Background: Proteasomes have six non-identical ATPases that move and unfold protein substrates.
Results: Mutating homologous substrate contact residues of each ATPase has diverse effects on degradation and cell growth.
Conclusion: Although homologous, each of the six has distinguishable functions.
Significance: Structurally similar elements within a complex molecular machine can evolve distinct tasks, diversifying the range of accessible cellular responses.
Keywords: ATP-dependent Protease, ATPases, Cell Growth, Mutagenesis, Proteasome, Protein Degradation, Yeast, Translocation, Unfolding, Proteolysis
Abstract
Degradation by proteasomes involves coupled translocation and unfolding of its protein substrates. Six distinct but paralogous proteasome ATPase proteins, Rpt1 to -6, form a heterohexameric ring that acts on substrates. An axially positioned loop (Ar-Φ loop) moves in concert with ATP hydrolysis, engages substrate, and propels it into a proteolytic chamber. The aromatic (Ar) residue of the Ar-Φ loop in all six Rpts of S. cerevisiae is tyrosine; this amino acid is thought to have important functional contacts with substrate. Six yeast strains were constructed and characterized in which Tyr was individually mutated to Ala. The mutant cells were viable and had distinct phenotypes. rpt3, rpt4, and rpt5 Tyr/Ala mutants, which cluster on one side of the ATPase hexamer, were substantially impaired in their capacity to degrade substrates. In contrast, rpt1, rpt2, and rpt6 mutants equaled or exceeded wild type in degradation activity. However, rpt1 and rpt6 mutants had defects that limited cell growth or viability under conditions that stressed the ubiquitin proteasome system. In contrast, the rpt3 mutant grew faster than wild type and to a smaller size, a defect that has previously been associated with misregulation of G1 cyclins. This rpt3 phenotype probably results from altered degradation of cell cycle regulatory proteins. Finally, mutation of five of the Rpt subunits increased proteasome ATPase activity, implying bidirectional coupling between the Ar-Φ loop and the ATP hydrolysis site. The present observations assign specific functions to individual Rpt proteins and provide insights into the diverse roles of the axial loops of individual proteasome ATPases.
Introduction
Proteasomes are multicomponent complexes that carry out regulated proteolysis in eukaryotic cells. They help to establish steady state levels of proteins, are key to clearing regulatory proteins whose function is no longer required, and destroy misfolded or damaged proteins that can cause cell pathology. The full complex, of ∼2.4 MDa, contains two kinds of subcomplexes (1). Protein degradation takes place within the catalytic complex, also termed 20S. This consists of a stack of four seven-member rings, of composition α1–7:β1–7:β1–7:α1–7, in which each of the seven individual α and β proteins is distinct but homologous (2). Together, these form a hollow barrel-shaped structure. Protein hydrolysis takes place at catalytic sites positioned between the pair of β rings. Because these catalytic sites are sequestered from the general cellular environment, substrate proteins must gain access to the interior for degradation to occur. Entry into the barrel lies through a pore centered at the axis of the α1–7 ring that caps each end of the barrel. The diameter of the pore is sufficient to accommodate an extended polypeptide but not a native folded protein. Degradation of such proteins therefore requires that they arrive at the proteasome and be grasped, unfolded, and threaded through the α ring pore.
These functions are performed by the regulatory complex, also termed 19S. It, in turn, is composed of two subcomplexes (3): the lid, which contains 13 proteins, and the base, with 10 proteins. Within the base are six distinct but homologous ATPases, whose action will be the focus of this paper. These ATPases, in yeast designated Rpt1p to -6p, form a ringlike heterohexamer (4) that docks with the α1–7 ring, in such a way that the two ring axes are nearly (but not precisely (5, 6)) co-axial. Together, these juxtaposed rings form a translocation channel. Fueled by ATP hydrolysis, the Rpt ring moves and unfolds substrate proteins and transfers them into the degradation chamber. These mechanisms have been studied in the compartmental proteases of bacteria, which are structurally much simpler than proteasomes (e.g. ClpXP, ClpAP, HslUV, and Lon) (7). These findings are likely to be informative in understanding how proteasomes work because there is a general similarity of architecture between the bacterial and eukaryotic complexes. For example, the ClpXP protease contains a proteolytic barrel formed by two heptameric rings of ClpP, and the translocase is a homohexamer of ClpX ATPase subunits, which mounts co-axially on the ClpP14 complex.
The mechanisms by which the proteasome unfolds and translocates substrate through its ATPase ring are still not well understood. Translocation and unfolding are thought to be coupled. Pulling by the ATPase ring on an accessible and unstructured portion of the substrate forces its folded domains to unravel (8–10). Pulling is carried out by a mobile loop in direct contact with substrate, called the Ar-Φ (aromatic-aliphatic) loop (11, 12), pointing toward the center of the Rpt channel. Its movements, coupled to the hydrolysis cycle of the ATPase of which it is a part, delivers a vectorial pulling force to substrate. In ClpAP, these loops contact substrate, and its tyrosine residue, the conserved Ar of the Ar-Φ loop, is essential for translocation and unfolding (13). In ClpXP, the conserved tyrosine residue was also shown to be responsible for mechanical work, gripping substrates during unfolding and translocation through a mechanism linked to ATP hydrolysis (14). In the proteasome, the Ar-Φ loops of the six ATPases contain the sequence (K/M)Y(V/L/I)G, where the aromatic-aliphatic pair (Ar-Φ) are Tyr-Val/Leu/Ile. Do the invariant tyrosines of these proteasome loops perform the work of unfolding? Is the work done by the six Rpt loop tyrosines identically allocated among them or distinguishably different? In this paper, we mutated tyrosine to alanine in each Rpt Ar-Φ loop and studied the effect of the mutation on cell proliferation, on the proteasome itself, and on its ability to degrade proteins. The Rpt proteins were found to have distinguishable and specific functions.
