Background: α-Catenin provides links for cadherin receptors to the actin cytoskeleton at cell-cell adherens junctions.
Results: Extensive α-catenin interactions with vinculin are displaced by the vinculin tail domain.
Conclusion: α-Catenin-vinculin interactions are stabilized by F-actin.
Significance: The data support a new model whereby vinculin activation at adherens junctions is sufficient to stabilize connections of α-catenin with the actin network.
Keywords: Cancer, Cell Adhesion, Crystal Structure, Cytoskeleton, Protein-Protein Interactions
Abstract
Adherens junctions (AJs) are essential for cell-cell contacts, morphogenesis, and the development of all higher eukaryotes. AJs are formed by calcium-dependent homotypic interactions of the ectodomains of single membrane-pass cadherin family receptors. These homotypic interactions in turn promote binding of the intracellular cytoplasmic tail domains of cadherin receptors with β-catenin, a multifunctional protein that plays roles in both transcription and AJs. The cadherin receptor-β-catenin complex binds to the cytoskeletal protein α-catenin, which is essential for both the formation and the stabilization of these junctions. Precisely how α-catenin contributes to the formation and stabilization of AJs is hotly debated, although the latter is thought to involve its interactions with the cytoskeletal protein vinculin. Here we report the crystal structure of the vinculin binding domain (VBD) of α-catenin in complex with the vinculin head domain (Vh1). This structure reveals that α-catenin is in a unique unfurled mode allowing dimer formation when bound to vinculin. Finally, binding studies suggest that vinculin must be in an activated state to bind to α-catenin and that this interaction is stabilized by the formation of a ternary α-catenin-vinculin-F-actin complex, which can be formed via the F-actin binding domain of either protein. We propose a feed-forward model whereby α-catenin-vinculin interactions promote their binding to the actin cytoskeleton to stabilize AJs.
Introduction
Cell-cell contacts require the formation and stabilization of multiprotein cell membrane complexes coined adherens junctions (AJs).2 These complexes are directed by homotypic interactions of single-pass transmembrane receptors (cadherins) that have tandem calcium binding ectodomains and short but functionally critical intracellular tail domains (1). Cadherin interactions promote interactions of their tail domains with β-catenin, an oncoprotein that both functions as a component of AJs and regulates the activity of the T-cell transcription factor (TCF) family of transcription factors (2, 3). Interestingly, the cadherin-β-catenin complex is hardwired as they bind to one another immediately following their synthesis and are co-transported to the cell membrane as a nascent complex (4). Once there, β-catenin binds to the cytoskeletal protein α-catenin to form the functional ternary cadherin-β-catenin-α-catenin complex, and all three components are required for development and tissue homeostasis (5–8). Stabilization of AJs had long been thought to rely on the interaction of this ternary complex with F-actin, where increased pools of α-catenin at nascent junctions lead to the formation of α-catenin homodimers that bind to F-actin via a C-terminal domain (9, 10). However, the isolated ternary complex, or these complexes in the context of isolated cell membranes, cannot bind to F-actin (11, 12). Further, in these experimental systems, F-actin binding of the ternary complex is not restored by the addition of the cytoskeletal proteins vinculin or α-actinin that themselves can bind to F-actin (2). Rather, the function of the ternary AJ complex was suggested to impair the activity of the F-actin nucleating, branching Arp2/3 complex (13, 14). Finally, to add more complexity, other α-catenin binding partners, specifically Eplin (15) or vinculin (16, 17), have been suggested to play important roles in stabilizing AJs, as for example, vinculin knockdown disables cadherin receptor-actin connections and provokes marked reductions in AJs (18).
