Background: Knowledge of dystrophin modifications at the protein molecular level is required to understand genotype/phenotype link in muscular dystrophies.
Results: A new mutation in DMD gene is accompanied with dystrophin structural and functional modifications.
Conclusion: This Becker muscular dystrophy is due to misfolding of the dystrophin rod domain.
Significance: We provide new insights in understanding the molecular bases of myopathy at the protein level.
Keywords: Biophysics, Dystrophin, Fluorescence, Molecular Modeling, Muscular Dystrophy, Protein Dynamics, Protein Misfolding, In-frame Single Mutation, Spectrin-like Repeats
Abstract
Mutations in the dystrophin gene without disruption of the reading frame often lead to Becker muscular dystrophy, but a genotype/phenotype correlation is difficult to establish. Amino acid substitutions may disrupt binding capacities of dystrophin and have a major impact on the functionality of this protein. We have identified two brothers (ages 8 and 10 years) with very mild proximal weakness, recurrent abdominal pain, and moderately elevated serum creatine kinase levels. Gene sequencing revealed a novel mutation in exon 11 of the dystrophin gene (c.1280T>C) leading to a L427P amino acid substitution in repeat 1 of the central rod domain. Immunostaining of skeletal muscle showed weak staining of the dystrophin region encoded by exons 7 and 8 corresponding to the end of the actin-binding domain 1 and the N-terminal part of hinge 1. Spectrofluorescence and circular dichroism analysis of the domain repeat 1-2 (R1-2) revealed partial misfolding of the L427P mutated protein as well as a reduced refolding rate after denaturation. Based on computational homology models of the wild-type and mutated R1-2, a molecular dynamics study showed an alteration in the flexibility of the structure, which also strongly affects the conformational space available in the N-terminal region of the fragment. Our results suggest that this missense mutation hinders the dynamic properties of the entire N-terminal region of dystrophin.
Introduction
Mutations in the dystrophin gene cause severe Duchenne (DMD)2 or the milder Becker muscular dystrophy (BMD) (1–3). The diagnosis of BMD can be made by molecular testing of the dystrophin gene, a muscle biopsy evaluated by immunostaining, clinical presentation, and family history. Although BMD is milder than DMD, the phenotype varies greatly from person to person and can involve skeletal muscle, heart, and central nervous system (4, 5). The dystrophin gene, which is 2.4 Mb and has 79 exons, is the largest known gene in humans. Dystrophin, the product of this gene, is a large cytoskeletal protein of 427 kDa situated along the inner side of the sarcolemma. It comprises four main domains (6–8). The N-terminal domain constitutes the first actin-binding domain with two calponin homology domains (CH). The major part of the protein is a long central filamentous domain comprising 24 repeating units of about 110 residues each, similar to spectrin repeats. This central domain has several known partners, such as neuronal nitric oxide synthase, filamentous actin, and membrane lipids (9). A Cys-rich domain near the C terminus interacts with the intrinsic sarcolemmal protein β-dystroglycan, and the C-terminal domain interacts with dystrobrevins and syntrophins.
Various types of dystrophin gene mutations have been reported (10–13). Large deletions and duplications are the most common, accounting for ∼75% of the total. Most of the remaining 25% are small insertions, deletions, exonic point mutations, or splice site mutations. The mutations most often seen in children with BMD are in-frame deletions that allow the expression of an internally truncated dystrophin. In contrast, those causing DMD tend to be out of frame, and very little or no dystrophin is expressed (3). This reading frame rule holds true for 93–96% of DMD or BMD cases with the exceptions being related to mutations affecting RNA splicing or alternative translation initiation (14–16). A genotype to phenotype correlation in BMD is likely to be related to the residual functions of the mutated dystrophin. In fact, truncated “mini-dystrophin” constructs are proposed as candidate forms of gene or cell therapy for DMD. To take up this challenge, protein structural studies of mutated or truncated forms of dystrophin will help to predict the functionality of such constructs (17–20).
In the present paper, we report a new point mutation in brothers presenting with a mild BMD phenotype. The mutation is located in exon 11, which encodes part of the first repeat of the dystrophin rod domain. We describe modified physical properties of this protein domain caused by a leucine to proline amino acid substitution and suggest a correlation between protein misfolding and dysfunction that leads to disease.
EXPERIMENTAL PROCEDURES
The study was exempted from human investigational research study by the Institutional Review Board. Genetic analysis of the DMD gene was performed by Athena Diagnostics (Worcester, MA) as a routine diagnostic test.
Muscle Histopathology
Immunofluorescence was carried out on unfixed cryosections of the patient's muscle biopsy as part of routine diagnostic testing at the University of Iowa. All of the anti-dystrophin protocols are 1-h incubation with primary antibodies and 30-min incubation with secondary antibodies. Detailed information about the primary antibodies is listed in supplemental Table 1. This panel of anti-dystrophin antibodies covers epitopes from the N terminus through the rod domain to the C terminus. Antibodies to the dystroglycans, sarcoglycans, utrophin, neuronal nitric-oxide synthase, spectrin, caveolin-3, and dysferlin were also used to evaluate the biopsy as described previously (21). The secondary antibodies are goat anti-mouse or goat anti-rabbit tagged with Alexa Fluor 488 or Alexa Fluor 594 (Molecular Probes, Invitrogen).
In Vitro Protein Biophysical Study
Materials
The pGEX-4T1 plasmid vector, GSTrapTM HP column were purchased from GE Healthcare. The E. coli BL21(DE3) bacteria were supplied by Invitrogen and restriction enzymes by New England Biolabs.