EXPERIMENTAL PROCEDURES
Construction of rpt Mutants
All plasmid constructions utilized standard molecular biology techniques (15). The identities of DNA fragments generated by PCR were verified by sequencing. The details of cloning procedures or the sequences of the primers used for PCR constructions are available upon request. Generally, the gene coding rpt mutants was inserted into a pRS306 plasmid. Standard protocols were used for maintenance and manipulation of Saccharomyces cerevisiae strains (16). Yeast strain YAH96 (MATα his3-Δ200 leu2-3,112lys2–801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS) was used as the genetic background for construction of rpt mutants by replacement of the wild type copy by the pop-in/pop-out technique (Table 1). Each mutant gene included a restriction site modification that allowed recognition of the mutated form of the rpt gene after colony PCR.
TABLE 1.
Genotype of rpt mutant strains
| Strain | Genotype |
|---|---|
| YAH96 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS |
| JE03 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS Rpt1::rpt1 (Y283A) |
| MHY292 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS Rpt2::rpt2 (Y256A) |
| MHY293 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS Rpt3::rpt3 (Y246A) |
| MHY294 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS Rpt4::rpt4 (Y255A) |
| MHY295 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11–3xFLAG-HIS Rpt5::rpt5 (Y255A) |
| MHY296 | MATα his3-Δ200 leu2-3,112lys2-801 trp1-1 ura3-52 rpn11::RPN11-3xFLAG-HIS Rpt6::rpt6 (Y222A) |
Analysis of Yeast Growth
Cells were grown to an A600 of 0.5. 10-ml cultures were harvested and resuspended in 500 μl of H2O, and 10-fold dilution series were applied to plates as described below. Cell growth in liquid YPD was also followed by a spectrophotometer at A600. Aliquots were taken to measure dry weight (17). Cell size was analyzed with a Coulter Counter Z1 DIT (Beckman). For that purpose, cells were fixed by the addition of 3.7% formaldehyde and briefly sonicated to disperse clumps, and the size distribution was determined by counting the number of cells that scored within size windows between 2 and 10 μm in diameter in 0.5-μm steps.
In Vivo Degradation Assay
FLAG-ornithine decarboxylase (ODC)3 protein was expressed from p414-ADH-FLAG-ODC plasmid in wild type or rpt mutant strains. Cells were grown to an A600 of 0.7, and FLAG-ODC abundance was measured by loading 40 μg of yeast extract on a 12% denaturing gel and revealed by Western blot with anti FLAG-HRP antibodies (1:10,000; Sigma).
Protein Purification
Substrates containing an Rpn10 domain were expressed in Escherichia coli BL21 Rosetta strains (Novagen) as N-terminal hexahistidine-tagged proteins. 35S-Radiolabeled substrates were prepared and purified as described (9). 26S proteasomes were purified from wild type (YAH96) or mutated strains (see Table 1) in which a 3xFLAG tag has been appended to the C terminus of the Rpn11 subunit of the proteasome. Cells were grown to an A600 of 3–4 in 1.5 liters of YPD, harvested, and washed once with 50 ml of ice-cold water. The cell pellet was then resuspended in an equal volume of proteasome buffer (50 mm Tris, pH 7.5, 25 mm NaCl, 5 mm MgCl2, 2 mm ATP, 10% glycerol) supplemented with 2× Complete EDTA-free protease inhibitor mixture (Roche Applied Science) and lysed by five passes through a Microfluidics microfluidizer at a pressure of 100 p.s.i. Lysates were clarified by centrifugation at 20,000 × g at 4 °C for 40 min. 26S proteasomes were affinity-purified by incubation with 1.2 ml of M2 anti-FLAG affinity gel (Sigma) for 45 min at 4 °C. Bound proteasome complexes were washed with 200 column volumes of proteasome buffer and eluted from the resin for 30 min in an equal volume of proteasome buffer supplemented with 0.2 mg/ml 3xFLAG peptide (Sigma). The eluates were concentrated on 100 kDa molecular mass cut-off Centricon concentrators. Proteasome composition was determined by denaturing gel electrophoresis followed by Coomassie staining. Complex assembly was assessed by native gel electrophoresis (18). Proteasome molarity was calculated using a molecular mass of 2,400 kDa.
In Vitro Degradation Assay
Degradation reactions containing 1 μm radiolabeled protein substrates were carried out at 30 °C by 50 nm 26S proteasome in a buffer (25 mm HEPES, pH 7.5, 100 mm KCl, 20 mm MgCl2, and 10% glycerol) containing 2 mm DTT, 5 mm ATP, and an ATP-regenerating system (7.5 mm phospho(enol)pyruvic acid, 1 mm NADH, and 0.5 unit of pyruvate kinase/lactate dehydrogenase mix (Sigma)). Time-dependent degradation was assessed by periodically removing 10-μl volumes from the reaction and precipitation in 140 μl of 20% TCA for 30 min on ice. After a 30-min centrifugation at 18,000 × g, the supernatant, containing the peptide products of proteolysis, was added to a scintillation vial for quantification of radioactivity. For each experiment, the total radioactivity present in substrate was determined by adding an aliquot of the degradation assay mixture to TCA, transferring the solution to a scintillation vial, and counting. Kinetic parameters were determined using SigmaPlot software.
