Background: Archaeal transcription is activated by a novel mechanism.
Results: The novel regulator PF1088 (TFB-RF1) is able to activate archaeal transcription by TFB recruitment.
Conclusion: Archaeal transcription can be activated by recruitment of not only TATA-binding protein but also TFB.
Significance: Exploring the hybrid transcription machinery in Archaea could reveal basic transcription mechanisms for all forms of life.
Keywords: Archaea, Gene Regulation, Transcription Factors, Transcription Initiation Factors, Transcription Regulation, TBP, TFB, Transcription Activation
Abstract
Archaeal promoters consist of a TATA box and a purine-rich adjacent upstream sequence (transcription factor B (TFB)-responsive element (BRE)), which are bound by the transcription factors TATA box-binding protein (TBP) and TFB. Currently, only a few activators of archaeal transcription have been experimentally characterized. The best studied activator, Ptr2, mediates activation by recruitment of TBP. Here, we present a detailed biochemical analysis of an archaeal transcriptional activator, PF1088, which was identified in Pyrococcus furiosus by a bioinformatic approach. Operon predictions suggested that an upstream gene, pf1089, is polycistronically transcribed with pf1088. We demonstrate that PF1088 stimulates in vitro transcription by up to 7-fold when the pf1089 promoter is used as a template. By DNase I and hydroxyl radical footprinting experiments, we show that the binding site of PF1088 is located directly upstream of the BRE of pf1089. Mutational analysis indicated that activation requires the presence of the binding site for PF1088. Furthermore, we show that activation of transcription by PF1088 is dependent upon the presence of an imperfect BRE and is abolished when the pf1089 BRE is replaced with a BRE from a strong archaeal promoter. Gel shift experiments showed that TFB recruitment to the pf1089 operon is stimulated by PF1088, and TFB seems to stabilize PF1088 operator binding even in the absence of TBP. Taken together, these results represent the first biochemical evidence for a transcriptional activator working as a TFB recruitment factor in Archaea, for which the designation TFB-RF1 is suggested.
Introduction
The transcription machinery in Archaea consists of a multisubunit RNA polymerase (RNAP)2 and three basal transcription factors, TATA-binding protein (TBP), transcription factor B (TFB), and transcription factor E (1–5). All of these components have closely related homologs in eukaryotes. TBP and TFB arrange the recognition of the archaeal promoter by binding to the TATA box and the TFB-responsive element (BRE), respectively (6–9). As in Eukarya, promoter-bound TBP and TFB recruit the RNAP. Transcription factor E promotes initiation of transcription on weak promoters under limiting concentrations of TBP and is also part of the archaeal elongation complex (10–12).
In contrast to the high similarity between the basal transcription machinery of the Archaea and Eukarya, the regulation of transcription fits more into the bacterial schema. All archaea encode a large number of proteins containing the helix-turn-helix DNA-binding domains, whose sequences closely resemble bacterial helix-turn-helix domains (13, 14). To date, a limited number of these archaeal regulators have been characterized in detail. The great majority analyzed so far act as transcriptional repressors (reviewed in Refs. 4 and 15–17). In most cases, the bound repressor prevents the recruitment of the RNAP, and in some cases, the binding of TBP or TFB (or both) is inhibited.
There are also some known instances of positive control in Archaea, with Ptr2 from Methanocaldococcus jannaschii being the most well characterized (18, 19). This Lrp family protein binds cooperatively to sequences upstream of the TATA box and stimulates transcription via recruitment of TBP. A similar mechanism is likely used by GvpE, a protein with a eukaryote-like basic leucine zipper DNA-binding domain that activates genes involved in gas vesicle production in halophiles (20). Additional examples of transcriptional activators exist, but the molecular details of the mechanism of activation are unknown in these cases. In Pyrococcus furiosus, SurR and TrmBL1 can act as transcriptional repressors as well as activators (21, 22). SurR is involved in the regulation of hydrogen and elemental sulfur metabolism, whereas TrmBL1 plays a role in the regulation of sugar metabolism.
To elucidate the potential of archaeal transcriptional regulators in more detail, we recently performed a computational screen to identify putative transcriptional regulators and their corresponding DNA-binding sites.3 Here, we report a detailed biochemical analysis of one of these predictions, the putative transcriptional regulator PF1088 and the corresponding binding site. Our data clearly demonstrate that this protein activates transcription via a novel mechanism involving the stimulation of TFB recruitment to an archaeal promoter.
EXPERIMENTAL PROCEDURES
Proteins and DNA Templates
The purification of RNAP and the transcription factors TBP and TFB was performed as described previously (24). For the purification of PF1088, the corresponding gene was cloned into the expression vector pET-14b (Novagen). The coding sequence was amplified by PCR from genomic DNA from P. furiosus using primers TAGGGAGATCATATGGAAGAAAT and TGGATCCTTAAAAATGTATTGCATCGATTACTGTCT. Amplification products were digested with BamHI and NdeI (underlined in the primer sequences) and ligated into vector pET-14b. Escherichia coli strain BL21(DE3)-CP (Stratagene) transformed with the corresponding plasmid was grown in 400 ml of LB medium containing ampicillin (100 μg/ml). Protein expression of PF1088 was induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. The bacterial culture was further incubated for additional 5 h. Cells were harvested and resuspended in 40 mm HEPES (pH 7.5), 20 mm imidazole, 0.5 m NaCl, and 15% glycerol. Cell lysis was done by the addition of lysozyme for 1 h at 37 °C following sonification. After centrifugation at 50,000 × g for 15 min, the soluble crude extract was exposed to 65 °C for 10 min to denature most of the heat-sensitive E. coli proteins. After a second centrifugation step at 50,000 × g for 60 min, the supernatant was applied to a 1-ml nickel column (HiTrap HP, GE Healthcare). Bound proteins were eluted with an increasing imidazole concentration up to 500 mm. Further purification was achieved by gel filtration chromatography (HiLoad 16/60 Superdex 75, GE Healthcare) with 25 mm HEPES (pH 7.5), 0.3 m NaCl, and 15% glycerol.