α-Catenin is a highly conserved, 906-residue cytoskeletal protein that is composed of four functional domains: (i) an N-terminal β-catenin interacting motif; (ii) a vinculin and α-actinin interacting domain; (iii) an l-afadin binding module that lies within a dimerization domain; and (iv) a C-terminal domain that can bind to both zonula occludens-1 (ZO-1) and F-actin (19). The crystal structure of the N terminus of α-catenin, and of this domain (residues 57–264) in complex with an α-helix of β-catenin, revealed that this region is a homodimer composed of two antiparallel four-helix bundle domains and that β-catenin binds to α-catenin via a helix exchange mechanism (20). In addition, the crystal structure of the “M (modulation) fragment” (residues 377–633) (9, 21), which harbors the binding site for l-afadin, established that this domain is composed of a tandem repeat of two antiparallel four-helix bundles that also form a homodimer and are similar in structure, as found in the N terminus of α-catenin.
Vinculin is a dynamic 117-kDa globular protein composed of five helix bundle domains that are held in a closed conformation via hydrophobic interactions of the N-terminal seven-helix bundle head (first vinculin head domain, Vh1) with the five-helix bundle tail (vinculin tail domain, Vt) (22, 23). These intramolecular interactions are severed by the binding of amphipathic α-helix vinculin binding sites (VBS) of talin or α-actinin to the N-terminal seven-helix bundle domain of vinculin, which bury into the four-helix bundle subdomain of Vh1 to form an entirely new five-helix bundle by a mechanism coined helix bundle conversion (22). Binding of these VBSs to vinculin first requires that they unravel from their buried positions in the helix bundles of talin or α-actinin, which is provoked by mechanical stress (24, 25).
Given the reported roles of vinculin in stabilizing AJs via its interactions with α-catenin (26), we solved the crystal structure of the vinculin Vh1 domain in complex with the vinculin binding domain (VBD) of α-catenin at 2.7 Å resolution. Remarkably, this structure reveals a novel mechanism of helix bundle interactions where α-catenin is bound to vinculin in an unfurled state allowing dimerization. Furthermore, this structure and binding studies support a model whereby simultaneous binding to F-actin functions to stabilize vinculin-α-catenin interactions and AJs.
EXPERIMENTAL PROCEDURES
Protein Preparation
α-Catenin (Open Biosystems; residues 82–906 and 144–906) was cloned into the pET24b expression vector (Novagen) using NheI and NotI restriction enzyme sites with an engineered proline residue at the end of the C terminus to obtain C-terminal hexahistidine-tagged proteins. The VBD (residues 277–382) was cloned into pGEX-6P-1 (BamHI/XhoI) (GE Healthcare) to result in precision protease-cleavable GST-tagged fusion protein. α-Catenin (residues 82–634 and 263–634) (NdeI/NotI) was cloned into a modified pET28 expression vector (Novagen) to obtain precision protease-cleavable N-terminal His8-tagged proteins. The Vh1-Vt chimera (residues 1–258 and 848–1066) was obtained through single-step cloning of the Vh1 and Vt domains into a modified pET28 vector to have an N-terminal His8-tagged protein. The individual Vh1 and Vt domains were obtained through PCR by using full-length vinculin cDNA as the template, having NdeI/BamHI and BamHI/NotI restriction sites at their 5′ and 3′ end, respectively. Subsequently, both the fragments were digested by restriction endonucleases, mixed with the NdeI/NotI-treated modified pET28 vector, and ligated to obtain the final plasmid containing the DNA encoding the Vh1-Vt chimera. All constructs were verified by sequencing. Full-length vinculin (residues 1–1066), vinculin head (VH) (1–843), Vt (879–1066), and Vh1 (1–252) were generated as described (23, 27, 28).
Proteins were expressed in Escherichia coli BL21(DE3). Cells were lysed in 20 mm Tris-HCl (pH 8), 500 mm NaCl, and 20 mm imidazole by sonication for 3 min with on/off cycles of 5 s and clarified by ultracentrifugation (100,000 × g; 45 min). Expressed proteins were purified using a HisTrap chelating nickel affinity column (GE Healthcare) and eluted over a gradient to 500 mm imidazole. Affinity tags were not cleaved for α-catenin 82–906 and 144–906 constructs. Further purification was performed on a Superdex 200 (GE Healthcare) in 20 mm Tris-HCl (pH 8), 150 mm NaCl, and 5 mm DTT and concentrated to about 25 mg/ml.