ATPase Assays
Proteasome ATPase activity was measured in a coupled assay in 25 mm HEPES (pH 7.5), 100 mm KCl, 20 mm MgCl2, 10% glycerol, 18 milliunits/ml lactate dehydrogenase/pyruvate kinase (Sigma), 7.5 mm phospho(enol)pyruvic acid, 1 mm NADH, 2 mm DTT, and 5 mm ATP. Proteasome concentration was 50 nm. ATPase activity was followed by loss of NADH absorbance at 340 nm. ATP consumption was calibrated using an NADH standard curve, performed with each assay. Reactions were carried out in 96-well plates in a Molecular Devices Spectra Max spectrophotometer.
RESULTS
Cell Growth
Strains with each of the six intended Tyr/Ala mutations were constructed and proved to be viable. These were constructed by integrating a copy of the mutant rpt gene at the homologous wild type locus and then selecting for excision of one of the two copies. If the mutant gene does not strongly impair growth, screening cells that have undergone deletion by homologous recombination should yield roughly similar numbers of excision events that remove the wild type or mutant copy. That was the case. Additionally, sequencing revealed no additional mutations in the mutant rpt other than that introduced to mutate tyrosine to alanine. Consequently, recovery and viability of the six mutant strains is unlikely to be dependent on secondary extra- or intragenic mutations.
We tested growth under a variety of conditions (Fig. 1A). Dilution assays on YPD plates revealed relatively minor effects upon incubation at 30 °C. rpt6 was most impaired in growth, and rpt1 less so. At 37 °C, a temperature that provides heat stress for yeast, rpt1 and rpt6 were most strongly impaired. Canavanine is an arginine analog that is incorporated into proteins and causes their misfolding, which stresses the ubiquitin-proteasome system. The presence of canavanine at 30 °C caused strong growth impairment in rpt6 and almost as strong an effect in rpt1. Last, we tested the effect of low dose cycloheximide. This inhibitor of protein synthesis has been observed to stimulate the growth or viability of cells with impairments of proteasome function (19). Without cycloheximide, rpt6 grew less well than wild type; in its presence, the growth of rpt6 was similar to that of wild type.
FIGURE 1.
Yeast growth. A, serial 10-fold dilutions of liquid cultures were spotted onto plates with various media and incubated as indicated. Can and CHX, canavanine and cycloheximide (0.5 μg/ml), respectively. B, growth of wild type or mutant rpt strains at 30 °C in YPD was followed by measuring culture turbidity at 600 nm. C, size distribution of wild type and rpt3 strains. Samples were removed from the cultures shown in B at 8 or 30 h, and the distribution of cell diameters was determined by a Coulter Counter.
Plate dilution assays provide a measure of growth and viability in a specific culture milieu. To test the generality of these results, cells also were grown in suspension YPD culture with vigorous aeration at 30 °C. Growth was monitored by turbidity at A600. Cell density exhibited the classic time-dependent growth phases: rapid fermentative growth, a pause and resumption of growth at a reduced rate, and finally slowly approaching a fixed terminal density as nutrient sources are depleted (Fig. 1B). The growth of the mutants, except rpt3 and rpt6, was not distinguishable from wild type. The rpt6 cells grew slowly. More striking was the pattern of rpt3, which appeared to grow faster than wild type; its doubling times were 1.5 and 6.8 h before and after the shift, compared with 1.7 and 14.0 h for wild type. Hemocytometer counts (data not shown) were concordant with the A600 turbidity data; by both measures, rpt3 cell number at terminal density was about 200–300% greater than wild type. However, at terminal density, rpt3 cellular dry weight, despite the much higher cell count, was only 40% greater than that of wild type. This comparison suggested that rpt3 completes its cell cycle faster and divides at a smaller size. To test that directly, we measured cell size by Coulter Counter (Fig. 1C). During both fermentative and aerobic growth, rpt3 cells were indeed smaller than wild type. The modal diameters of rpt3 versus wild type in fermentative growth were 3.25 and 3.75 μm, respectively. The corresponding modal sizes after cells reached stationary phase were 3.25 and 4.25 μm. Linear dimension ratios must be cubed to infer relative volume. rpt3 volume is 65% of wild type during exponential growth and 45% in lag phase. Fast growth to small size has been extensively described in fungal mutants and in S. cerevisiae has been denoted as the whi phenotype.
Ubiquitin Accumulation
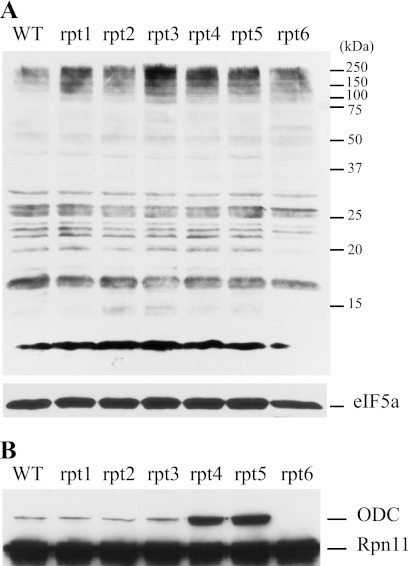
Cells with impairments in proteasome function typically accumulate a population of high molecular weight ubiquitin-conjugated proteins (20). We therefore tested cell extracts to assess the relative amount of such ubiquitin-modified proteins. Western blotting (Fig. 2A) revealed that, compared with wild type, all of the mutants contained increased amounts of high molecular weight ubiquitin-conjugated proteins. Among the mutants, rpt3, rpt4, and rpt5 were most markedly elevated. Qualitatively similar results were seen in three independent experiments.