The promoter region of pf1089 was amplified by PCR using primers 1089Fd150 (5′-CAAACCTAAGCTCTGAACTACAGAG-3′) and 1089Rd150 (5′-TAAATGCCCGTATATTTAGGTGC-3′). The resulting fragment was cloned into the SmaI restriction site of the pUC19 vector to create pf1089_MUR9. The gdhC20 plasmid was used for control experiments (25). The mutated promoter templates pf1089Mu1_MUR10, pf1089Mu2_MUR11, gdhC20Mu4_MUR12, and gdhC20Mu5_MUR13 were constructed following the QuikChange II site-directed mutagenesis protocol (Stratagene). The pf1089Mu1 mutation was generated using primers BRE-Mu-1089F (5′-CCTAAGCTCTGAACTACAGAGTTTGCGAAAAGTATAAATACCTCTACCTCC-3′) and BRE-Mu-1089R (5′-GGAGGTAGAGGTATTTATACTTTTCGCAAACTCTGTAGTTCAGAGCTTAGG-3′). The pf1089Mu2 and the gdhC20Mu4 mutations were generated using primer pairs TATA-Mu-1089F (5′-CTACAGAGTTTGACCTGAGTTTATATACCTCTACCTCCATTTAA-3′)/TATA-Mu-1089R (5′-TTAAATGGAGGTAGAGGTATATAAACTCAGGTCAAACTCTGTAG-3′) and gdhF (5′-CAAAAGGATTTCCACTCTTGTTTACACCTGGCTTTATATAGGCTATTGCCC-3′)/BRE-Mu-gdhR (5′-GGGCAATAGCCTATATAAAGCCAGGTGTAAACAAGAGTGGAAATCCTTTTG-5′). For the mutant gdhC20Mu5, the 5′-phosphorylated primers Bdst-BRE-Mu-gdhF (5′-GAGTTTCACCTGGCTTTATATAGGCTATTGCCCAAAAATGTATCG-3′) and Bdst-BRE-Mu-gdhR (5′-TGTAGTTCAGAGCTTTTGTTTATTTGATTAGGCTCAAAGAATC-3′) and pUC19/gdhC20 as a template were used in combination with the Phusion® site-directed mutagenesis kit protocol (Finnzymes). The resulting plasmids were verified by sequencing analysis.
Electrophoretic Mobility Shift Assay
DNA templates were obtained from genomic DNA by PCR amplification with the corresponding primer pairs. One of the two primers was labeled at the 5′-end with 6-FAM (6-carboxyfluorescein) or Cy5. 5 nm DNA was incubated with 250 nm recombinant PF1088 in a 15-μl reaction volume containing the following buffer: 10% glycerol, 80 mm HEPES (pH 7.5), 5 mm MgCl2, 0.2 mm EDTA, 0.5 m KCl, 40 μg/ml BSA, and 50 μg/ml HindIII-digested λ DNA as a competitor. The reactions were incubated for 20 min at 70 °C and analyzed using a nondenaturing 5% polyacrylamide gel. After electrophoresis, the DNA fragments were visualized with a Fujifilm FLA-5000 fluorescence imager.
DNase I Footprints
13 nm template DNA and 1.45 μm PF1088 were incubated under the conditions used for the EMSAs. 0.01 unit of DNase I (1 μl; Fermentas) was added for 1–5 min at 37 °C, and the reaction was terminated by the addition of 95% formamide. After the addition of 0.3 m sodium acetate and 2 mg/ml glycogen (final concentrations), the DNA was precipitated with ethanol and resuspended in 3 μl of formamide buffer. A DNA sequencing ladder was generated using the same primer as a molecular mass standard. Samples were loaded onto a 4.5% denaturing polyacrylamide gel and analyzed using an ABI 377 DNA sequencer.
Hydroxyl Radical Footprints
15.6 nm template DNA and 3.48 μm PF1088 were incubated in 25 μl of radical reaction buffer (40 mm HEPES (pH 7.5), 0.1 mm EDTA, 0.15 m KCl, and 2.5 mm MgCl2) together with HindIII-digested λ DNA (1 ng/μl) as a competitor for 20 min at 70 °C. 5 mm Fe(II)(NH4)2(SO4)2·(H2O)6, 10 mm EDTA, 0.1 m sodium ascorbate, and 10% H2O2 were added to start the cleavage reaction. Note that it is important to add H2O2 last because this starts the generation of hydroxyl radicals. After 1–2 min of incubation at 70 °C, the reaction was stopped by the addition of 75 μl of radical stop buffer (13 mm Tris-HCl (pH 8.0), 4 mm EDTA, 1.5 mm thiourea, 0.3% SDS, and 1.3% glycerol). The DNA fragments were precipitated with ethanol and analyzed as described above.