Cloning
The plasmid pTG11025, kindly provided by Dr. S. Braun (Transgene, Strasbourg, France), is an E. coli plasmid that carries the cDNA encoding the full-length Dp427m isoform of human dystrophin (NCBI Nucleotide Data Base NM-004006). Recombinant dystrophin rod domain wild-type tandem repeat R1-2 was cloned downstream of the sequence of glutathione S-transferase (GST) into pGEX-4T1 vector, using the BamHI restriction site of the preScission thrombin recognition sequence and XhoI restriction site. The L427P mutation was introduced by site-directed mutagenesis carried out by PCR using complementary oligonucleotide primers containing a CCG codon according to E. coli codon usage in place of the patient mutated CCA codon. In addition, this codon allows a new EcoRI restriction site for mutant selection. The cloned fragments were verified by sequencing using the dideoxy chain termination method.
Boundaries of Wild-type and Mutated Proteins
Boundaries of recombinant R1-2 were extended by assuming the original alignment of spectrin-type triple-helical repeats in dystrophin as proposed previously (22). The two recombinant proteins were extended by six additional residues at the C terminus to improve solubility and stability as reported previously (23, 24). The sequence range is E-338 to R-563, and two GS residues were residual at the N terminus after thrombin cleavage.
Preparation of Wild-type and Mutated Proteins
Plasmid constructs were used to transform into the E. coli BL21(DE3) strain, and cultures were then grown to an A600 nm of 0.5 at 37 °C in LB broth supplemented with 100 μg/ml ampicillin. Expression of GST fusion proteins was induced by addition of 0.5 mm isopropyl-β-d-thiogalactopyranoside. After 4 h, bacteria were harvested by centrifugation at 4,000 × g for 15 min at 4 °C. As described previously, pellets from 500-ml cultures were resuspended in 20 ml of ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl) and incubated with 0.5 mg·ml−1 lysozyme and 40 μg·ml−1 DNase on ice for 30 min before being broken up by sonication. After centrifugation at 10,000 × g for 20 min at 4 °C, filter-clarified extracts were loaded onto a preequilibrated 5-ml GSTrapTM FF column at a flow rate of 0.25 ml·min−1. The column was washed with 10 column volumes of lysis buffer at a flow rate of 1 ml·min−1, and 50 NIH units of thrombin were then added. On-column cleavage took place for 48 h at 4 °C, and fractions of 5 ml were collected in the washing buffer. Fractions were concentrated on 10K Centricon, and the final purification steps consisted of ion exchange chromatography on HiTrap Q FF (GE Healthcare). Protein concentration was assessed using absorbance at 280 nm and purity by SDS-PAGE.
Circular Dichroism (CD) Measurements
CD spectra were acquired at 20 °C with a path length of 0.2 cm on a JASCO J-815 spectropolarimeter, equipped with a Peltier device for temperature control. From the molar ellipticity at 222 nm [θ222], the mean residue ellipticity [θ]MRW was calculated using the mean residue molar concentration of the proteins, and the percentage of α-helix was obtained using a 100% α-helix value of −36,000 degrees·cm2·dmol−1 at 222 nm as described previously (25). The thermal unfolding of each construct was monitored by CD at 222 nm at concentrations of 2.5 μm in TNE buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0,1 mm EDTA) with temperature increasing 1 degree/min from 15 to 85 °C. As refolding after heating was observed, denaturation was demonstrated to be reversible (see “Results”), CD signals were fitted to a two-state transition with the following equation
where y is the CD signal at 222 nm at temperature T, αN and αD are the intercepts of the native and denatured states, respectively, and βN and βD are the slopes of the native and denatured states, respectively; ΔGun is the free energy of unfolding and R is the gas constant in cal·mol−1·K−1. This equation can be rewritten to facilitate nonlinear regression analysis by SigmaPlot 10.0 in Windows as follows (25, 26),
 |
where Tm is the melting temperature and ΔT is the width of the unfolding transition.
Steady-state Fluorescence Measurements
Tryptophan fluorescence spectra of native R1-2 WT and R1-2 L427P proteins at 1 μm in TNE buffer were recorded at 295-nm excitation wavelength (bandwidth, 1 nm) on a Fluorolog 3 spectrofluorometer (Horiba Jobin-Yvon, France), using 10 × 4-mm quartz cuvette at 20 °C. After appropriate correction, we obtained the emission spectra of nondenatured or urea-denatured proteins. For urea denaturation equilibrium experiments, the proteins were previously incubated for 2 h in TNE with 0.25–8 m urea. The concentration of urea was determined by refractive index measurements. After fluorescence spectra acquisition, we obtained the maximum wavelength (λmax) and intensity of fluorescence from the emission spectra. The λmax, y, was then plotted as a function of urea concentration, U, and fitted to a one-transition, two-state reversible process as described previously (25) and given in the following equation,
 |
where α is the intercept of the native (N) and denatured (D) states λmax, β the slope of the native (N) and denatured (D) states, U50% is the urea concentration at the mid-point of denaturation, m is the slope of the transition between the native and denatured states, R is the gas constant, 1.987 cal·mol−1·K−1, and T the temperature in K. The curves were fitted by nonlinear regression with Sigma plot 10.0 in Windows. The term mU50% in Equation 3 is equal to the free energy of denaturation in the absence of urea, ΔGun.
For 8-anilino-1-naphthalenesulfonic acid (ANS) fluorescence measurements, proteins were diluted at 1 μm in TNE containing 20 μm ANS. ANS was excited at 350 nm and emission scanned from 460 to 620 nm. All background spectra were subtracted.
