF; 2.05 Å; 32.3; 1.7 Å; 31%), and Pseudomonas aeruginosa (PDB ID 1SQ4; 2.7 Å; 28.3; 2.5 Å; PDB ID 26%). Highly conserved residues are shown in red and boxed in blue. Strictly conserved residues are shown on a red background. The secondary structural elements defined in a ligand-free form are shown for the corresponding AtUGlyAH sequences, with the N and C domains in red and blue, respectively. Black asterisks and triangles represent residues involved in the Mn2+ coordination shell and the (S)-ureidoglycine binding site, respectively. Residues involved in the interactions between monomers within the layer are indicated by blue rectangles; open and closed rectangles represent residues from different monomers. Cyan circles indicate residues involved in inter-layer interactions between monomers located vertically, and magenta circles indicate those located diagonally across the layers. This figure was prepared using ESPript (35). B, shown is the monomer structure, with an orientation identical to that of the monomer in gray in the left panel of C. The presence of a Mn2+ ion is indicated by a black circle. Two residues, Thr-138 and Ser-139, are disordered in the loop between β6 and β7. C, shown is a surface representation of an octamer in two orientations. A side view, with each monomer in a different color, is presented in the left panel, whereas the right panel shows a top view of the layer.
The crystal structure of monomeric AtUGlyAH, which contains the ordered residues Pro-39 to Leu-298, is composed of 19 β-strands and 4 short 310-helices (Fig. 1A). Its structure is divided into two structurally similar β-barrel domains, called N and C domains, with an N-terminal segment protruding from the rest of the structure (Fig. 1B). Each domain represents a typical cupin fold of a β-barrel, in which two layers of β-sheet are packed in a parallel manner with a funnel-like structure (31). Specifically, the N domain includes β3 to β11. Six strands are arranged in the spatial order β3, β4, β10, β5, β8, and β11, with an anti-parallel orientation. They form a central face of a β-barrel fold, whereas the outer face of the fold consists of β9, β6, and β7, with their orientations parallel to the central β-sheet. The C domain also exhibits a similar architecture in which N-terminal anti-parallel β1 and β2 strands form part of the central anti-parallel β-sheet that comprises β1, β2, β17, β14, β19, β13, and β12 in a spatial order. Three strands, β16, β15, and β18, line along the outer surface of the β-barrel fold. Therefore, the AtUGlyAH monomer is a bicupin molecule and is stabilized by packing the central β-sheets from each domain in a parallel manner (Fig. 1B). These interactions result in 2830 Å2 of buried surface area and a root mean square deviation (r.m.s.d.) of 1.9 Å for the structurally equivalent 91 Cα atoms between the 2 cupin folds.
The AtUGlyAH functional unit is assembled from two layers of tetramers, each layer with a dorsal and a ventral surface (Fig. 1C). Within a layer, four monomers are related by a non-crystallographic four-fold symmetry along the vertical axis running through the center of the octamer, as shown by the top view of a dorsal surface. These four monomers are structurally identical within 0.34–0.42 Å r.m.s.d. for 258 Cα atoms. The two layers are stabilized by interacting through the ventral surface but are offset from the vertical axis by ∼22.5° of rotation (Fig. 1C). Monomers in different layers are related by a 2-fold symmetry along the two horizontal axes, which are orthogonal to each other. Therefore, a 422-symmetry along a vertical axis and two horizontal axes, respectively, fully describes the molecular architecture of octameric AtUGlyAH using a monomer as a protomer.