For crystallizations, cell pellets of His-tagged Vh1 and GST-tagged α-catenin VBD were lysed together in 20 mm Tris-HCl (pH 8) and 150 mm NaCl and purified on a GST-Sepharose column (GE Healthcare). The complex was eluted with 15 mm reduced glutathione, GST cleaved with PreScission protease in 20 mm Tris-HCl (pH 8), 150 mm NaCl, 1 mm EDTA, and 1 mm DTT, reloaded onto a GST-Sepharose column to remove the cleaved GST, and further purified on a preparative Superdex-200 26/60 column (GE Healthcare) in 20 mm Tris-HCl and 150 mm NaCl. The complex was concentrated to 27 mg/ml.
α-Catenin-Vinculin Crystallization
Vh1-VBD complex crystals were obtained by hanging drop vapor diffusion equilibration against a reservoir condition containing 12% (w/v) polyethylene glycol 3350 and 0.3 m sodium acetate. The crystals were flash-frozen in liquid nitrogen and cryoprotected with 25% (v/v) glycerol.
X-ray Data Collection, Reduction, Structure Determination, and Crystallographic Refinement
x-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline BM22 at the Advanced Photon Source, Argonne National Laboratory and integrated and scaled using autoPROC (29), which uses XDS (X-ray Detector Software) (30) and SCALA (31) as the data reduction engine. The Vh1 domain of the vinculin-IpaA complex (32) was used as the search model to obtain molecular replacement solution with MOLREP (33). The difference electron density map calculated after initial rigid body refinement followed by restrained refinement with BUSTER (34) showed clear electron density for all four α-catenin α-helices, which enabled the manual building of the remaining model using COOT (Crystallographic Object-oriented Toolkit) (35). The final refinement statistics are detailed in Table 1.
TABLE 1.
X-ray data reduction and crystallographic refinement statistics
| X-ray data reduction statistics | |
| Space group | P21212 |
| Unit cell dimensions (a, b, c) | 57.2 Å, 73.9 Å, 107.4 Å |
| Resolution (last shell) | 107.43 Å–2.65 Å (2.8 Å–2.65 Å) |
| Total measurements | 98,915 |
| Number of unique reflections (last shell) | 13,785 (1,970) |
| Wavelength | 1 Å |
| R-mergea (last shell) | 0.068 (0.485) |
| Average I/σ(I) (last shell) | 21.2 (3.8) |
| Completeness (last shell) | 1 (1) |
| Redundancy (last shell) | 7.2 (7.3) |
| Crystallographic refinement statistics | |
| Resolution (last shell) | 50 Å–2.66 Å (2.87 Å–2.66Å) |
| No. of reflections (working set) | 12,951 |
| No. of reflections (test set) | 686 |
| R-factorb (last shell) | 0.193 (0.202) |
| Rfreec (last shell) | 0.229 (0.293) |
| No. of amino acid residues | 341 |
| No. of protein atoms | 2,678 |
| No. of solvent molecules | 82 |
| No. of acetate atoms | 36 |
| Average B-factor | |
| Protein | 64.8 Å2 |
| Solvent | 56.6 Å2 |
| Acetate | 70.4 Å2 |
| r.m.s.d.d from ideal geometry | |
| Bond lengths | 0.01 Å |
| Bond angles | 1.06° |
| Ramachandran plot statistics | |
| Most favored | 98.8% |
| Additional allowed | 1.2% |
| Generously allowed | 0.0% |
| Disallowed | 0.0% |
a R-merge = .
b R-factor = where 〈Fcalc〉 denotes the expectation of Fcalc(hkl) used in defining the likelihood refinement target.
c The free R-factor is a cross-validation residual calculated by using about 5% reflections, which were randomly chosen and excluded from the refinement.
d r.m.s.d., root mean square deviation.