Refolding and Unfolding Kinetics by Stopped-flow Fluorescence Spectroscopy
Stopped-flow data were recorded on a BioLogic SFM-3, MOS-250 instrument (Grenoble, France). The dead time of the stopped-flow device was 2.2 ms. For refolding measurements, stock solutions of 8 m urea-treated proteins were prepared for 2 h in TNE buffer. The refolding reactions were initiated at 25 °C by 10-fold dilution of the urea-treated proteins in TNE buffer. The final protein concentrations were 0.5 μm. The time-dependent fluorescence of tryptophan changes was monitored at excitation and emission wavelengths of 295 and 345 nm, respectively. Curves were derived from the averages of at least 10 individual kinetic data points after subtraction of the background urea buffer signal. Using Biokine software (BioLogic), the experimental results were fitted with exponential functions to obtain the rate constants.
Molecular Models and Dynamics Simulations
A homology model of the R1-2 tandem repeat was obtained previously via the I-Tasser server using a spectrin repeat structural pattern (27). The mutant form L427P of the protein was modeled by the leucine residue substitution with the Yasara program (28). Each protein was described by parameters of the Yamber3 force field, placed in an explicit solvent box using the TIP3P water model, and neutralized (0.9% NaCl). Both systems were simulated under periodic boundary and in the NVT ensemble (298 K). Structures were relaxed during a 1-ns molecular dynamics simulation, providing a stable root mean square deviation over the last 100 ps. Production trajectories of 12 ns were collected at 2-ps intervals for each of the two structural models (R1-2 WT and L427P mutant). Secondary structure measurements were performed using the DSSP program (29). Mean molecular surfaces and molecular hydrophobicity potential were provided by the Platinum webserver.
RESULTS
Clinical and Genetic Findings
The index case is a 10-year-old male who was evaluated initially by a pediatric gastroenterologist because of severe recurrent abdominal pain that started at 8 years of age. The investigations did not reveal any specific gastrointestinal disease, but transaminases were found to be elevated (alanine aminotransferase, 119 IU, normal 30–65 IU; and aspartate aminotransferase, 81 IU, normal 15–37 IU) along with elevated creatine kinase (CK; 1,632 IU, normal 35–250 IU). The elevated CK triggered evaluation by a child neurologist who noted moderate enlargement of the gastrocnemius muscles and heel cord tightness. The patient had trouble stooping all the way to the ground as well as recovering from a lunge position without using a Gower's maneuver, particularly after multiple trials. Motor testing suggested a mild weakness in hip flexors with Medical Research Council scale of 4. Although the medical history did not suggest any perinatal or developmental problems, a school performance evaluation suggested a form of learning disability with dyslexia. A three-generation pedigree was unremarkable for neuromuscular disease. However, shortly following the patient's diagnosis, his 8-year-old brother presented with recurrent abdominal pain and greatly elevated CK (3,440 IU/liter).
A negative DMD deletion and duplication analysis was followed by complete sequencing of the gene, which identified a novel nucleotide transition T>C at position 1280. The Leiden Muscular Dystrophy Pages mutation data base does not include this missense mutation. This mutation in codon 427 results in the amino acid substitution leucine to proline (L427P). The same genetic variant of the dystrophin gene was subsequently identified in the index patient's 8-year-old-brother as well as their mother.
Muscle Histology
Routine hematoxylin and eosin staining of a biopsy from the left vastus lateralis muscle showed only mild variation in fiber size due to scattered moderately atrophic and a few hypertrophic muscle fibers (Fig. 1). However, no necrotic and only rare regenerating muscle fibers were seen. Internal nuclei were mildly increased, and rare muscle fibers had clefts or splitting. No endomysial fibrosis and no lymphocytic inflammation were seen in the specimen. Immunofluorescence staining showed greatly reduced to absent immunoreactivity near the N terminus with antibodies recognizing epitopes encoded by exons 7 and 8 (see laser confocal fluorescence microscopy panels in Fig. 1). Dystrophin immunostaining abnormalities were absent or only mild using antibodies to other epitopes. Other proteins of the dystrophin-glycoprotein complex (dystroglycans and sarcoglycans), caveolin-3, and dysferlin stained normally; utrophin was not detected at the sarcolemma (data not shown).
FIGURE 1.
Muscle biopsy evaluation. The upper left panel is a representative image from an H&E stained cryosection photographed using the 20× objective. There are several atrophic fibers and a few mildly hypertrophic fibers. Scale bar, 50 μm. Immunofluorescence images compare a pediatric, nondystrophic control biopsy with the BMD patient's biopsy. The antibodies are described in supplemental Table 1. Each image from the BMD patient illustrates the same region of the biopsy; the lowercase a identifies the same muscle fiber in each serial cryosection. The dystrophin regions recognized by each anti-dystrophin antibody are printed beside each side-by-side pair of control BMD images. All immunofluorescence images were obtained on a Zeiss 710 laser confocal microscope using a 20× objective. Scale bar, 100 μm.
Structural Analysis of Mutated Repeat 1-2 of Dystrophin
Protein Characterization
Fig. 2A shows a schematic representation of human dystrophin and an enlargement of the region from the CH2 of the actin-binding domain 1 to the start of repeat 3 together with the respective encoding exons. The exact sequence of this region together with the exon boundaries are given in supplemental Fig. S1. An alignment of this region for other species shows that the Leu residue in position 427 is highly conserved in dystrophin from mammals and is also present in chicken, zebra fish, and Drosophila. In the same position, a hydrophobic residue is also found in Caenorhabditis elegans (Fig. 2B). The L427P mutation found in our patients is located in helix C of the repeat R1 and is located at a d position known to stabilize hydrophobic helix-helix interacting clusters in the spectrin like triple coiled-coils (22, 27, 30). The mutation introduces a Pro residue that is known to be a helix breaker (31–33).
FIGURE 2.