Interactions between Monomers in Intra- and Inter-layers
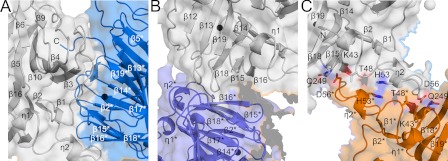
Within each layer, monomeric AtUGlyAH is packed against its neighbors, whose positions differ by 90° of rotation via hydrophobic interactions. These interactions are asymmetrical in that distinct regions from neighboring monomers participate in the packing. Specifically, residues in the loop regions of the protruding N-terminal segment and those connecting β2 and β3 and connecting β3 and β4 mediate largely hydrophobic interactions with other loop residues, mainly in the C domain β13*-β14* and β17*-β18* of the adjacent monomer (hereafter, an asterisk denotes an element or a residue of a neighboring monomer) (Fig. 2A and supplemental Fig. S3A). In addition, the C-terminal residues Asn-296* to Leu-298* are plugged into the concave surface formed by the residues in the vicinity of the N domains β3 and β4. Several hydrogen bonds were characterized in this packing within a layer (supplemental Fig. S3A). A 4-fold arrangement of monomers in each layer generates four independent but structurally identical interfaces; each interface exhibits a buried surface area of 1069 Å2 corresponding to ∼8.8% of the surface area of the monomer.
FIGURE 2.
Interactions of monomers within and between layers. Color code and orientation for each monomer in these figures are identical to those in the left panel of Fig. 1C, and residues involved in these interactions are indicated with symbols as in Fig. 1A. A stereo view of these figures is shown in supplemental Fig. S3. A, interactions of two neighboring monomers within the layer are shown. B, two monomers located vertically in different layers are presented. Residues, such as Phe-230, Ile-251, Trp-258, Trp-276, Ala-278, Ala-279, and Leu-280 and equivalent residues from another monomer are clustered into this region to mediate hydrophobic interactions as well as hydrogen bonding between the main chain Gly-228 and Gly-281*. C, interactions occurring between two monomers located diagonally across the layers are shown. In particular, Lys-43, Thr-48, and Gln-249 participate in hydrogen bonding with Asp-56*, whereas His-53 in the middle of the protruding region mediates a stacking interaction with the equivalent His-53* of a different monomer.
More extensive, diverse packings were identified in the interactions between the two layers, with a total buried surface area of 4677 Å2. The inter-layer interactions are symmetrical, in which structurally and sequentially identical residues are involved in the packing of monomers in different layers. Two types of interactions are observed (Fig. 1C); one is between monomers located vertically in different layers, and the other is between monomers located diagonally across the layers. Specifically, residues in the outer surface of the C domain cupin fold, including β16, β15, and β18, are major structural elements in stabilizing monomers orientated vertically in different layers (Fig. 2B and supplemental Fig. S3B). In the other type of interaction, the protruding N-terminal region containing Pro-39 to Ser-63 as well as residues on the outer surface of the C domain cupin fold in the vicinity of β16 and β15 are packed in a 2-fold symmetrical manner against the equivalent elements of a monomer in a different layer (Fig. 2C and supplemental Fig. S3C). Overall, these interactions serve as key structural features in stabilizing the two layers, in particular by inserting the protruding N-terminal segment into the concave surface of another monomer.
Active Site in Ligand-free Form of AtUGlyAH
In the monomer, each N and C domain with a funnel-like β-barrel structure exhibits two possible openings, one at each end of the long axis of the funnel (Fig. 1B). However, various interactions in monomers and octamers effectively seal off these openings, except for one end in the C domain. Specifically, a smaller opening near β3 and β4 in the N domain is occluded by the C-terminal segment from the adjacent monomer within the layer (Fig. 2A and supplemental Fig. S3A). The entrance to a larger opening in the N domain is closed off by several layers of hydrophobic residues whose side chains point into the cavity from the central and the outer side β-strands in a cupin fold (supplemental Fig. S4A). In contrast, the inside cavity of the C domain is readily accessible from the surface of the enzyme through the larger opening, with its location in the direction of the dorsal surface in the layer (Fig. 1B). A smaller end of the C domain funnel points toward the center of the octamer but is sealed off by the presence of β1 and β2 (Fig. 1B).