Size Exclusion and Light Scattering
Light scattering (LS) data were collected by size exclusion chromatography using a Superdex 200, 10/300, HR column (GE Healthcare) that was connected to an Agilent 1200 high performance liquid chromatography (HPLC) system (Agilent Technologies, Wilmington, DE) equipped with an autosampler. Elution was monitored using a photodiode array UV-visible detector (Agilent Technologies), a differential refractometer (OptiLab rEx, Wyatt Technology Corp., Santa Barbara, CA) and static and dynamic, multiangle laser LS detector (HELEOS II with Quasi-Elastic-Light-Scattering capability, Wyatt Technology). The size exclusion chromatography-UV/LS/refractive index system was equilibrated in 20 mm Tris (pH 8) and 150 mm NaCl, with a flow rate of 0.5 ml/min. For data collection and analysis, two software packages were used. The ChemStation software (Agilent Technologies) controlled the operation of the HPLC and data collection from the UV-visible detector, whereas the ASTRA software (Wyatt Technology) was used for data collection from the refractive index and LS detectors and to record the 280-nm UV trace from the photodiode array detector. ASTRA software was used to determine the weight average molecular masses across the entire elution profile in 1-s intervals from static LS measurements as described (36).
Native Gel and F-actin Co-sedimentation Assays
All binding studies were performed in 20 mm Tris-HCl (pH 8), 150 mm NaCl, and 5 mm DTT. VH-Vt or VH-α-catenin (82–906, 82–634, or 263–634) complexes were obtained by incubating equimolar amounts (15 μm) of proteins for 10 min at room temperature. Complexes were then incubated with Vt or