In Vitro Transcription Assay
In vitro transcription reactions were performed in principle as described (26). For basal transcription, reaction mixtures were assembled in 25 μl (final volume) of transcription buffer (40 mm NaHEPES (pH 7.3), 0.1 mm EDTA (pH 8), 0.1 mg/ml BSA, 12.5 mm MgCl2, 250 mm KCl, 20 mm NaCl, 20 mm imidazole, and 20 mm potassium glutamate). 4 nm template DNA was combined with 95 nm TBP, 30 nm TFB, and 11 nm RNAP. Transcription was initiated by the addition of ATP, CTP, and GTP to a final concentration of 40 μm and UTP to a final concentration of 2.68 μm, including 2 μCi of [α-32P]UTP (3000 Ci/mmol). After incubation at 80 °C for 15 min, the transcripts was analyzed as described (26).
Primer Extension Analysis
Primer extension experiments were carried out with RNA from the in vitro transcription reaction of template pf1089 and the 5′-FAM-labeled primer Fd50_FAM (5′-TATTAAAGGGAATTGTTGATACTCTTAAG-3′). After denaturing RNA and primers at 70 °C for 5 min, dNTPs and reverse transcriptase (RNase H−, Promega) were added in the recommended buffer and incubated at 50 °C for 60 min. The cDNA was then purified by ethanol precipitation, resuspended in 4 μl of loading buffer (1:4 dextran blue/formamide), and analyzed on a denaturing urea-4.5% polyacrylamide gel together with sequencing reactions performed with the same primer. The products were analyzed using an ABI 377 sequencer.
RESULTS
Electrophoretic Mobility Shift Analyses of PF1088
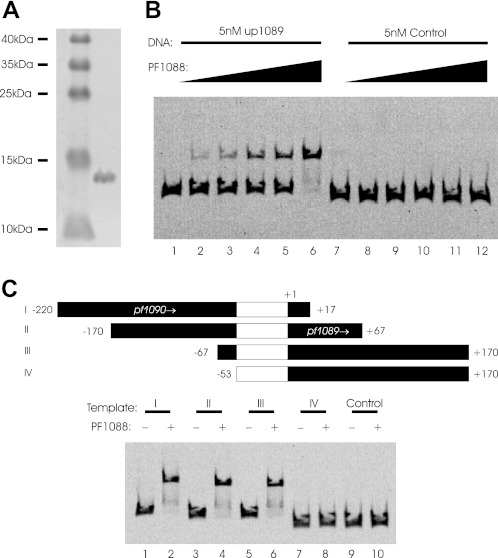
The PF1088 protein of P. furiosus was identified as a putative transcriptional regulator using a bioinformatic approach.3 To study the function of this putative regulator in more detail, we expressed the protein in E. coli using an N-terminal His6 tag for purification of the protein. The purified protein had an apparent molecular mass of 13.8 kDa under denaturing conditions (Fig. 1A). The DNA-binding properties of PF1088 were first characterized using electrophoretic mobility shift assays. As this gene is most likely located within an operon structure together with pf1089, we used the upstream sequence of pf1089 for binding experiments. Using 5 nm upstream DNA of pf1089, we obtained a specific DNA-protein complex in the presence of 50–250 nm PF1088 (Fig. 1B, lanes 1–6). Experiments with control DNA exhibited no specific binding of PF1088 (Fig. 1B, lanes 7–12).
FIGURE 1.
Electrophoretic mobility shift assays of PF1088. A, Coomassie Blue-stained 15% SDS-polyacrylamide gel of purified N-terminally His-tagged PF1088 with molecular mass standards. B, electrophoretic mobility shift assays on a nondenaturing 5% polyacrylamide gel. The assays contained 6-FAM-labeled DNA fragments (5 nm) either with pf1089 upstream sequence or, as a negative control, pf1189 upstream sequence. The concentration of PF1088 was varied from 50 nm (lanes 2 and 8) to 250 nm (lanes 6 and 12) in 50 nm steps. C, EMSA with PCR-amplified templates with different upstream sequences of pf1089. The upstream and downstream positions relative to the transcription start site are indicated in the upper part of the figure as black boxes for each template. The putative promoter and regulation region is indicated within the templates as white boxes. The absence (−) or presence (+) of 300 nm PF1088 in the reactions is indicated.
To localize the specific binding site of the putative transcriptional regulator in more detail, we prepared PCR-amplified templates with different upstream regions from positions −220 to −53 with respect to the transcription start site of pf1089 (determined in Fig. 3B). The upstream region could be deleted up to position −67 without any changes in the binding properties of PF1088 (Fig. 1C, compare lanes 1 and 6). In contrast, further deletion of the upstream region up to position −53 resulted in loss of the electrophoretic mobility shift (Fig. 1C, lanes 7 and 8). This indicates that the upstream boundary of the binding site of the transcriptional regulator PF1088 is located in the upstream sequence of pf1089 between positions −53 and −67.
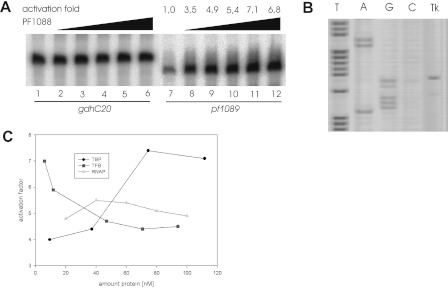
FIGURE 3.