Schematic representation of dystrophin. A, localization of the corresponding exons 6–14 and mutation L427P in repeat 1 in the central rod domain. CH2, calponin-like domain; R, repeats from central domain; H, connecting hinges. The red square indicates the position of the L427P mutation in repeat 1. B, amino acid sequence alignment of the end of the loop connecting helices B-C and N-terminal part of helix C of repeat R1 of dystrophin rod domain from various species. GenBank accession numbers are indicated. The mutated leucine is in a highly conserved region and corresponds to an hydrophobic residue at a d position in a heptad motif characteristic of dystrophin triple-coiled coils (22).
The wild-type and mutated proteins were obtained in comparable purity as assessed by Coomassie Blue-stained SDS-polyacrylamide gels (Fig. 3A). N-terminal sequencing confirmed the expected cleavage by thrombin for both proteins. CD spectra (Fig. 3B) display the typical features of proteins with a predominant α-helix folding as reported previously for dystrophin repeats, i.e. two minima troughs at 222 and 208 nm. However, the spectra from the wild-type and mutated proteins are different. First, the values at 222 nm are half as deep in the mutated compared with the wild type, indicating 73 and 45% of helicity for the wild-type and mutated proteins, respectively. This indicates that the mutated protein is in a different native state compared with the wild-type protein. Second, the ratio of the [222]/[208] values is about 1 for the wild-type protein whereas it is about 0.85 for the mutated protein. Taken together, this indicates that the coiled-coil would not be fully formed in the mutated protein compared with the wild type.
FIGURE 3.
Biochemical characterization of R1-2 WT and R1-2 L427P. A, analysis by SDS-PAGE and Coomassie Blue staining. Proteins were obtained by conventional site-directed mutagenesis methods followed by expression in E. coli as GST-tagged proteins and affinity chromatography on glutathione-Sepharose. Both purified proteins were obtained by an additional ion exchange chromatography step. B, CD spectra of R1-2 WT and R1-2 L427P. Concentration of proteins was 2.5 μm. C, steady-state tryptophan fluorescence spectra of R1-2 WT and R1-2 L427P. Concentration of proteins was 1 μm. Excitation wavelength is 295 nm. D, fluorescence emission spectra of 20 μm ANS in the presence of 1 μm R1-2 WT and R1-2 L427P. Excitation wavelength is 350 nm.
As shown in Fig. 3C, only a slight increase of tryptophan fluorescence intensity was observed for the mutated protein, with no change at the 338-nm maximal emission wavelength. The extent of exposure of hydrophobic surfaces in the R1-2 WT and R1-2 L427P was measured by their ability to bind to the fluorescent dye ANS. Fig. 3D shows that the fluorescence parameters of ANS, intensity and maximal emission wavelength, are in agreement with an extended hydrophobic surface area for the mutated protein compared with the wild type. One-dimensional proton NMR spectra corroborated these observations (supplemental Fig. S2). Several resonances at high field and the dispersion of the amide protons are indicative of a folded wild-type protein. In contrast, the spectrum of the mutated protein rather corresponds to a different folding state of the protein as indicated by the broad NH resonances and the absence of high field signals.
Thermal Denaturation
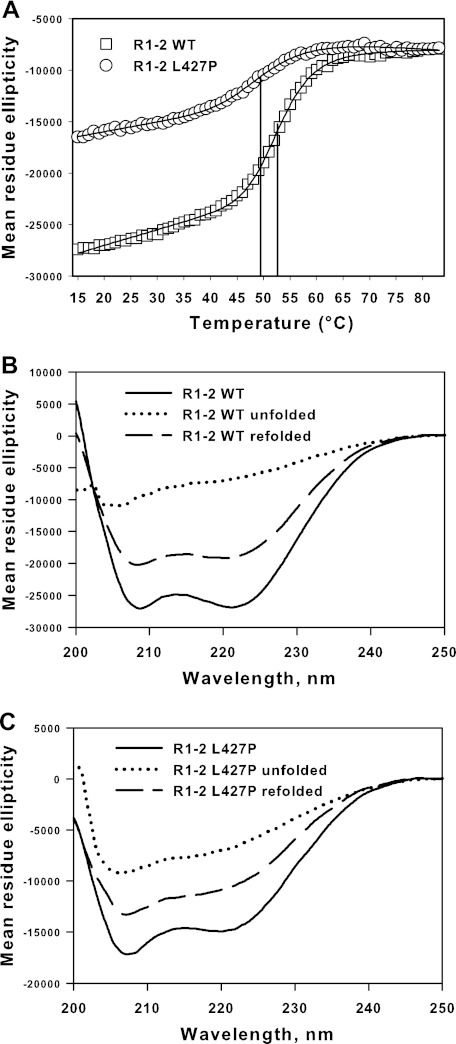
Thermal denaturation was followed by CD at 222-nm wavelength. The two proteins were denatured in a one-step mechanism (Fig. 4A). However, it should be noted that because the mutated protein has a less helical content than the wild type protein, the starting mean residue ellipticity values of CD at 222 nm are far from identical, in agreement with the spectra first obtained. Spectra acquired at the end of the heating procedure showed that the proteins are all denaturated (Fig. 4, B and C). As large refolding of about 75–80% of ellipticity after heating was observed for the two proteins (Fig. 4, B and C) the equations describing the thermodynamic process of denaturation can be used. Therefore, the temperatures at mid-denaturation were determined to be 53.4 and 50 °C for the wild-type and the mutated protein, respectively (Fig. 4A). This indicates a lower thermal stability associated with the mutation.
FIGURE 4.
Unfolding and refolding of R1-2 WT and R1-2 L427P. A, thermal unfolding as measured by CD spectroscopy at 222 nm. Concentration of proteins was 2.5 μm. The mean residue ellipticity is expressed as a function of temperature increases. The denaturation of R1-2 (Tm 53.4 °C) and R1-2 L427P (Tm 50.0 °C) follows a single-step process. The temperatures at mid-denaturation are indicated by vertical lines. B and C, R1-2 WT (B) and R1-2 L427P (C) CD spectra before and after 15 min at 20 °C refolding time following heating at 80 °C.