α-catenin, respectively, at equimolar and 5-fold excess concentration for an additional 10 min. The samples were analyzed by PhastGel using native PhastGel buffer strips (homogeneous 20% for Fig. 4, C–E or 10–15% gradient for Figs. 4, A and B, and 5) and stained with Coomassie Blue. Similarly, VBS (residues 332–353) was added to preformed Vh1-Vt complexes at 5-fold excess concentration. F-actin pulldown assays were carried out as described (37).
FIGURE 4.
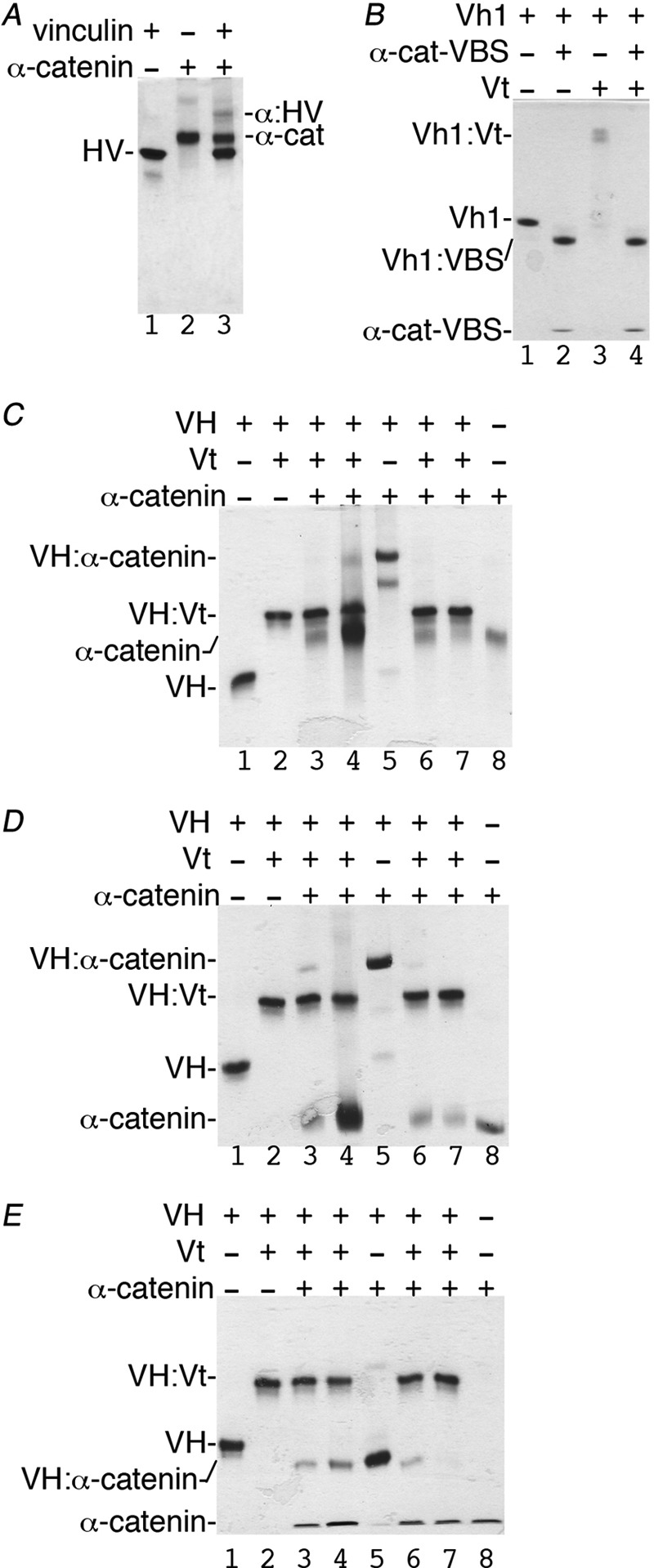
Vt has higher affinity for VH when compared with α-catenin. A, native gel shift assay shows that some full-length human vinculin (HV, residues 1–1066, lane 1) binds to full-length α-catenin (α-cat, residues 82–906, lane 2; complex, α-HV, lane 3). B, the minimal α-catenin VBS (α-cat:VBS; residues 332–353) displaces Vt from the Vh1-Vt complex (lane 3) to form a new Vh1-α-catenin-VBS complex (Vh1:VBS, lanes 2 and 4) that migrates farther than Vh1 alone (lane 1). C–E, reciprocal competition assays by titrating equimolar concentrations (15 μm) or 5-fold excess concentrations (75 μm) of α-catenin (lanes 3 and 4) residues 82–906 (C), residues 82–634 (D), or 263–634 (E) to VH-Vt versus titration of increasing amounts of Vt to VH in complex with α-catenin (lanes 6 and 7) residues 82–906 (C), 82–634 (D), or 263–634 (E) as analyzed by native gel shift mobility assays. VH alone (lane 1), VH-Vt (lane 2), VH-α-catenin (lane 5), and α-catenin (lane 8) are also shown. As is evident, all three α-catenin constructs fail to displace Vt from preformed VH-Vt complexes (lanes 3 and 4). In contrast, Vt readily displaces all three α-catenin constructs from preformed VH-α-catenin complexes (lanes 6 and 7). VH, vinculin head domain (residues 1–843); Vt, vinculin tail domain (residues 879–1066).