Transcriptional analysis of PF1088. A, the function of PF1088 was analyzed by in vitro transcription experiments. The templates used for the experiments were gdhC20 (lanes 1–6) as a control and the promoter region of pf1089 (lanes 7–12). To analyze the influence of PF1088, an increasing amount of the protein was added to the reactions (46 nm, lanes 2 and 8; 93 nm, lanes 3 and 9; 139 nm, lanes 4 and 10; 186 nm, lanes 5 and 11; and 232 nm, lanes 6 and 12). The -fold transcription activation was quantified using a Fujifilm FLA-5000 fluorescence imager. B, primer extension with in vitro synthesized RNA of template pf1089 (Tk lane). The in vitro RNA was synthesized using the conditions described for basal transcription but without radiolabeled UTP and in the presence of 150 nm PF1088. The DNA sequencing reactions were generated with the same primer used in the primer extension experiment. The corresponding sequence lanes on the left are labeled with the complementary bases. The determined transcription start site is indicated by +1 in Fig. 2C. C, in vitro transcription experiments with different protein concentrations of TBP (circles; 10–115 nm), TFB (squares; 5–95 nm), and RNAP (triangles; 20–100 nm). For each concentration, the -fold activation was quantified, and the results are summarized in the diagram presented.
Mapping of Binding Site with Footprinting Techniques
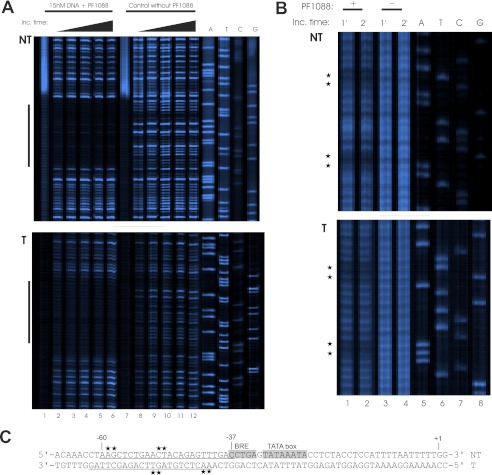
To determine the binding site in the upstream region of pf1089 more exactly, we applied DNase I footprinting experiments using fluorescently labeled DNA fragments of the non-template and template strands. As shown in Fig. 2A (upper panel), PF1088 protected a region from −60 to −37 on the non-template strand from DNase I digestion (compare lanes 2–6 with control lanes 8–12). On the template strand, the binding of PF1088 was in the range of positions −62 to −41 (Fig. 2A, lower panel).
FIGURE 2.
Identification of DNA-binding site of PF1088. DNase I and hydroxyl radical footprinting experiments were performed to identify the binding site of PF1088. A, the results from DNase I footprinting analysis of PF1088 at the non-template strand (NT; upper panel) and template strand (T; lower panel) are shown. The 6-FAM-labeled DNA fragments (15 nm) were incubated with 0.01 unit of DNase I for an increasing amount of time from 1 (lanes 2 and 8) to 5 min (lanes 6 and 12). The presence or absence of 1.4 μm PF1088 in the reaction is indicated at the top of the figure. DNA fragments were analyzed on a denaturing polyacrylamide gel using a sequencing reaction of the corresponding DNA as a size marker. The protected region of the DNA sequence is indicated on the left with black bars. B, the hydroxyl radical footprints of the non-template strand (upper panel) and template strand (lower panel) are shown. The incubation (Inc.) time of each reaction and the presence (+) or absence (−) of PF1088 are indicated at the top. Analysis of the DNA fragments was performed as described for the DNase I footprints. C, the summarized footprinting data of both strands are shown. The positions are indicated relative to the transcription start site. The TATA box and the corresponding BRE are highlighted in gray. The results from the DNase I footprinting experiments are underlined, and the positions identified with hydroxyl radical footprints are marked with asterisks.
For a more precise localization of the binding site, we performed hydroxyl radical footprinting assays. Hydroxyl radicals have the advantage of cleaving DNA without any sequence preference, and the small radicals allow a much higher resolution of DNA-protein contacts in comparison with DNase I (27). We used the same DNA fragments as used for the DNase I footprints. To our knowledge, these are the first experiments using hydroxyl radical footprints with fluorescently labeled DNA fragments. The problem is that the radicals also attack the chromatophore, and therefore, the strength of the signals goes down in comparison with the DNase I footprints. The results of the hydroxyl radical footprinting experiments are shown in Fig. 2B. Positions −49/−50 and −58/−59 on the non-template strand (Fig. 2B, upper panel) and positions −41/−42 and −50/−51 on the template strand (lower panel) were protected from cleavage (compare lanes 1 and 2 with control lanes 3 and 4). The results of the DNase I and hydroxyl radical footprinting experiments are summarized in Fig. 2C. The indicated boundary positions are relative to the mapped start site of transcription (Fig. 3B). As archaeal promoter elements are usually located about −37 to −25 nucleotides upstream of the transcription start site, we highlighted the putative BRE and the TATA box in Fig. 2C in gray (9, 28, 29). Taken together, the footprinting data clearly indicate that the binding site of PF1088 is located directly upstream of the archaeal promoter, consistent with the assumption that this protein is most likely an activator of archaeal transcription.
PF1088 Is a Transcriptional Activator
To investigate the effects of PF1088 on transcription, we employed an in vitro transcription system reconstituted from highly purified RNAP from P. furiosus and recombinant transcription factors TBP and TFB (26). Using the upstream sequence of pf1089 as a template in in vitro transcription reactions, we confirmed that this gene can be weakly transcribed in our system and that the addition of PF1088 increases the transcription product (Fig. 3A, compare lanes 7 and 8). Quantification of the transcripts with increasing amounts of PF1088 revealed an ∼3.5–7-fold activation. As expected, using the gdh promoter as a template, the addition of increasing amounts of PF1088 had no effect on transcription efficiency (Fig. 3A, lanes 1–7). These data clearly demonstrate that the transcriptional regulator PF1088 is able to activate transcription specifically at the pf1089 promoter.