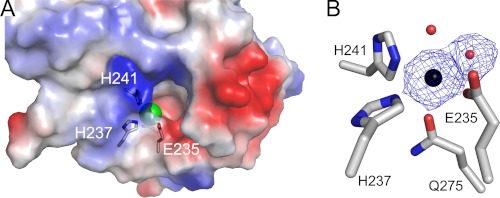
Consistent with the requirement of Mn2+ ion for catalytic activity (9), the location of the active site in AtUGlyAH was indicated by the presence of a metal binding site in the C domain. Indeed, crystallization of AtUGlyAH was performed with 1 mm MnCl2. The metal binding site is embedded in the middle of a funnel-like cavity in the C domain, which is located at 14 Å deep from the surface (Fig. 3A). Four residues from the central side and the outer side of a β-barrel are involved in the metal coordination shell with one or two water molecules depending on the subunit (Fig. 3B). Specifically, Glu-235 and His-237 in the long loop between β13 and β14, which transverses and folds back between the central and outer side of the β-barrel, as well as His-241 in the central β14 and Gln-275 in the outer side β18 are part of an octahedral coordination with the bound metal. In this coordination, the metal ion lies in the center of an equatorial plane formed by Glu-235, His-237, His-241, and a water molecule across from His-237 (Fig. 3B and supplemental Table S3A). Gln-275 and another water molecule trans to Gln-275 serve as the axial ligands, which are normal to the equatorial plane; the electron density for the axial water molecule is relatively disordered in different monomers. Under our crystallization condition, the metal ion was identified as Mn2+ ion by inductively coupled plasma-atomic emission spectroscopy (supplemental Table S4). It is noticeable that two water molecules ligating with the Mn2+ ion are near the entrance, and therefore, their locations likely serve as the binding site for the incoming (S)-ureidoglycine substrate molecule.
FIGURE 3.
Active site in the C domain cupin fold of the ligand-free form AtUGlyAH. A, surface representation of the opening in the C domain of monomer is shown. In this orientation, the N-terminal protruding region is located on the back side. The active site residues are indicated with the bound Mn2+ ion in green. B, shown are the Mn2+ binding residues, with the Mn2+ ion in a black sphere and water molecules indicated by red circles. The Mn2+ ion and water molecule trans to His-237 are overlaid with a 2Fo − Fc electron density map contoured at 3.0σ and 1.0σ, respectively. The axial water molecule across from Gln-275 is present in some monomers, and its density is relatively disordered but obvious at 0.8 σ of the 2Fo − Fc electron density map. Stereochemical details of the Mn2+ binding site are shown in supplemental Table S3.
Binding Mode of (S)-Ureidoglycine in Binary Complex
The structure of AtUGlyAH in complex with (S)-ureidoglycine was determined in a soaking experiment. We produced (S)-ureidoglycine, which is commercially unavailable, by an enzyme reaction using allantoate amidohydrolase with allantoate as the substrate ( supplemental Figs. S1 and S2A). By using the x-ray data from a crystal of AtUGlyAH soaked with (S)-ureidoglycine, an extra electron density was identified in the proximity of the bound Mn2+ ion (Fig. 4A and supplemental Fig. S4B). The density was assigned as that of (S)-ureidoglycine, given that allantoate amidohydrolase produces (S)-ureidoglycine in a stereospecific manner (9) and only the (S)-configuration of ureidoglycine fits the electron density identified in the binary complex. We also modeled the observed density with ureidoglycolate, the product of UGlyAH (Scheme 1); however, only the (R)-configuration fits the density, which is inconsistent with the previous assignment of absolute stereochemistry of the ureidoglycolate enantiomer produced by UGlyAH (32). Therefore, it is likely that low catalytic activity of crystalline AtUGlyAH relative to that in solution as well as the cryogenic conditions during x-ray data collection stabilizes chemically labile (S)-ureidoglycine rather than catalyzing the formation of (S)-ureidoglycolate.