Urea Unfolding and Refolding
The equilibrium denaturation was studied using tryptophan fluorescence. When denatured in 8 m urea, the intensity of tryptophan fluorescence increased, and the maximal emission wavelength was 348 nm for both proteins (Fig. 5A). The tryptophan fluorescence data following urea denaturation indicate that R1-2 wild-type and R1-2 L427P proteins are unfolded in a single-step process with identical urea concentrations of 4.10 m at mid-denaturation (Fig. 5B).
FIGURE 5.
A, steady-state tryptophan fluorescence spectra of R1-2 WT and R1-2 L427P in TNE buffer or in TNE buffer containing 8 m urea. Concentration of proteins was 1 μm. Excitation wavelength is 295 nm. B, urea-induced equilibrium denaturation curves for R1-2 wild type and L427P. Proteins were equilibrated at 25 °C in appropriate urea final concentrations, and changes in maximal emission wavelength were measured. C, refolding kinetics observed by stopped-flow spectrofluorescence following denaturation in 8 m urea. R1-2 WT and R1-2 L427P unfolded in 8 m urea were diluted 10-fold at 25 °C in urea-free buffer. Measurements were performed by monitoring fluorescence emission at 345 nm after excitation at 295 nm. Stopped-flow data used for rate constants calculation were the averages of at least 10 individual kinetics. Experimental signal, fitted curves (continuous line for R1-2 WT, and dotted line for R1-2 L427P) and residuals (upper) are shown. Both refolding fit well with a two-exponential model.
Refolding kinetics of the two proteins were then monitored by stopped-flow tryptophan fluorescence. These proteins were first unfolded in 8.0 m urea and then rapidly diluted to a final urea concentration of 0.8 m. The refolding data fitted well to a two-exponential process for both proteins. The fastest phase accounted for 92 and 68% of the amplitude with calculated rate constants of 23 s−1 and 11 s−1 for R1-2 WT and R1-2 L427P, respectively. The slowest phase accounted for 8 and 32% of the amplitude with rate constants of 0.02 s−1 and 0.06 s−1 for R1-2 WT and R1-2 L427P, respectively (Fig. 5C). This indicates a slower refolding for the mutant compared with the wild type.
Molecular Modeling
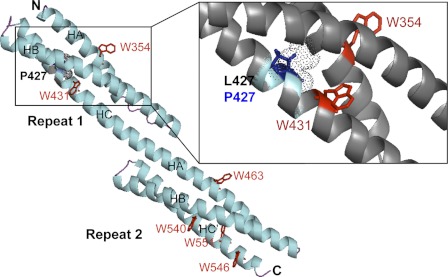
The models for the R1-2 WT and L427P were built using I-Tasser and Yasara, respectively. The two proteins exhibit the triple-helical coiled-coil three-dimensional structure typical of the spectrin-like repeats (Fig. 6). The two proteins contained six tryptophan residues, two of them being in repeat 1 and four of them in repeat 2. The Trp-431 situated in the helix C of repeat 1 is 4 residues from the mutation site (Fig. 6, zoom). Therefore, its environment can be modified by the mutation, and this can explain the increase in intrinsic tryptophan fluorescence observed in the mutated protein compared with the wild-type protein. In fact, the nonnative state of the mutated protein due to a modification of the folding around the mutation site likely exposes the Trp-431 to a more hydrophilic environment leading to the observed increase of its fluorescence. The environment of the other tryptophan residues is not modified, and the averaged fluorescence of the six tryptophan residues is only slightly modified.
FIGURE 6.
Molecular model of the R1-2 protein (methods as in Ref. 27. The tandem repeat is constituted by the two repeats R1 and R2, each of them comprising three helices, HA, HB, and HC for R1 and HA′, HB′, and HC′ for R2. The N and C termini are denoted N and C, respectively The tryptophan residues are denoted as red W, and the L427 site of the mutation is shown in Repeat 1. The region of the mutation is enlarged on the right, showing that the Trp-431 is very near the mutation pointing into the interior of the molecule in the wild-type model. Leucine 427 (dotted gray sphere) points to the interior of the molecule. Therefore, the mutation L427P leads to the proline residue (blue sticks) pointing also to the interior with putative helix breaking; this likely is able to destabilize the folding of these three helices.
Molecular Simulations
As expected from experimental structures and theoretical observations (31–33), the proline insertion caused by the L427P mutation induces R1 helix C destabilization, related to a short helix bend and a break in the H-bonding chain defining the helical structure. Atomic fluctuations shown by the root mean square fluctuation of the 12 ns of molecular dynamics trajectories are clearly increased at the mutation spot (Fig. 7A). The a and d hydrophobic positions of the heptad pattern specific to coiled-coil superstructures remain effective around the mutation. However, the total molecular surface increases by 4%, and the hydrophobic surface contribution moves from 31 to 33%, indicating that the coiled-coil arrangement in the R1 L427P is less stable than the wild-type R1. The main impact of the mutation on the R1-2 dynamics profile concerns the stiffening of the linker region supported by the important freezing in the mobility of the A/B loop. This modification in the flexibility of the coiled-coil structure also strongly affects the conformational space available to the N-terminal region of the R1-2 protein and the relative position of R1 repeat toward the R2 repeat (Fig. 7B). Principal component analysis showed that the first and the third most contributing components to be global motion are the most strongly modified by the mutation (supplemental Fig. S3).
FIGURE 7.