To localize the binding site of PF1088 with regard to the promoter of the template pf1089, we used a primer extension experiment with in vitro RNA synthesized in the presence of PF1088 (Fig. 3B). The mapped transcript started with a guanine nucleotide, and a preceding thymine is in perfect agreement with the consensus sequence of an archaeal initiator element (28, 29).
The transcription start site and the derived promoter sequence of pf1089 are indicated in the DNA sequence of Fig. 2C. The mapped position of the transcription start site confirmed that the binding site of PF1088 is located directly upstream of the archaeal promoter and is therefore in perfect agreement with the finding that this protein is a transcriptional activator.
To gain further insight into the mechanism of activation, we performed in vitro transcription experiments with varying concentrations of RNAP, TBP, and TFB. For each experiment, the -fold activation was determined. Fig. 3C summarizes the results. Varying the concentration of RNAP from 20 to 100 nm did not dramatically change the -fold activation (Fig. 3C, triangles). As expected, reduced concentrations of TBP decreased the -fold activation because, in this case, the binding of TBP is most likely a rate-limiting step (Fig. 3C, circles). The most interesting result from these experiments is that limited TFB concentrations are particularly sensitive to PF1088 activation, whereas increasing concentrations reduce the level of activation (Fig. 3C, squares). Therefore, it is tempting to speculate that the presence of PF1088 stimulates binding of TFB to the promoter.
PF1088 Promotes Binding of TFB
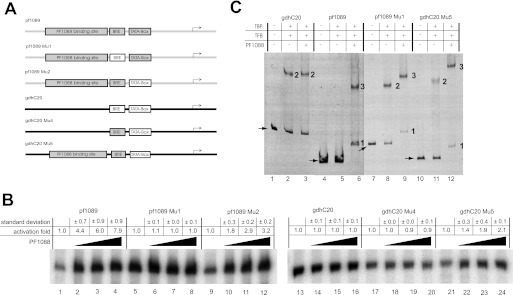
The BRE located directly upstream of the TATA box plays an important role in the recruitment of TFB to the DNA-TBP complex (9). Recently, the sequence 5′-VRAAA-3′ was proposed as a consensus sequence for the BRE in Pyrococcus (30). As the most likely corresponding BRE (5′-CCTGA-3′) of the pf1089 promoter shows some distinct deviations from the consensus sequence, we assume that this element has a reduced affinity for TFB recruitment. To test this hypothesis, mutational analysis of the pf1089 promoter elements was performed by introducing promoter elements of the strong gdh promoter (Fig. 4A).
FIGURE 4.
Transcription activation mechanism of PF1088. A, schematic drawing of the mutated templates used for in vitro transcription and EMSA experiments. Template pf1089 and the corresponding mutants are shown in gray, and template gdhC20 and the corresponding mutants are shown in black. The important elements (PF1088-binding site, BRE, and TBP) of the mutated templates are also indicated in gray if they originated from template pf1089 (white elements are from template gdhC20). The position of the transcription start site is labeled with arrows. B, in vitro transcription with the mutated templates pf1089Mu1, pf1089Mu2, gdhC20Mu4, gdhC20Mu5, and pf1089 as well as gdhC20 as a control. The influence of PF1088 was analyzed with three different concentrations (58 nm, lanes 2, 6, 10, 14, 18, and 22; 116 nm, lanes 3, 7, 11, 15, 19, and 23; and 232 nm, lanes 4, 8, 12, 16, 20, and 24). The -fold activation with S.D. are indicated at the top. Each experiment was repeated three times. C, EMSAs with the mutated sequences pf1089Mu1, gdhC20Mu5, and pf1089 as well as gdhC20 as a control. The arrows indicate free DNA. The numbers indicate DNA-PF1088 (1), DNA-TBP-TFB (2), and DNA-TBP-TFB-PF1088 (3) complexes. The presence of 465 nm TBP, 59 nm TFB, or 483 nm PF1088 is indicated at the top.
The exchange of the weak BRE of the pf1089 promoter with the strong BRE of the gdh promoter with the BRE consensus sequence 5′-CGAAA-3′ resulted in a strong increase in transcription of the template pf1089Mu1, even in the absence of the activator PF1088 (Fig. 4B, compare lanes 1 and 5). Furthermore, the addition of PF1088 did not increase the strength of the transcription signal of the pf1089Mu1 template (Fig. 4B, lanes 6–8). Transcription experiments performed with the gdh promoter with the weak BRE site from the pf1089 promoter (template gdhC20Mu4) showed a reduced activity in comparison with the corresponding wild-type gdh promoter (Fig. 4B, lanes 13 and 17). As expected, the presence of PF1088 had no influence on transcription of this template (Fig. 4B, lanes 18–20) because the binding site of PF1088 is missing. The introduction of the PF1088-binding site into the gdh template (template gdhC20Mu5) enabled PF1088-dependent activation of transcription by up to 2-fold (Fig. 4B, lanes 21–24). This -fold activation is in a similar range to that obtained for the pf1089 template with the strong TATA box from the gdh promoter (pf1089Mu2) (Fig. 4B, lanes 9–12). We suggest that, in these cases, the presence of the strong TATA box shifted the rate-limiting step in transcription initiation from TBP binding to TFB recruitment, making activation of these mutants still possible. In contrast, the binding of TBP to DNA is the rate-limiting step for the mutant with the weak TATA box, and therefore, no activation was observed.