FIGURE 2.
Impairment of degradation in vivo. Shown is accumulation of polyubiquitinated protein (A) and FLAG-ODC (B). Lysates were prepared from the indicated strains, separated by SDS-PAGE on a 12% gel, and analyzed by immunoblotting with an anti-ubiquitin antibody (or with antibody to eIF5a as loading control) (A) or anti-FLAG (B). Anti-FLAG recognize both FLAG-ODC and Rpn11-FLAG proteins.
Degradation of Model Substrate in Vivo
To test the ability of mutant cells to degrade a proteasome substrate, we assessed the steady-state level of mouse ODC. This heterologous protein was chosen for the following reasons. Its proteasomal degradation is independent of ubiquitination (21). This removes secondary changes in ubiquitin pools and their partitioning as a factor in turnover. Proteasomes and their associated proteins perform editing of ubiquitin conjugates (22). Using a substrate that does not depend on ubiquitin reduces the effects of such potential changes and focuses experimental observations on determinants of processing rates. ODC is a tightly folded protein (23). It therefore provides a stringent test of whether mutations impair translocation and unfolding (24). Finally, mouse ODC has a very short half-life in yeast, providing a sensitive test of any change in its turnover (25). FLAG-tagged ODC was expressed, and its relative level was determined by anti-FLAG Western blotting (Fig. 2B). rpt4 and rpt5 had strikingly increased amounts of ODC, compared with wild type or with rpt1, rpt2, and rpt3, which were similar to wild type. In rpt6, no ODC was detected under the conditions used. Proteasomal degradation is by far the dominant determinant of ODC steady-state and the principal mechanism of its metabolic regulation (26). These data are therefore consistent with the conclusion that the rpt4 and rpt5 cells have a reduced rate of ODC turnover, whereas rpt6 has increased degradation compared with wild type.
Abundance and Structural Integrity of rpt Mutant Proteasomes
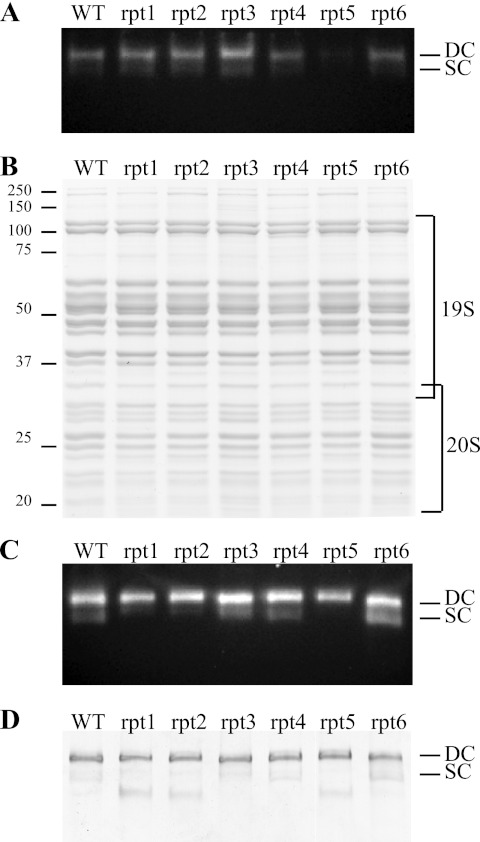
Based on the nature of the rpt mutants and the data presented thus far, it is plausible that these have an impairment in the ubiquitin-proteasome system and that such defects may be localized to proteasomes. To assess this, we fractionated cellular extracts on native gels and used an in-gel assay that depends on hydrolysis of a fluorogenic peptide by proteasomes (Fig. 3A). These gels resolved proteasomes that are singly capped (one regulatory complex per 20S catalytic complex) from those that are doubly capped (two regulatory complexes per 20S catalytic complex). In all of the extracts, the doubly capped form predominated, whereas lesser amounts of the singly capped form were also evident. Strikingly and reproducibly in multiple independent experiments, less rpt5 was observed to be present or active in crude extracts compared with all of the other rpt mutants and wild type. However, the mobility of the rpt5 proteasomes was not obviously different. Because of the relatively weak rpt5 signal, the presence or absence of the singly capped form in crude extracts could not be determined. Furthermore, Rpn11 is present in our strains with a FLAG tag, which is used for affinity purification (see below). The tagged form of Rpn11 is evident in the denaturing SDS-PAGE Western blot of ODC abundance (Fig. 2A) and there acts as a loading control. The amount of Rpn11p recovered from crude extracts is not diminished by the rpt5 mutation, suggesting that the reduced amount of rpt5 proteasomes observed by native gel functional analysis results from 26S disassembly associated with the cell breakage conditions used to prepare crude extracts for native gel analysis.
FIGURE 3.