Cα root mean square fluctuation over 12-ns molecular dynamics trajectories. A, α-helices delimitation (27) for the R1 (A, B, and C) and R2 (A′, B′, and C′) repeats and mutation location (*) are reported along the x axis in correspondence with the residue numbers in the dystrophin sequence. Cα root mean square fluctuation (in Å) is plotted for each residue of R1-2 WT (black) and R1-2 L427P (green). B, flexibility variations are mapped on the mutant structure. The mutation spot is identified with a sphere. Higher rigidity (blue) or higher flexibility (red) spots of L427P mutant by comparison with the WT structure are shown on the molecular models, respectively.
DISCUSSION
Clinical variability in dystrophinopathy-related BMD ranges between a severe, Duchenne-like phenotype and a late onset mild disease. In-frame deletions in the mid-portion of dystrophin gene are responsible for most of the milder cases, but the size of the deletion does not appear to be a key factor in determining the severity of disease (4, 34, 35). Serum CK levels also show little correlation with the severity of clinical symptoms. Severe, even lethal cardiomyopathy may develop in patients with relatively minimal skeletal muscle weakness, although the genotype correlation has not been determined for these cases (36). Central nervous system involvement and gastrointestinal manifestations even in the early disease stages are more typical of DMD than BMD (37, 38). Our index case presented with subtle proximal skeletal muscle weakness and no cardiac involvement. It was not certain whether his recurrent abdominal pain was related to gastrointestinal hypomotility or other dysfunctions. The serum CK elevation and histopathology of the muscle biopsy were milder than is typical of BMD, but both dystrophin immunostaining and DMD gene sequencing confirmed this diagnosis.
A phenotype-genotype correlation is difficult to establish in BMD. In-frame deletions constitute the majority of the mutations, and it appears that mutations of the N terminus of dystrophin affecting the actin-binding domain 1 (39, 40) or the Cys-rich domain (41) are more severe than the others. However, deletions around exons 45–55 are in a large number of cases accompanied by mild skeletal muscle disease but a possibly severe cardiomyopathy. Large deletions in the central rod domain (between exons 45 and 59) have been seen in late onset BMD (2). By combining genetic and protein information, it appears that there is a strong association between specific structural modifications of the mutated dystrophin and the age of onset of the cardiomyopathy (42).
Point mutations of the dystrophin gene are less common than large deletions and duplications. Mutated dystrophin is expressed in skeletal muscle of cases with missense point mutations, but to date there are very few publications that evaluate potential genotype-phenotype correlations. Previous studies of point mutations within the actin-binding domain of patients with either DMD or BMD phenotypes have concluded that the dystrophin produced is mostly misfolded, rather unstable and prone to aggregation (39, 40). We recently showed that a double point mutation in the central rod domain repeat R23 of a DMD patient leads to a substantially lower thermal and chemical stability as well as slower refolding of the mutated protein compared with the wild-type protein (25).
In this work, the novel c.1280T>C DMD mutation introduces a proline residue in the helix C of repeat 1. Proline is a residue known as a helix breaker, even though it does not always preclude the helical structure (31–33) and is found in some helices of wild-type dystrophin repeats (22). In the present case, the data demonstrate that the helical content of the mutated R1-2 fragment is dramatically decreased compared with the wild-type protein, showing that the mutated protein does not retain a folded state compared with the wild-type protein. This is consistent with the hydrophobic surface changes detected by ANS fluorescence. The mutation also affects thermal unfolding and urea refolding kinetics. The molecular modeling and dynamics presented here are in agreement with these experimental observations and clearly show that the single mutation L427P has striking local and global effects on the dynamic and mechanical behavior of the filamentous structure of the tandem repeat R1-2 of dystrophin. Taken together, these results demonstrate that the proline residue induces a break in the helix which in turn destabilizes the entire helix toward the C terminus of the molecule. The dystrophin protein as expressed in the patient's skeletal muscle is found to be subsarcolemmal just as in control muscle. However, the protein is poorly visualized by immunofluorescence using antibodies that recognize peptides encoded by exons 7 and 8. Given that the area bearing the L427P mutation is encoded by exon 11, the lack of staining of the full-length dystrophin in the patient's biopsy is likely due to conformational changes in hinge 1 consecutive to the rigidity changes we observed in the N-terminal part of R1-2.
It thus can be concluded that, even though full-length dystrophin is detected in the patient, the missense mutation modifies peptide stability and leads to misfolding of the N-terminal and proximal rod domains. This in turn has a negative effect on the mechanical or binding functions of dystrophin leading to BMD.
Supplementary Material
Acknowledgments
We thank IFR 140 (Rennes, France) for spectroscopic facilities, Arnaud Bondon for NMR studies, Aurélie Nicolas for sequence alignment, Christophe Tascon for protein purification, Terese Nelson for muscle biopsy immunostaining, Joel Carl for assembling the images to create Fig. 1, and Graham Parker of Wayne State University for helpful suggestions in editing the manuscript.
This work was supported by the Association Française contre les Myopathies and Iowa Wellstone Muscular Dystrophy Cooperative Research Center Grant NS053672.

This article contains supplemental Table S1, Figs. S1–S3, and additional references.
- DMD
- Duchenne muscular dystrophy
- ANS
- 8-anilino-1-naphthalenesulfonic acid
- BMD
- Becker muscular dystrophy
- CH
- calponin homology
- CK
- creatine kinase
- IU
- international units.