To gain a further understanding of the underlying mechanisms, we performed additional experiments analyzing a subset of these mutated templates by EMSAs. We found a striking difference between the strong gdh and weak pf1089 promoters. In the case of the pf1089 promoter, no stable DNA-TBP-TFB shift could be observed (Fig. 4C, compare lanes 2 and 5). The addition of PF1088 did not change the composition of the DNA-TBP-TFB complex on the gdh promoter but allowed the formation of specific DNA-PF1088 as well as DNA-TBP-TFB-PF1088 complexes on the pf1089 template (Fig. 4C, lanes 3 and 6). (The sizes of the DNA fragments are different, and therefore, these distinct complexes were located within the gel at different positions.) Using pf1089Mu1 with the strong BRE as a template resulted in a specific TBP-TFB shift (Fig. 4C, lane 8). This suggests that the sequence of the BRE provides a significant contribution to the stability of the TBP-TFB complex. The addition of PF1088 led to the formation of a further retarded band, indicating the formation of a stable TBP-TFB-PF1088 complex (Fig. 4C, lane 9). Similar behavior was observed for the gdhC20Mu5 template (Fig. 4C, lanes 10–12). The additional complexes seen in lanes 9 and 12 represent DNA-bound PF1088.
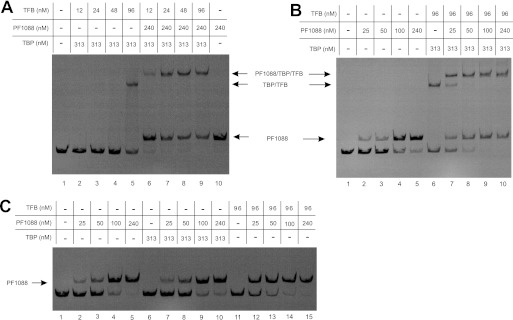
As the mutated template pf1089Mu1 with the strong BRE allowed the analysis of stable TBP-TFB complexes, we used this template for further EMSA experiments (Fig. 5). For the formation of a stable DNA-TBP-TFB complex, a TFB concentration of 96 nm was required (Fig. 5A, lane 5). In the presence of the regulator, a similar amount of shifted complexes was observed at a 4-fold reduced TFB concentration (24 nm), but the shifted complexes showed lower mobility due to the additional binding of PF1088 (Fig. 5A, compares lanes 5 and 7). These data suggest that PF1088 stimulates binding of TFB to the BRE. In the next step, we analyzed whether a promoter-bound TBP-TFB complex is also able to promote the recruitment of PF1088. In the absence of TBP and TFB, 240 nm PF1088 was an almost saturating concentration (Fig. 5B, lane 5). In the presence of TBP and TFB, only 50 nm was required for almost saturating complex formation (Fig. 5B, lane 8), indicating that bound TBP-TFB stimulates regulator recruitment. To provide conclusive evidence that PF1088 is able to interact with TFB and not with TBP, we analyzed the influence of TBP and TFB in separate experiments (Fig. 5C). The presence of 313 nm TBP had no influence on the binding of PF1088 (Fig. 5C, compare lanes 2–5 with lanes 7–10). In contrast, TFB stimulated the binding of PF1088 even in the absence of TBP (compare lane 7 in Fig. 5B with lane 12 in Fig. 5C). This finding strongly confirms the conclusion that TFB is able to interact with PF1088. Taken together, the transcriptional and electrophoretic mobility shift analyses of the promoter mutants demonstrate that the bound transcriptional regulator PF1088 is able to activate transcription by compensating for the negative effects of a weak BRE.
FIGURE 5.
Electrophoretic mobility shift assays. All experiments were performed using 3 nm pf1089Mu1 Cy5-labeled DNA fragments. The presence of TBP, TFB, or PF1088 is indicated at the top. The corresponding complexes are labeled with arrows, and the proteins bound to DNA are indicated.
DISCUSSION
The data presented clearly demonstrate that PF1088 is a sequence-specific transcriptional activator that is able to facilitate TFB binding to the DNA-TBP complex. For efficient activation, the PF1088 protein must be bound to the corresponding binding sequence directly upstream of a weak BRE. In contrast, for promoters harboring a strong BRE, PF1088-induced activation is dispensable. These results, along with the findings that the binding site of PF1088 is located directly upstream of the BRE and that the presence of TFB stimulates DNA binding of PF1088 (Fig. 5C), indicate that PF1088 most likely stabilizes bound TFB by a direct protein-protein interaction. This hypothesis is further supported by the result that it was not possible to detect a stable DNA-TBP-TFB gel shift complex when the pf1089 promoter was used as a template in the DNA binding experiments (Fig. 4C, lane 5). A stable complex could be obtained only in the presence of PF1088 or with a mutated strong BRE sequence (Fig. 4C). From these results, one can also conclude that the BRE is not only necessary to determine the direction of transcription (9, 31) but also plays a major role in the stability of the DNA-TBP-TFB complex. Crystal structure analyses with DNA-TBP-TFB complexes and photo-cross-linking experiments indicated that TFB interacts with promoter DNA not only in the BRE region upstream of the TATA box but also downstream near the transcription start site (32–34). The results mentioned above suggest that this downstream interaction of TFB has only a minor influence on DNA-TBP-TFB complex stability. Furthermore, the interaction between TFB and sequences downstream of the TATA box seems not to be highly sequence-specific because no consensus sequences have been identified in this region.
A similar activation mechanism was recently suggested for the araS promoter of Sulfolobus solfataricus (35). Using a gene reporter system, Peng et al. (35) demonstrated that in vivo activation of the araS promoter, consisting of a TATA box and a BRE with strong deviations from the Sulfolobus consensus sequence, strictly depends on the presence of the ara-box as an upstream binding sequence. Furthermore, substitution of the endogenous BRE with a BRE consensus sequence dramatically increases the basal promoter activity in the absence of the ara-box. These in vivo results are in agreement with our biochemical analysis. As the ara-box is also located directly upstream of the BRE, we suggest that the hitherto unknown ara-box-binding protein and PF1088 are members of a group of archaeal activators we refer to as TFB recruitment factors, and for the Pyrococcus regulator identified here, we suggest the designation TFB-RF1.