Proteasome integrity, composition, and function. A, proteasome integrity after subcellular fractionation. Lysates were prepared from the indicated strains, and 50 μg were loaded on a 4% native gel. Proteasomes were visualized by incubating the gel with N-succinyl-LLVY-7-amino-4-methylcoumarin peptide, a fluorogenic proteasome substrate. B, 12% SDS-PAGE and Coomassie staining of 6 μg of purified wild type and mutated proteasomes. C and D, 4% native gel analysis of 2 μg of purified wild type and mutated proteasomes, revealed by peptidase activity (C) or by Coomassie staining (D). DC and SC, doubly capped and singly capped proteasomes, respectively.
To further determine the composition of the proteasomes and to perform biochemical experiments, we used the FLAG tag on the Rpn11p protein for affinity purification. Proteasomes of high purity were recovered. SDS-PAGE analysis of these revealed no marked differences between wild type and mutants in the conventional pattern of proteins (Fig. 3B). The nature of the affinity purification and the position of the tag utilized should recover doubly and singly capped proteasomes as well as free regulatory complexes. Native activity gels of the purified proteasomes (Fig. 3C) revealed results congruent with those observed in crude extracts. However, as equal amounts of purified proteasome were loaded, rpt5 was no longer underrepresented. Additionally, the cleaner backgrounds and larger amounts of the active protein more readily revealed variations in the ratio of singly to doubly capped forms. In all cases the doubly capped form predominated, but this was the exclusive form observed with the purified rpt5 proteasomes. Additionally, rpt1 and rpt2 showed ratios of the two forms that more strongly favored doubly capped proteasomes. After Coomassie staining (Fig. 3D), we observed that the rpt5 singly capped form was replaced by a non-active form corresponding in mobility to free 19S. We cannot confidently ascribe these different ratios among the rpt mutants to conditions of assembly present in cells or as a consequence of purification. In any case, the overall data support the conclusion that wild type and mutant purified proteasomes have a similar structure (forms other than doubly capped 26S are barely detectable by Coomassie staining), and their biochemical activities can be appropriately compared.
ATPase Activity of Purified Proteasomes
Unfolding and translocation of proteins by the proteasome depend on ATP. ATP binding (by the Walker A motif) and hydrolysis (Walker B motif) have been shown in other ATPases to produce conformational changes that are propagated to effecter elements; their action in turn moves or disassembles substrates. These general findings assure that the Ar-Φ loops move in coordination with one or more steps of the ATP binding and hydrolysis cycle. Because changes in these structures are coupled, mutating the Rpt Ar-Φ loops could affect ATP hydrolysis. We tested this by measuring the ATPase activity of the mutants (Fig. 4). Quantitative differences were observed. In rpt4 proteasomes, the rate of ATP hydrolysis decreased by 30%. Surprisingly, in all of the other rpt mutants, activity increased compared with wild type. In the most extreme case, rpt1 activity increased by about 120%. These findings suggest that Ar-Φ loops have, in the absence of substrate, a regulatory function that constrains the tempo of ATP hydrolysis.
FIGURE 4.
ATPase activity of the proteasome. ATPase activity was measured in the presence of 50 nm proteasome and in excess of ATP (5 mm). The catalytic constant (kcat) is the mean of three or four experiments. Error bars, S.D.
Degradation Kinetics of Model Substrates
Aside from its apparent regulatory effects on ATPase activity, the Ar-Φ loops and their conserved tyrosines act to contact substrate proteins, propelling their translocation and unfolding. These processes can be rate-limiting, especially for tightly folded substrate proteins (9). We therefore compared the ability of the wild type and rpt mutant proteasomes to degrade proteins. We have previously (9) described and characterized ubiquitin-independent proteasome substrates that contain, at their N-terminal extremity, Rpn10p (denoted R) and an unstructured C-terminal extension (denoted ext). Rpn10p is a native proteasome protein that binds substrates; the unstructured extension provides a region at which proteasome insertion and translocation are initiated (10). Two structurally similar substrates, R-X-ext, were used, where X is either the very stable I27 domain of titin or a mutant (27) destabilized by a single-residue V13P mutation, I27V13P. Degradation kinetics of these two substrates were previously characterized; they have, respectively, 40- and 8-min turnover times (9). We asked whether the rpt mutants are affected in their capacity to degrade these proteins and if the stability of the I27 domain protein can increase this effect. Kinetics were measured in saturating concentrations of substrates. The catalytic constants obtained are plotted in Fig. 5 (compared with wild type); absolute values are listed in Table 2. Generally the ratio of activity between the two substrates, R-I27-ext and R-I27V13P-ext, is similar to that in wild type, with the first of these 4–5 times more slowly processed. However, the absolute values of turnover times differed in almost all cases from wild type. Turnover rate increased about 60% for rpt1 and rpt2 (only 20% for rpt2 with the R-I27V13P-ext substrate). In marked contrast, rpt3, rpt4, and rpt5 had decreased activity; these declined 50–80% compared with wild type. No marked effect was seen in rpt6. A strong effect was thus observed for almost all of these mutants; its extent and direction depended on which Rpt protein was mutated.
FIGURE 5.
In vitro degradation of substrates. Initial rates were determined for 9 min in the presence of 50 nm proteasome and were initiated with 1 μm substrate, either R-I27-ext or R-I27V13P-ext. The activity of the mutant proteasomes is represented as a percentage compared with the wild type proteasomes for each substrate. For wild type proteasomes, the value of the catalytic constant (min−1) was 0.036 ± 0.001 and 0.159 ± 0.008 for R-I27-ext and R-I27V13P-ext, respectively. Error bars, S.D. of four independent experiments.