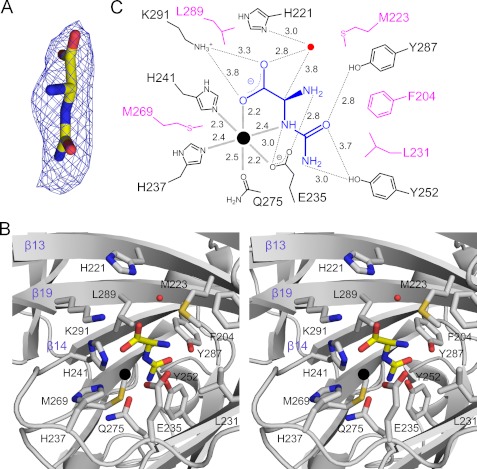
FIGURE 4.
The binding mode of (S)-ureidoglycine in the binary complex. A, the modeled (S)-ureidoglycine is overlaid with an Fo − Fc electron density map contoured at 2.5σ. B, the active site in the binary complex is shown in a stereoscopic view, with neighboring residues less than 5.0 Å from the bound (S)-ureidoglycine. C, shown is a schematic representation of the interactions around the Mn2+ ion and (S)-ureidoglycine. Residues involved in hydrophobic interactions and hydrogen bonding are indicated in magenta and black, respectively, with the interatomic distance values averaged among 16 monomers. For clarity, the following possible hydrogen bonds were not indicated: the ring nitrogen of His-241 to the carboxyl group of (S)-ureidoglycine within 3.2 Å, the side-chain carbonyl oxygen of Gln-275 to the amino group of the ureido group within 2.8 Å, and the two hydroxyl groups in Tyr-252 and Tyr-287 within 3.7 Å.
There were no significant changes in conformation between the ligand-free form and the binary complex, with an r.m.s.d. of less than 0.5 and 0.32 Å for 258 and 2064 Cα atoms in the monomer and octamer, respectively. When bound to the Mn2+ coordination shell, (S)-ureidoglycine is in an almost planar conformation (Fig. 4, A and B); the carboxyl and the ureido groups are in an anti conformation along the bond between the chiral carbon atom and the nitrogen atom in the amide bond, with a dihedral angle of 162° (±16.0) throughout the 16 monomers. The amino group and hydrogen attached to the chiral carbon atom are the only ones out of the plane. The substrate (S)-ureidoglycine is almost parallel to the axial plane of the Mn2+ coordination shell (Fig. 4B); the ureido group is located at the innermost part of the active site, whereas the carboxylate group points toward an entrance to the active site. Specifically, the binding of (S)-ureidoglycine is achieved by replacing the two water molecules (one each for the axial and equatorial ligands in the Mn2+ coordination shell of the ligand-free form). One of the oxygen atoms in the carboxylate group and the nitrogen atom in the amide bond then take the positions of the axial and equatorial ligands, respectively (Fig. 4B), without distorting the geometry of the octahedral coordination with the bound metal (supplemental Table S3B).
The binding of (S)-ureidoglycine in the complex is further stabilized by hydrogen bonding along the molecule (Fig. 4, B and C). Specifically, the carboxyl group of (S)-ureidoglycine mediates several interactions within 3.0 Å, i.e. a possible charge neutralization with Lys-291 and hydrogen bonding with an equatorial ligand His-241. The side chain of an equatorial ligand Glu-235 also stabilizes the binding of the amino group within a distance of 2.8 Å and the nitrogen atom in the amide bond within a distance of 3.0 Å, allowing the (S) configuration of ureidoglycine. The ureido group is also within a hydrogen-bonding distance of 2.8–3.0 Å from Tyr-252, Tyr-287, and an axial ligand Gln-275, facilitating the binding of the ureido group in the innermost part of the active site. In addition, hydrophobic residues comprise a part of the active site within 5.0 Å from the bound substrate and position the incoming (S)-ureidoglycine into the productive binding mode for catalysis. These residues include Phe-204, Met-223, and Leu-231 near the ureido group and Met-269 and Leu-289 near the amide bond (Fig. 4, B and C). It is noteworthy that the Mn2+ binding residues and those hydrophilic residues near the (S)-ureidoglycine binding site are invariant among UGlyAHs and its structural homologs (Fig. 1A), suggesting the functional roles of these residues in catalysis. In particular, His-221, which is conserved in AtUGlyAH and its E. coli ortholog (Fig. 1A; PDB ID 1RC6), is located on the side of the hydrogen attached to the chiral carbon atom, 5.5 Å from the chiral carbon atom.