REFERENCES
- 1. Koenig M., Beggs A. H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C. R., Lindlöf M., Kaariainen H. (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am. J. Hum. Genet. 45, 498–506 [PMC free article] [PubMed] [Google Scholar]
- 2. Bushby K. M., Gardner-Medwin D., Nicholson L. V., Johnson M. A., Haggerty I. D., Cleghorn N. J., Harris J. B., Bhattacharya S. S. (1993) The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. II. Correlation of phenotype with genetic and protein abnormalities. J. Neurol. 240, 105–112 [DOI] [PubMed] [Google Scholar]
- 3. Monaco A. P, Bertelson C. J, Liechti-Gallati S., Moser H., Kunkel L. M. (1988) An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 2, 90–95 [DOI] [PubMed] [Google Scholar]
- 4. Norman A. M., Thomas N. S., Kingston H. M., Harper P. S. (1990) Becker muscular dystrophy: correlation of deletion type with clinical severity. J. Med. Genet. 27, 236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beggs A. H., Hoffman E. P., Snyder J. R., Arahata K., Specht L., Shapiro F., Angelini C., Sugita H., Kunkel L. M. (1991) Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am. J. Hum. Genet. 49, 54–67 [PMC free article] [PubMed] [Google Scholar]
- 6. Campbell K. P., Kahl S. D.(1989) Association of dystrophin and an integral membrane glycoprotein. Nature 338, 259–262 [DOI] [PubMed] [Google Scholar]
- 7. Chamberlain J. S., Corrado K., Rafael J. A., Cox G. A., Hauser M., Lumeng C. (1997) Interactions between dystrophin and the sarcolemma membrane. Soc. Gen. Physiol. Ser. 52, 19–29 [PubMed] [Google Scholar]
- 8. Hanft L. M., Bogan D. J., Mayer U., Kaufman S. J., Kornegay J. N., Ervasti J. M. (2007) Cytoplasmic γ-actin expression in diverse animal models of muscular dystrophy. Neuromuscul. Disord. 17, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Rumeur E., Winder S. J., Hubert J. F. (2010) Dystrophin: more than just the sum of its parts. Biochim. Biophys. Acta 1804, 1713–1722 [DOI] [PubMed] [Google Scholar]
- 10. Aartsma-Rus A., Van Deutekom J. C., Fokkema I. F., Van Ommen G. J., Den Dunnen J. T. (2006) Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34, 135–144 [DOI] [PubMed] [Google Scholar]
- 11. Taylor P. J., Maroulis S., Mullan G. L., Pedersen R. L., Baumli A., Elakis G., Piras S., Walsh C., Prósper-Gutiérrez B., De La Puente-Alonso F., Bell C. G., Mowat D. R., Johnston H. M., Buckley M. F. (2007) Measurement of the clinical utility of a combined mutation detection protocol in carriers of Duchenne and Becker muscular dystrophy. J. Med. Genet. 44, 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flanigan K. M., Dunn D. M., von Niederhausern A., Soltanzadeh P., Gappmaier E., Howard M. T., Sampson J. B., Mendell J. R., Wall C., King W. M., Pestronk A., Florence J. M., Connolly A. M., Mathews K. D., Stephan C. M., Laubenthal K. S., Wong B. L., Morehart P. J., Meyer A., Finkel R. S., Bonnemann C. G., Medne L., Day J. W., Dalton J. C., Margolis M. K., Hinton V. J., United Dystrophinopathy Project Consortium, and Weiss R. B. (2009) Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum. Mutat. 30, 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Desguerre I., Christov C., Mayer M., Zeller R., Becane H. M., Bastuji-Garin S., Leturcq F., Chiron C., Chelly J., Gherardi R. K. (2009) Clinical heterogeneity of Duchenne muscular dystrophy (DMD): definition of subphenotypes and predictive criteria by long-term follow-up. PLoS One 4, e4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tuffery-Giraud S., Béroud C., Leturcq F., Yaou R. B., Hamroun D., Michel-Calemard L., Moizard M. P., Bernard R., Cossée M., Boisseau P., Blayau M., Creveaux I., Guiochon-Mantel A., de Martinville B., Philippe C., Monnier N., Bieth E., Khau Van Kien P., Desmet F. O., Humbertclaude V., Kaplan J. C., Chelly J., Claustres M. (2009) Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledge base. Hum. Mutat. 30, 934–945 [DOI] [PubMed] [Google Scholar]
- 15. Gurvich O. L., Maiti B., Weiss R. B., Aggarwal G., Howard M. T., Flanigan K. M. (2009) DMD exon 1 truncating point mutations: amelioration of phenotype by alternative translation initiation in exon 6. Hum. Mutat. 30, 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flanigan K. M., Dunn D. M., von Niederhausern A., Soltanzadeh P., Howard M. T., Sampson J. B., Swoboda K. J., Bromberg M. B., Mendell J. R., Taylor L. E., Anderson C. B., Pestronk A., Florence J. M., Connolly A. M., Mathews K. D., Wong B., Finkel R. S., Bonnemann C. G., Day J. W., McDonald C., United Dystrophinopathy Project Consortium, and Weiss R. B. (2011) Nonsense mutation-associated Becker muscular dystrophy: interplay between exon definition and splicing regulatory elements within the DMD gene. Hum. Mutat. 32, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harper S. Q., Hauser M. A., DelloRusso C., Duan D., Crawford R. W., Phelps S. F., Harper H. A., Robinson A. S., Engelhardt J. F., Brooks S. V., Chamberlain J. S. (2002) Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 8, 253–261 [DOI] [PubMed] [Google Scholar]
- 18. Banks G. B., Chamberlain J. S. (2008) The value of mammalian models for Duchenne muscular dystrophy in developing therapeutic strategies. Curr. Top. Dev. Biol. 84, 431–453 [DOI] [PubMed] [Google Scholar]