Ptr2 is the first example of an archaeal transcriptional activator whose mechanism of activation was elucidated by a detailed biochemical in vitro analysis. This protein belongs to the Lrp family, binds cooperatively to specific sequence elements upstream of TATA boxes, and stimulates in vitro transcription at weak promoters (18). In these cases, the binding of TBP to the TATA box is the rate-limiting step, and bound Ptr2 provides an additional interaction platform for TBP and therefore helps to recruit TBP to the TATA box (18, 19). The detailed biochemical analysis of this activator clearly demonstrates that this protein works with a different activation mechanism and therefore, according to the classification suggested above, should be referred to as a TBP recruitment factor. A similar mechanism is most likely used by the transcriptional activator GvpE in Halobacterium salinarum (20). Whether other transcriptional activators of the Lrp family, such as LysM and S. solfataricus LrpB, also use this form of activation is currently not known (36, 37). Further studies exploring the mechanisms of transcription activation in Archaea should shed light on this field. In this context, it would also be interesting to know whether transcriptional activators exist that are able to promote the recruitment of both TBP and TFB. Interaction with both basal transcription factors was recently described for a repressor involved in the regulation of nitrogen metabolism in Methanosarcina mazei Gö1 (38). Meanwhile, more and more members are being identified for an additional group of archaeal transcriptional regulators that can act as both repressors and activators depending on the location of the binding site, including TrmBL1, Tgr, and SurR (21, 22, 39). In these cases, the mechanism of activation has yet to be elucidated.
The protein described here is highly conserved within the Thermococcales, although its physiological function is still unclear. Furthermore, the structure of the corresponding protein in Pyrococcus horikoshii (77% amino acid identity) has been previously published, but without any additional information about the function of the protein (40). The protein belongs to the ArsR family of transcriptional regulators (40), a family of metalloregulators that are widespread in bacteria (reviewed in Ref. 41). Many of these proteins are involved in metal homeostasis in the cell, either by controlling the level of biologically important metal ion concentrations in the cell or by inducing the expression of proteins necessary for the export of toxic metals. The main features of this family are a winged helix-turn-helix motif and a metal-sensing domain that has evolved to bind different metals. In EMSA experiments, it was shown that the presence of metal ions like Zn2+ and Ni2+ did not influence the binding behavior of PF1088 (data not shown). Therefore, we assume that PF1088 performs a different function in Pyrococcus. The regulator itself seems to be co-transcribed with pf1089, whose function is also unknown. Furthermore, under the conditions we used for growth of Pyrococcus on starch or pyruvate, it was not possible to detect the expression of the pf1089 promoter by primer extension experiments (Fig. 3B). To elucidate the physiological role of this activator and the corresponding targets of regulation, overexpression of PF1088 is in progress using a recently developed genetic system for P. furiosus (23).
This work was supported by Deutsche Forschungsgemeinschaft Grant TH 422/13-1.
M. T. Weirauch, S. M. Ochs, W. Hausner, M. Thomm, and T. M. Lowe, manuscript in preparation.
- RNAP
- RNA polymerase
- TBP
- TATA-binding protein
- TFB
- transcription factor B
- BRE
- TFB-responsive element.
REFERENCES
- 1. Grohmann D., Hirtreiter A., Werner F. (2009) Molecular mechanisms of archaeal RNA polymerase. Biochem. Soc. Trans. 37, 12–17 [DOI] [PubMed] [Google Scholar]
- 2. Thomm M., Hausner W. (2007) In Archaea: Evolution, Physiology and Molecular Biology (Garrett R., Klenk H.-P., eds) pp. 185–198, Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
- 3. Bartlett M. S. (2005) Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 8, 677–684 [DOI] [PubMed] [Google Scholar]
- 4. Geiduschek E. P., Ouhammouch M. (2005) Archaeal transcription and its regulators. Mol. Microbiol. 56, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 5. Bell S. D., Jackson S. P. (2001) Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4, 208–213 [DOI] [PubMed] [Google Scholar]
- 6. Hausner W., Wettach J., Hethke C., Thomm M. (1996) Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 271, 30144–30148 [DOI] [PubMed] [Google Scholar]
- 7. Qureshi S. A., Bell S. D., Jackson S. P. (1997) Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 16, 2927–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qureshi S. A., Jackson S. P. (1998) Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1, 389–400 [DOI] [PubMed] [Google Scholar]
- 9. Bell S. D., Kosa P. L., Sigler P. B., Jackson S. P. (1999) Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. U.S.A. 96, 13662–13667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell S. D., Brinkman A. B., van der Oost J., Jackson S. P. (2001) The archaeal TFIIEα homolog facilitates transcription initiation by enhancing TATA box recognition. EMBO Rep. 2, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanzelka B. L., Darcy T. J., Reeve J. N. (2001) TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183, 1813–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grünberg S., Bartlett M. S., Naji S., Thomm M. (2007) Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 282, 35482–35490 [DOI] [PubMed] [Google Scholar]