Functional Analysis
We produced (S)-ureidoglycine in situ by employing the two preceding enzymes, allantoinase (14) and allantoate amidohydrolase (11), with allantoin as the substrate (supplemental Figs. S1 and S2B). We also used a coupled reaction in which (S)-ureidoglycolate produced by AtUGlyAH was oxidized into oxalurate by an NAD-dependent E. coli (S)-ureidoglycolate dehydrogenase, with concomitant formation of NADH (supplemental Figs. S1 and S2C). Therefore, the steady-state kinetic parameters for various AtUGlyAH mutant enzymes were measured by monitoring the increase in absorbance at 340 nm caused by the formation of NADH.
A total of eight residues were selected for site-directed mutagenesis (Fig. 4C); their kinetic parameters are listed in Table 1 (supplemental Fig. S5). The residues involved in the Mn2+ coordination shell were sensitive to mutation. Mutant enzymes such as E235A, H237A, H241A, and Q275A were catalytically incompetent, suggesting that these residues are essential for coordinating with the Mn2+ ion. It is noticeable that E235Q mutant is an essentially inactive enzyme (Table 1) but is capable of binding the Mn2+ ion as much as does the WT enzyme (supplemental Table S4). We did not expect to find that the Km value of Y252F was comparable, but its kcat value was reduced by ∼10-fold compared with that of WT enzyme. However, enzyme activity was essentially abolished in the Y287F and Y287A mutants, suggesting that the side chain hydroxyl group in Tyr-287, but not Tyr-252, plays an essential role in catalysis and/or substrate binding. The Km value of K291R was ∼2-fold higher, and its kcat value was ∼65% that of the WT enzyme, whereas enzyme activity was completely absent in the K291A mutant. This suggests that the positive charge at Lys-291 plays a pivotal role in substrate binding, possibly by neutralizing the negative charge of the carboxyl group of (S)-ureidoglycine (Fig. 4, B and C). Unlike other mutant enzymes, initial velocity measurements of the H221A mutant were unsuccessful, mainly due to nonlinear changes in absorbance as a function of reaction time under various assay conditions. However, its activity was estimated to be similar to that of Y252F (supplemental Fig. S5D). Because all AtUGlyAH mutant enzymes used in this functional analysis were estimated to be in a native conformation based on circular dichroism spectra that was similar to the WT enzyme (supplemental Fig. S6), a lack of activity in some of these mutants is related to the functional features of those residues, not due to a possible conformation change by mutation.
TABLE 1.
Kinetic parameters of AtUGlyAH and its mutants
Mutants as follows were inactive: E235A, E235Q, H237A, H241A, Q275A, Y287A, Y287F, K291A. The initial velocity measurements of the H221A mutant were unsuccessful, mainly due to non-linear changes in absorbance as a function of reaction time under various assay conditions.
| Km | kcat | kcat/Km | |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| WT | 1.77 (0.104)a | 761 (14.1) | 429 |
| Y252F | 2.30 (0.0910) | 73 (0.73) | 32 |
| K291R | 3.46 (0.196) | 493 (10.9) | 143 |
a Values in parentheses are S.E. estimated by the program SigmaPlot.