TABLE 2.
Degradation kinetic parameters
Shown is the catalytic constant (min−1) obtained for each mutated proteasome (50 nm) in the presence of 1 μm substrate. Four independent experiments were performed.
| WT | rpt1 | rpt2 | rpt3 | rpt4 | rpt5 | rpt6 | |
|---|---|---|---|---|---|---|---|
| R-I27-ext | 0.036 ± 0.001 | 0.059 ± 0.004 | 0.062 ± 0.005 | 0.018 ± 0.002 | 0.014 ± 0.002 | 0.021 ± 0.001 | 0.0298 ± 0.0001 |
| R-I27V13P-ext | 0.159 ± 0.008 | 0.279 ± 0.009 | 0.19 ± 0.01 | 0.078 ± 0.003 | 0.028 ± 0.003 | 0.068 ± 0.005 | 0.176 ± 0.009 |
DISCUSSION
The Ar-Φ loops present in the ATPases are held to be responsible for protein substrate unfolding and translocation to the 20S degradation chamber of the proteasome. A conserved tyrosine residue has previously been shown in the bacterial ClpXP and ClpAP ATPases to be involved in such activities. In each of the six different proteasome ATPase subunits, Rpt1p to -6p, we individually mutated the Ar-Φ loop tyrosine to alanine. All mutant proteasomes kept their structural integrity and mostly formed a doubly capped 26S complex both in unfractionated extracts and after purification.
In yeast and presumably in other eukaryotes, all of the Rpt ATPases are essential; deletion of any is lethal. Here we made not deletions but focally directed single-residue mutations. All of the mutants were viable, and all six had a non-trivial effect in one or more assays related to proteasome function. If each Rpt and its Ar-Φ loop carries out quantitatively equal and functionally identical tasks, the residual function associated with mutating any one would be at least ⅚ that of wild type, a quantitatively small change. Instead, each mutant was markedly impaired according to one or another assay used, and different assays revealed distinct Rpt subsets as of primary importance.
Coordinated position-dependent firing of ATP hydrolysis is a common feature of AAA ATPases (e.g. see Ref. 28) and may form the basis of asymmetry of function in proteasomes. A recent paper (29) demonstrated that incapacitating ATP hydrolysis by mutating rpt3 alone, in proteasomes that were otherwise unchanged, abolished global ATPase activity, a result consistent with coordination of hydrolysis. Rubin et al. (30) individually mutated the Walker A motif to prevent ATP binding by each Rpt protein. The resulting cells had distinct phenotypes, depending on which of the six Rpts was targeted. The degree of impairment ranged from indiscernible to lethal. This implied that individual Rpts may have specific functions. Goldberg and colleagues (31) have shown that a proteasome binds fewer than six ATPs and favor a model in which there is a dynamic cycle resulting in coordinated binding and hydrolysis, such that pairs of sites cyclically contain ATP, ADP, or no nucleotide. Cross-link data (32) imply that the Rpt6 side of the ring is invariant in its docking to the α ring but that the opposite side of the Rpt ring can assume alternate docking interactions, suggesting that the non-Rpt6 side of the ring, centered on Rpt5, may be more dynamic. Last, recent cryoelectron microscopy papers (5, 6) on fungal 26S structure describe ATPase asymmetries that were observed as non-planer positioning of individual Rpts within the heterohexameric ring of the proteasome.
These prior findings provide a context for considering our experimental data. Three assays were used to test the efficacy of degradation: 1) the steady-state level of high molecular weight ubiquitin conjugates; 2) degradation of ODC, an enzyme of short half-life (∼15 min in yeast) whose turnover is ubiquitin-independent; and 3) turnover rates using purified proteasomes and a pair of model substrates. The two are structurally identical except for a single residue mutation that renders its I27 folded domain mechanically less stable and makes degradation 4–5-fold faster (9).
The data from these assays are summarized in Table 3, along with additional data from the mutants. In this summary presentation, semiquantitative comparisons are referenced to the wild type. It is immediately apparent that the six mutants divide into two classes. rpt3, rpt4, and rpt5 mutants are less active in degradation. The others are not substantially affected or are more active than wild type. All three assays give nearly concordant results, consistent with the conclusion that all are measuring related aspects of the same function. Certain of the assays gave results that were not fully aligned. For example, rpt1 and rpt2 are biochemically more active but not more effective than wild type in clearing ODC from cells, whereas rpt3, which clusters in vitro with rpt4 and rpt5, clears ODC nearly as well as wild type and much more effectively than rpt4 and rpt5. Each assay constitutes a specific test, not a general test of proteasome activity, and such divergences among them are to be expected. More diverse assays will be needed to unravel the basis of such distinctions.
TABLE 3.