- 19. Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P., Garralda M. E., Rutherford M., McCulley C., Popplewell L., Graham I. R., Dickson G., Wood M. J., Wells D. J., Wilton S. D., Kole R., Straub V., Bushby K., Sewry C., Morgan J. E., Muntoni F. (2009) Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 8, 918–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Odom G. L., Banks G. B., Schultz B. R., Gregorevic P., Chamberlain J. S. (2010) Preclinical studies for gene therapy of Duchenne muscular dystrophy. J. Child Neurol. 25, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 21. Moore S. A., Shilling C. J., Westra S., Wall C., Wicklund M. P., Stolle C., Brown C. A., Michele D. E., Piccolo F., Winder T. L., Stence A., Barresi R., King N., King W., Florence J., Campbell K. P., Fenichel G. M., Stedman H. H., Kissel J. T., Griggs R. C., Pandya S., Mathews K. D., Pestronk A., Serrano C., Darvish D., Mendell J. R. (2006) Limb-girdle muscular dystrophy in the United States. J. Neuropathol. Exp. Neurol. 65, 995–1003 [DOI] [PubMed] [Google Scholar]
- 22. Winder S. J., Gibson T. J., Kendrick-Jones J. (1995) Dystrophin and utrophin: the missing links! FEBS Lett. 369, 27–33 [DOI] [PubMed] [Google Scholar]
- 23. Kahana E., Gratzer W. B. (1995) Minimum folding unit of dystrophin rod domain. Biochemistry 34, 8110–8114 [DOI] [PubMed] [Google Scholar]
- 24. Legardinier S., Raguénès-Nicol C., Tascon C., Rocher C., Hardy S., Hubert J. F., Le Rumeur E. (2009) Mapping of the lipid-binding and stability properties of the central rod domain of human dystrophin. J. Mol. Biol. 389, 546–558 [DOI] [PubMed] [Google Scholar]
- 25. Legardinier S., Legrand B., Raguénès-Nicol C., Bondon A., Hardy S., Tascon C., Le Rumeur E., Hubert J. F. (2009) A two-amino acid mutation encountered in Duchenne muscular dystrophy decreases stability of the rod domain 23 (R23) spectrin-like repeat of dystrophin. J. Biol. Chem. 284, 8822–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kusunoki H., Minasov G., Macdonald R. I., Mondragón A. (2004) Independent movement, dimerization and stability of tandem repeats of chicken brain α-spectrin. J. Mol. Biol. 344, 495–511 [DOI] [PubMed] [Google Scholar]
- 27. Legrand B., Giudice E., Nicolas A., Delalande O., Le Rumeur E. (2011) Computational study of the human dystrophin repeats: interaction properties and molecular dynamics. PLoS One 6, e23819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krieger E., Darden T., Nabuurs S. B., Finkelstein A., Vriend G. (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57, 678–683 [DOI] [PubMed] [Google Scholar]
- 29. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 30. Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci. 21, 375–382 [PubMed] [Google Scholar]
- 31. Sankararamakrishnan R., Vishveshwara S. (1993) Characterization of proline-containing α-helix (helix F model of bacteriorhodopsin) by molecular dynamics studies. Proteins 15, 26–41 [DOI] [PubMed] [Google Scholar]
- 32. Cordes F. S., Bright J. N., Sansom M. S. (2002) Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323, 951–960 [DOI] [PubMed] [Google Scholar]
- 33. Rey J., Deville J., Chabbert M. (2010) Structural determinants stabilizing helical distortions related to proline. J. Struct. Biol. 171, 266–276 [DOI] [PubMed] [Google Scholar]
- 34. Hart K. A., Hodgson S., Walker A., Cole C. G., Johnson L., Dubowitz V., Bobrow M. (1987) DNA deletions in mild and severe Becker muscular dystrophy. Hum. Genet. 75, 281–285 [DOI] [PubMed] [Google Scholar]
- 35. Palmucci L., Doriguzzi C., Mongini T., Restagno G., Chiadò-Piat L., Maniscalco M. (1994) Unusual expression and very mild course of Xp21 muscular dystrophy (Becker type) in a 60-year-old man with 26 percent deletion of the dystrophin gene. Neurology 44, 541–543 [DOI] [PubMed] [Google Scholar]
- 36. Yokota R., Shirotani M., Kouchi I., Hirai T., Uemori N., Ohta Y., Mitsui Y., Hattori R. (2004) Subclinical Becker's muscular dystrophy presenting with severe heart failure. Intern. Med. 43, 204–208 [DOI] [PubMed] [Google Scholar]
- 37. Jaffe K. M., McDonald C. M., Ingman E., Haas J. (1990) Symptoms of upper gastrointestinal dysfunction in Duchenne muscular dystrophy: case-control study. Arch. Phys. Med. Rehabil. 71, 742–744 [PubMed] [Google Scholar]
- 38. Staiano A., Del Giudice E., Romano A., Andreotti M. R., Santoro L., Marsullo G., Rippa P. G., Iovine A., Salvatore M. (1992) Upper gastrointestinal tract motility in children with progressive muscular dystrophy. J. Pediatr. 121, 720–724 [DOI] [PubMed] [Google Scholar]
- 39. Singh S. M., Kongari N., Cabello-Villegas J., Mallela K. M. (2010) Missense mutations in dystrophin that trigger muscular dystrophy decrease protein stability and lead to cross-β aggregates. Proc. Natl. Acad. Sci. U.S.A. 107, 15069–15074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Henderson D. M., Lee A., Ervasti J. M. (2010) Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 9632–9637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bies R. D., Caskey C. T., Fenwick R. (1992) An intact cysteine-rich domain is required for dystrophin function. J. Clin. Invest. 90, 666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaspar R. W., Allen H. D., Ray W. C., Alvarez C. E., Kissel J. T., Pestronk A., Weiss R. B., Flanigan K. M., Mendell J. R., Montanaro F. (2009) Analysis of dystrophin deletion mutations predicts age of cardiomyopathy onset in Becker muscular dystrophy. Circ. Cardiovasc. Genet. 2, 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.