DISCUSSION
The presence of a gene encoding the UGlyAH enzyme and its biochemical function in the ureide pathway were only recently identified in both A. thaliana and E. coli (9, 10). However, the Research Centers for Structural Genomics had earlier determined the crystal structure of YlbA, an E. coli ortholog of UGlyAH (Fig. 1A; PDB ID 1RC6), and three structural homologs from other bacteria (Fig. 1A; PDB IDs 1SFN, 1SEF, and 1SQ4). These four proteins exhibit structural features highly homologous with those of AtUGlyAH; a bicupin monomer and an overall octameric conformation, with Z-scores of 28.3–34.8, sequence identities of 26–40%, and r.m.s.d. values of 1.4–2.5 Å for the monomer, as defined by the DALI program (33). Although the metal binding site was not recognized in those structures, high similarities in both structure and sequence, including residues for metal coordination and the substrate binding site (Fig. 1A), suggest that these three structural homologs are also members of the UGlyAH family. In particular, the cupin fold in this family of proteins is associated with a broad range of enzyme activities and is widely distributed in the protein kingdom (31).
Structural and functional analysis of AtUGlyAH revealed several functional consequences. First, AtUGlyAH is a Mn2+-dependent enzyme (9). It is evident from the structure of the binary complex that the Mn2+ ion acts as a molecular anchor for the substrate; it positions the incoming (S)-ureidoglycine into the productive binding mode for catalysis (Fig. 4B). Second, the resulting binding mode serves as the molecular basis of the catalytic enantioselectivity. More specifically, a molecular face with a hydrogen atom attached to the chiral carbon atom is vulnerable to enzyme catalysis. Of the neighboring residues, Tyr-287 and Lys-291 are essential for enzyme activity, possibly by dictating the orientation of the ureido and carboxyl groups of the substrate, respectively (Table 1 and Fig. 4B). Binding of the (S) configuration of ureidoglycine is further stabilized by Glu-235 due to possible interactions between the amino group of a reaction center carbon and the carboxyl group of Glu-235 (Fig. 4C). In addition, the side chain carboxyl group on Glu-235 appears to be crucial in catalysis because the E235Q mutant still containing Mn2+ ion (supplemental Table S4) is catalytically incompetent.
We considered potential catalytic mechanisms of AtUGlyAH by examining structural and functional analyses as well as inversion of configuration at the chiral carbon center between (S)-ureidoglycine and (S)-ureidoglycolate (Scheme 1) (9). A possible direct attack of the general base-catalyzed water molecule at the chiral carbon appears to be highly unlikely; such an Sn2-type reaction requires a water molecule that presents on the side opposite to the leaving amino group, with a geometrical requirement for an inline attack on the chiral carbon center. The presence of the water molecule is implausible, mainly due to steric hindrance in that area which is densely populated with His-241, Leu-289, and Lys-291 (Fig. 4B). Alternatively, allanturate (5) could be an intermediate in catalysis (supplemental Fig. S7). Glu-235, with the probable low pK value of the hydrogen on the Mn-bound amide nitrogen, could facilitate abstraction of the proton from the amide nitrogen and subsequent transfer of this proton to the amino leaving group in the form of ammonia, which is analogous to the proposed function of the metal-bound aspartate in adenine deaminase (34). Subsequently, the resulting planar intermediate allanturate could be subject to a nucleophilic attack by the general base-catalyzed water molecule, yielding (S)-ureidoglycolate. Specifically, it is likely that the re face of the reaction center carbon in the intermediate remains occluded by Glu-235, as observed in the binary complex (Fig. 4B). Therefore, the general base-catalyzed water molecule could reach the intermediate only from the si face of the planar intermediate, which is consistent with a previous observation of an inversion of configuration at the chiral carbon atom after catalysis (Scheme 1) (9).