Summary of mutant phenotypes
The strength of effect is scored semiquantitatively with respect to wild type. 0 is used to represent phenotype or activity similar to wild type, and the number of positive or negative signs represents the extent of deviation from wild type. Mutants are tabulated in an order that draws attention to their functional clustering.
| ATP activity, ATPase | Degradation assay |
Cell growth |
|||||
|---|---|---|---|---|---|---|---|
| R-I27-ext | R-I27V13P-ext | ODC | Ubiquitin | 37 °C | Canavanine | ||
| rpt3 | ++ | −− | −− | − | −−− | 0 | 0 |
| rpt4 | −− | −−− | −−− | −−− | −−− | 0 | 0 |
| rpt5 | ++ | −− | −−− | −−− | −−− | 0 | 0 |
| rpt1 | +++ | +++ | +++ | 0 | −− | −−− | −−− |
| rpt2 | ++ | +++ | + | 0 | − | 0 | 0 |
| rpt6 | ++ | 0 | 0 | ++ | −− | −−− | −−− |
Changes in degradation, either increased or decreased, were similar in direction and extent for I27 (more stable form) and I27V13P (less stable form). The most divergent case is rpt4, which could in part be ascribed to measurement error, because differential effects on the two substrates may be least accurately measured in this low specific activity case. Because I27 turnover is rate-limited by unfolding and I27V13P turnover is rate-limited by translocation (9), these data are consistent with the conclusion that translocation and unfolding are concomitant processes, both of which are similarly impaired or augmented by the individual loop Tyr to Ala mutations.
The loop Tyr is also important for ATPase activity. Mutating any of the loop Tyr residues changed ATPase activity, increasing it in all but rpt4. In ClpXP, similar mutations also led to a higher rate of ATP hydrolysis (14). What could account for such effects? ATP binding and hydrolysis have been shown in other ATPases to produce conformational changes that are propagated to effecter elements; their action in turn moves or disassembles substrates. This mechanism assures that the Rpt Ar-Φ loops move in coordination with one or more steps of the ATP binding and hydrolysis cycle. Because changes in these structures are coupled, mutating the Rpt Ar-Φ loops could reciprocally affect ATP hydrolysis. In five of the six cases we examined, a Tyr to Ala substitution increased activity.
The effects on ATPase activity could be due to changes in constraints on Ar-Φ loop trajectories. Reduced motional constraint, the most likely outcome of excising the bulky hydroxyphenol of the Tyr side chain, could thus increase activity. According to this scheme, the Tyr to Ala substitution would, in rpt4 uniquely, increase motional constraint, reducing ATP hydrolysis. These inferences assume interactions that occur in cis within an individual Rpt, but more complex mechanisms can be readily imagined. Alternate possibilities include altered loop trajectories that distort the action of neighboring Rpts, a trans-acting effect. This last effect could be observed for ClpX ATPase (14). Mutating the Ar-Φ loop in one ATPase subunit, modified such that it cannot hydrolyze ATP, increased the overall rate of ATP hydrolysis by the hexamer. Interpretation of ATPase data is limited by the unknown nature and extent of coordination of firing among AAA ATPases (33, 34).
We observed that Ar-Φ loop mutations cause changes of ATPase activity in the absence of substrate. Hence, these Tyr residues constrain and control activity independent of their interactions with substrate. The diverse effects of mutating individual loop Tyr residues are most plausibly explained according to two mechanisms: 1) the Ar-Φ loop Tyr of Rpts drives translocation of substrates, and 2) the loop Tyr residues function to modulate ATP hydrolysis. The experimental data support both mechanisms, but distinct Rpts allocate these functions differently. Because both of these factors are at work and work differently in the various Rpts, there is no evident correlation between alterations in degradation and in ATP hydrolysis rate.
As summarized in Table 3, introducing mutations in rpt3, rpt4, and rpt5 impaired degradation, but homologous substitutions in the others did not. In eukaryotes, the Rpt proteins form an ordered hexameric ring, a trimer of dimers: (transRpt1:cisRpt2)(transRpt6:cisRpt3)(transRpt4:cisRpt5) (4). cis and trans designate which of two alternate conformations is assumed by an Rpt N-terminal element (11, 35); dimer pairs form between specific Rpts of different conformation. The mutants most strongly impaired in degradation, rpt3, rpt4, and rpt5, are adjacent in the ring. We can reject two hypotheses suggested by known asymmetries: 1) the activity differences do not correspond to cis versus trans conformational state, and 2) one side of the ATPase ring is more fixedly associated with the subjacent 20S α ring, that which is centered on Rpt6p (32). The observed activity differences do not depend on whether the mutated Rpts occupy the apparently more fixed side of the ring or lie instead on the opposite side of the ring, centered on Rpt5p.
However, the impaired degradation of rpt3, rpt4, and rpt5 does correlate with a reported Rpt ring asymmetry inferred from cryoelectron microscopy (5, 6). In one such model, the Rpts are not coplanar with the 20S α ring but have a spiral staircase configuration, with Rpt3p highest (most distant from the α ring) and Rpt2p lowest; Rpt6p is tilted and connects the top and bottom of the staircase. Ar-Φ loops positioned higher in the staircase have a greater influence on degradation, perhaps because they lie nearest the site of first arrival of the unstructured extension, which must invade the regulatory complex to initiate translocation (9, 10). Insertion must depend first on unpowered stochastic events, because power cannot be delivered before the extension arrives at the ATPase ring. Such stochastic movements may preferentially engage Ar-Φ loops lying closest to the entry site.