- 13. Aravind L., Koonin E. V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 27, 4658–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu W., Vierke G., Wenke A. K., Thomm M., Ladenstein R. (2007) Crystal structure of the archaeal heat shock regulator from Pyrococcus furiosus: a molecular chimera representing eukaryal and bacterial features. J. Mol. Biol. 369, 474–488 [DOI] [PubMed] [Google Scholar]
- 15. Bell S. D. (2005) Archaeal transcription regulation: variation on a bacterial theme? Trends Microbiol. 13, 262–265 [DOI] [PubMed] [Google Scholar]
- 16. Thomm M. (2007) In Archaea: Molecular and Cellular Biology (Cavicchioli R., ed) pp. 139–157, ASM Press, Washington, D.C [Google Scholar]
- 17. Peeters E., Charlier D. (2010) The Lrp family of transcriptional regulators in archaea. Archaea 2010, 750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ouhammouch M., Dewhurst R. E., Hausner W., Thomm M., Geiduschek E. P. (2003) Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 100, 5097–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ouhammouch M., Langham G. E., Hausner W., Simpson A. J., El-Sayed N. M., Geiduschek E. P. (2005) Promoter architecture and response to a positive regulator of archaeal transcription. Mol. Microbiol. 56, 625–637 [DOI] [PubMed] [Google Scholar]
- 20. Teufel K., Pfeifer F. (2010) Interaction of transcriptional activator GvpE with TATA box-binding proteins of Halobacterium salinarum. Arch. Microbiol. 192, 143–149 [DOI] [PubMed] [Google Scholar]
- 21. Lipscomb G. L., Keese A. M., Cowart D. M., Schut G. J., Thomm M., Adams M. W., Scott R. A. (2009) SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulfur metabolism in Pyrococcus furiosus. Mol. Microbiol. 71, 332–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S. J., Surma M., Hausner W., Thomm M., Boos W. (2008) The role of TrmB and TrmB-like transcriptional regulators for sugar transport and metabolism in the hyperthermophilic archaeon Pyrococcus furiosus. Arch. Microbiol. 190, 247–256 [DOI] [PubMed] [Google Scholar]
- 23. Waege I., Schmid G., Thumann S., Thomm M., Hausner W. (2010) Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Environ. Microbiol. 76, 3308–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., Thomm M., Cramer P. (2009) RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 [DOI] [PubMed] [Google Scholar]
- 25. Spitalny P., Thomm M. (2003) Analysis of the open region and of DNA-protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 278, 30497–30505 [DOI] [PubMed] [Google Scholar]
- 26. Hethke C., Bergerat A., Hausner W., Forterre P., Thomm M. (1999) Cell-free transcription at 95°: thermostability of transcriptional components and DNA topology requirements of Pyrococcus transcription. Genetics 152, 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. (1987) Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 155, 537–558 [DOI] [PubMed] [Google Scholar]
- 28. Hausner W., Frey G., Thomm M. (1991) Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNAVal gene of Methanococcus vannielii. J. Mol. Biol. 222, 495–508 [DOI] [PubMed] [Google Scholar]
- 29. Hain J., Reiter W. D., Hüdepohl U., Zillig W. (1992) Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 20, 5423–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van de Werken H. J., Verhees C. H., Akerboom J., de Vos W. M., van der Oost J. (2006) Identification of a glycolytic regulon in the archaea Pyrococcus and Thermococcus. FEMS Microbiol. Lett. 260, 69–76 [DOI] [PubMed] [Google Scholar]
- 31. Littlefield O., Korkhin Y., Sigler P. B. (1999) The structural basis for the oriented assembly of a TBP-TFB-promoter complex. Proc. Natl. Acad. Sci. U.S.A. 96, 13668–13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsai F. T., Sigler P. B. (2000) Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 19, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartlett M. S., Thomm M., Geiduschek E. P. (2004) Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 279, 5894–5903 [DOI] [PubMed] [Google Scholar]
- 34. Renfrow M. B., Naryshkin N., Lewis L. M., Chen H. T., Ebright R. H., Scott R. A. (2004) Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J. Biol. Chem. 279, 2825–2831 [DOI] [PubMed] [Google Scholar]
- 35. Peng N., Xia Q., Chen Z., Liang Y. X., She Q. (2009) An upstream activation element exerting differential transcription activation on an archaeal promoter. Mol. Microbiol. 74, 928–939 [DOI] [PubMed] [Google Scholar]
- 36. Brinkman A. B., Bell S. D., Lebbink R. J., de Vos W. M., van der Oost J. (2002) The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J. Biol. Chem. 277, 29537–29549 [DOI] [PubMed] [Google Scholar]
- 37. Peeters E., Albers S. V., Vassart A., Driessen A. J., Charlier D. (2009) Ss-LrpB, a transcriptional regulator from Sulfolobus solfataricus, regulates a gene cluster with a pyruvate ferredoxin oxidoreductase-encoding operon and permease genes. Mol. Microbiol. 71, 972–988 [DOI] [PubMed] [Google Scholar]
- 38. Weidenbach K., Ehlers C., Kock J., Schmitz R. A. (2010) NrpRII mediates contacts between NrpRI and general transcription factors in the archaeon Methanosarcina mazei Gö1. FEBS J. 277, 4398–4411 [DOI] [PubMed] [Google Scholar]
- 39. Kanai T., Akerboom J., Takedomi S., van de Werken H. J., Blombach F., van der Oost J., Murakami T., Atomi H., Imanaka T. (2007) A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J. Biol. Chem. 282, 33659–33670 [DOI] [PubMed] [Google Scholar]
- 40. Okada U., Sakai N., Yao M., Watanabe N., Tanaka I. (2006) Structural analysis of the transcriptional regulator homolog protein from Pyrococcus horikoshii OT3. Proteins 63, 1084–1086 [DOI] [PubMed] [Google Scholar]
- 41. Osman D., Cavet J. S. (2010) Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27, 668–680 [DOI] [PubMed] [Google Scholar]