However, this analysis does not reveal any obvious general base to deprotonate the incoming water molecule in the immediate vicinity of the chiral carbon atom of (S)-ureidoglycine (Fig. 4, B and C) but provides two candidates, His-221 and Tyr-287. Both are located on the side opposite the leaving amino group of the substrate, 5.3–5.4 Å from the chiral carbon atom. The presence of a water molecule connecting His-221 and the chiral carbon atom (Fig. 4C) as well as the low catalytic activity of the H221A mutant (supplemental Fig. S5D) raises the possibility that His-221 acts as the proposed general base. However, His-221 is not a strictly conserved residue in the UGlyAH family (Fig. 1A), in which the mechanistic features should remain identical. In contrast, Tyr-287 is invariant in the family, but its side chain hydroxyl group should be deprotonated to act as a general base. The following observations suggest that a hydrogen bonding network between Tyr-252 and Tyr-287 as well as the ureido group might participate in the deprotonation of Tyr-287 and facilitate its functional role in catalysis; (i) the Y287F mutant is an inactive enzyme, (ii) the two hydroxyl groups in Tyr-287 and Tyr-252 are separated by ∼3.7 Å, and (iii) the hydroxyl group in Tyr-252 is involved in catalysis rather than substrate binding, given that the Y252F mutation largely affects the kcat, but not the Km, value. Therefore, Tyr-287 is a candidate general base. However, Tyr-287 became almost buried after binding of (S)-ureidoglycine. Lack of the amino group in the intermediate could cause its binding mode to differ from that in the enzyme-substrate complex, possibly by inclining toward Glu-235, exposing Tyr-287 to the solvent. Further investigation is necessary for conclusive identification of the general base in AtUGlyAH.
In this study we determined a crystal structure of the ligand-free form of AtUGlyAH and its complex with (S)-ureidoglycine. The structures revealed the overall features of both the monomeric and octameric conformations, which are applicable to other UGlyAH family proteins, as well as the binding mode of (S)-ureidoglycine in the Mn2+ coordination shell. Our enzyme assay, which employed an E. coli (S)-ureidoglycolate dehydrogenase as a coupled enzyme, allows for measurement of the steady-state kinetic parameters of AtUGlyAH. These results provide molecular insights into the structure of UGlyAH and early events in the catalytic release of ammonia from the nitrogen-rich metabolite, which is the crucial step of the ureide pathway.
Supplementary Material
Acknowledgments
We thank Woo-Suk Jung for his contribution to the initial stage of research. We also thank Peter Tipton for his suggestion for the proposed mechanism and Hanjin Oh for performing the circular dichroism measurements.
This work was supported by grants from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center no. PJ008059), Rural Development Administration, and from National Research Foundation (R11-2008-062-01002-0 and 2010-0025883) funded by the Ministry of Education, Science, and Technology, Republic of Korea.

This article contains supplemental Tables S1–S4 and Figs. S1–S7.
The atomic coordinates and structure factors (codes 4E2Q and 4E2S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UGlyAH
- (S)-ureidoglycine aminohydrolase
- AtUGlyAH
- UGlyAH from A. thaliana
- EcAAH
- allantoate amidohydrolase from E. coli
- r.m.s.d.
- root mean-square deviation.