Defective growth phenotypes are anti-correlated with measures of degradation efficacy (summarized in Table 3). At elevated temperature (37 °C) and on medium with canavanine, rpt1 and rpt6 were growth-deficient, yet these showed little evidence of impairment in assays of degradation; rpt1, in fact, was the most robust mutant in assays of degradation. Cell growth depends preeminently on coordinating metabolism with diverse biosynthetic requirements, particularly DNA and protein synthesis. Excess proteasome-mediated proteolysis could disrupt such coordination, causing problems of growth or survival. The case is rather different for rpt3. Its faster growth and smaller cell size more resemble a choice of lifestyle than a molecular catastrophe. A host of single gene mutations have been described to cause the whi growth phenotype (36); these typically cause cells to initiate DNA synthesis at smaller cell size. A much studied case is the WHI1-1 allele (37); it encodes a form of the G1 cyclin Cln3p that is abnormally stable and thus prolongs G1 cyclin activation. It is likely that the rpt3 growth phenotype is based on distorted degradation kinetics of one or more proteins involved in cell cycle regulation.
Ar-Φ loop mutations were assessed using assays that are broad (cell growth, ubiquitin conjugates) or focally directed (degradation of model substrates). These measurements, taken together, revealed that each of the six loops has a unique activity profile (summarized in Table 3). Fully probing the nature and mechanisms of their differentiated functions will require further investigations. For example, we have shown that substrate polypeptide chain composition can alter the ability of the proteasome to pull and unfold (24). Do different Rpt loops have distinct roles in engaging polypeptides that bear different amino acid side chains? This question is addressable using model substrates. Additionally, we have inferred that the loop tyrosines must also have interactions within the proteasome itself that modulate ATP hydrolysis. These interactions may be independent of those with substrates, or perhaps there is cross-talk, such that loops connect with proteasomes versus substrates in a manner that is competitive and potentially regulatory.
Acknowledgments
We thank George Oster (University of California, Berkeley) for helpful discussions and Allen Henderson for providing antibody to eIF5a.
This work was supported, in whole or in part, by National Institutes of Health Grant GM45335 (to P. C.).
- ODC
- ornithine decarboxylase.
REFERENCES
- 1. Glickman M. H., Rubin D. M., Fried V. A., Finley D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell Biol. 18, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 3. Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 4. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases. Implications for proteasome structure and assembly. Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sauer R. T., Baker T. A. (2011) AAA+ proteases. ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 [DOI] [PubMed] [Google Scholar]
- 8. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 9. Henderson A., Erales J., Hoyt M. A., Coffino P. (2011) Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 286, 17495–17502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeuchi J., Chen H., Coffino P. (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., Horwich A. L. (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 14. Martin A., Baker T. A., Sauer R. T. (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 16. Guthrie C., Fink G. R., eds. (2002) Guide to Yeast Genetics and Molecular Biology, Academic Press, Inc., San Diego [Google Scholar]
- 17. Pringle J. R., Mor J. R. (1975) Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 11, 131–168 [DOI] [PubMed] [Google Scholar]
- 18. Elsasser S., Schmidt M., Finley D. (2005) Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 19. Gerlinger U. M., Gückel R., Hoffmann M., Wolf D. H., Hilt W. (1997) Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol. Biol. Cell 8, 2487–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hilt W., Enenkel C., Gruhler A., Singer T., Wolf D. H. (1993) The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. J. Biol. Chem. 268, 3479–3486 [PubMed] [Google Scholar]
- 21. Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 22. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 23. Kern A. D., Oliveira M. A., Coffino P., Hackert M. L. (1999) Structure of mammalian ornithine decarboxylase at 1.6 Å resolution. Stereochemical implications of PLP-dependent amino acid decarboxylases. Structure 7, 567–581 [DOI] [PubMed] [Google Scholar]
- 24. Hoyt M. A., Zich J., Takeuchi J., Zhang M., Govaerts C., Coffino P. (2006) Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 25, 1720–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoyt M. A., Zhang M., Coffino P. (2003) Ubiquitin-independent mechanisms of mouse ornithine decarboxylase degradation are conserved between mammalian and fungal cells. J. Biol. Chem. 278, 12135–12143 [DOI] [PubMed] [Google Scholar]
- 26. van Daalen Wetters T., Macrae M., Brabant M., Sittler A., Coffino P. (1989) Polyamine-mediated regulation of mouse ornithine decarboxylase is posttranslational. Mol. Cell. Biol. 9, 5484–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H., Carrion-Vazquez M., Oberhauser A. F., Marszalek P. E., Fernandez J. M. (2000) Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 7, 1117–1120 [DOI] [PubMed] [Google Scholar]
- 28. Joly N., Buck M. (2010) Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism. J. Biol. Chem. 285, 15178–15186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee S. H., Moon J. H., Yoon S. K., Yoon J. B. (2012) Stable incorporation of ATPase subunits into 19S regulatory particle of human proteasome requires nucleotide binding and C-terminal tails. J. Biol. Chem. 287, 9269–9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith D. M., Fraga H., Reis C., Kafri G., Goldberg A. L. (2011) ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian G., Park S., Lee M. J., Huck B., McAllister F., Hill C. P., Gygi S. P., Finley D. (2011) An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 18, 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 [DOI] [PubMed] [Google Scholar]
- 34. Yu J., Moffitt J., Hetherington C. L., Bustamante C., Oster G. (2010) Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 400, 186–203 [DOI] [PubMed] [Google Scholar]
- 35. Djuranovic S., Hartmann M. D., Habeck M., Ursinus A., Zwickl P., Martin J., Lupas A. N., Zeth K. (2009) Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell 34, 580–590 [DOI] [PubMed] [Google Scholar]
- 36. Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M. (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 [DOI] [PubMed] [Google Scholar]
- 37. Sudbery P. E., Goodey A. R., Carter B. L. (1980) Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature 288, 401–404 [DOI] [PubMed] [Google Scholar]