REFERENCES
- 1. Atkins C. A., Smith P. M. C. (2000) in Prokaryotic Nitrogen Fixation: A Model System for Analysis of a Biological Process (Triplett E. W., ed.) pp. 559–587, Horizon Scientific Press, Wymondham, UK [Google Scholar]
- 2. Smith P. M., Atkins C. A. (2002) Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 128, 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boldt R., Zrenner R. (2003) Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant 117, 297–304 [DOI] [PubMed] [Google Scholar]
- 4. Werner A. K., Witte C. P. (2011) The biochemistry of nitrogen mobilization. Purine ring catabolism. Trends Plant Sci. 16, 381–387 [DOI] [PubMed] [Google Scholar]
- 5. Vogels G. D., Van der Drift C. (1976) Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40, 403–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loh K. D., Gyaneshwar P., Markenscoff Papadimitriou E., Fong R., Kim K. S., Parales R., Zhou Z., Inwood W., Kustu S. (2006) A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. U.S.A. 103, 5114–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim K. S., Pelton J. G., Inwood W. B., Andersen U., Kustu S., Wemmer D. E. (2010) The Rut pathway for pyrimidine degradation. Novel chemistry and toxicity problems. J. Bacteriol. 192, 4089-4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramazzina I., Folli C., Secchi A., Berni R., Percudani R. (2006) Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat. Chem. Biol. 2, 144–148 [DOI] [PubMed] [Google Scholar]
- 9. Serventi F., Ramazzina I., Lamberto I., Puggioni V., Gatti R., Percudani R. (2010) Chemical basis of nitrogen recovery through the ureide pathway. Formation and hydrolysis of S-ureidoglycine in plants and bacteria. ACS Chem. Biol. 5, 203–214 [DOI] [PubMed] [Google Scholar]
- 10. Werner A. K., Romeis T., Witte C. P. (2010) Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 6, 19–21 [DOI] [PubMed] [Google Scholar]
- 11. Agarwal R., Burley S. K., Swaminathan S. (2007) Structural analysis of a ternary complex of allantoate amidohydrolase from Escherichia coli reveals its mechanics. J. Mol. Biol. 368, 450–463 [DOI] [PubMed] [Google Scholar]
- 12. Cendron L., Berni R., Folli C., Ramazzina I., Percudani R., Zanotti G. (2007) The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation. J. Biol. Chem. 282, 18182–18189 [DOI] [PubMed] [Google Scholar]
- 13. Kim K., Park J., Rhee S. (2007) Structural and functional basis for (S)-allantoin formation in the ureide pathway. J. Biol. Chem. 282, 23457–23464 [DOI] [PubMed] [Google Scholar]
- 14. Kim K., Kim M. I., Chung J., Ahn J. H., Rhee S. (2009) Crystal structure of metal-dependent allantoinase from Escherichia coli. J. Mol. Biol. 387, 1067–1074 [DOI] [PubMed] [Google Scholar]
- 15. Ramazzina I., Cendron L., Folli C., Berni R., Monteverdi D., Zanotti G., Percudani R. (2008) Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases. J. Biol. Chem. 283, 23295–23304 [DOI] [PubMed] [Google Scholar]
- 16. Hennebry S. C. (2009) Evolutionary changes to transthyretin. Structure and function of a transthyretin-like ancestral protein. FEBS J. 276, 5367–5379 [DOI] [PubMed] [Google Scholar]
- 17. French J. B., Ealick S. E. (2010) Structural and mechanistic studies on Klebsiella pneumoniae 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase. J. Biol. Chem. 285, 35446–35454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G. E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lamberto I., Percudani R., Gatti R., Folli C., Petrucco S. (2010) Conserved alternative splicing of Arabidopsis transthyretin-like determines protein localization and S-allantoin synthesis in peroxisomes. Plant Cell 22, 1564–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 0.99, Schrödinger, LLC, New York [Google Scholar]
- 30. Muratsubaki H., Satake K., Enomoto K. (2006) Enzymatic assay of allantoin in serum using allantoinase and allantoate amidohydrolase. Anal. Biochem. 359, 161–166 [DOI] [PubMed] [Google Scholar]
- 31. Agarwal G., Rajavel M., Gopal B., Srinivasan N. (2009) Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PLoS ONE 4, e5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gravenmade E. J., Vogels G. D., Van der Drift C. (1970) Hydrolysis, racemization and absolute configuration of ureidoglycolate, a substrate of allantoicase. Biochim. Biophys. Acta 198, 569–582 [DOI] [PubMed] [Google Scholar]
- 33. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 34. Kamat S. S., Bagaria A., Kumaran D., Holmes-Hampton G. P., Fan H., Sali A., Sauder J. M., Burley S. K., Lindahl P. A., Swaminathan S., Raushel F. M. (2011) Catalytic mechanism and three-dimensional structure of adenine deaminase. Biochemistry 50, 1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.