FIGURE 5.
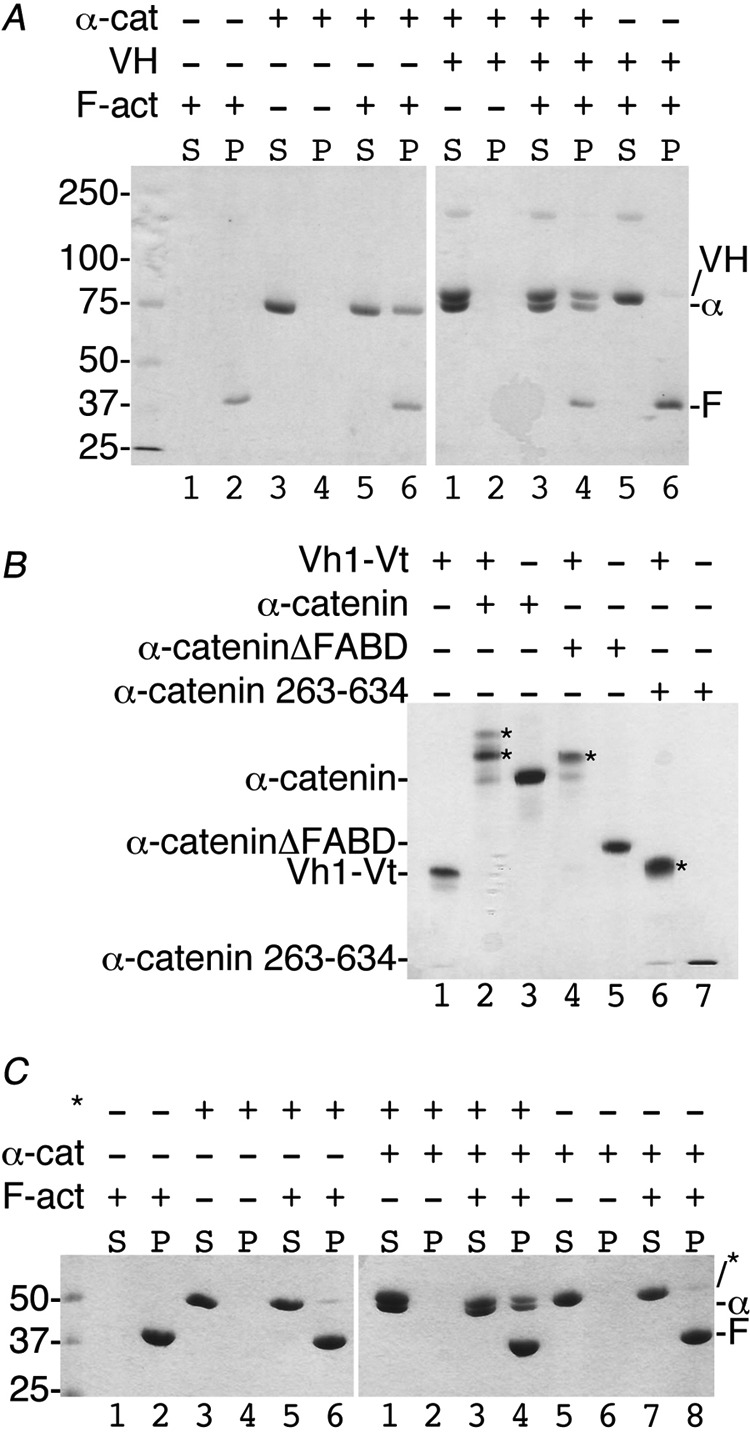
Vinculin-α-catenin complex co-sediments with F-actin via F-actin binding domain (FABD) of either α-catenin or vinculin. A, the α-catenin-VH complex binds F-actin via the α-catenin FABD. About 30 μm of either vinculin or α-catenin was used in the corresponding reaction mixtures. Left gel, lanes 1 and 2, F-actin is found in the pellet (P). Lanes 3 and 4, α-catenin does not aggregate and remains in the soluble fraction (S). Lanes 5 and 6, α-catenin binds to F-actin. Right gel, lanes 1 and 2, the VH-α-catenin complex does not aggregate. Lanes 3 and 4, the VH-α-catenin complex binds to F-actin via the α-catenin FABD. Lanes 5 and 6, VH does not bind to F-actin. Molecular weight markers are shown for the left gel. α-cat or α, α-catenin (residues 144–906); VH, vinculin head domain (residues 1–843); F-act or F, F-actin. B, the Vt2 domain seems to hinder closed, inactive vinculin to bind to α-catenin. The addition of α-catenin (residues 82–906, 20 μm, lane 2), α-cateninΔFABD (residues 82–634, 20 μm, lane 4), or α-catenin 263–634 (20 μm, lane 6) to the Vh1-Vt chimera (residues 1–258 and 848–1066, 15 μm) lacking the Vt2 domain results in α-catenin-vinculin complex formation (indicated by asterisks). Binding in each case was saturable with no unbound α-catenin remaining. C, the α-cateninΔFABD-vinculin complex binds F-actin via Vt. Left gel, lanes 1 and 2, F-actin is found in the pellet (P). Lanes 3 and 4, the Vh1-Vt chimera (asterisks) does not aggregate and remains in the soluble fraction (S). Lanes 5 and 6, the Vh1-Vt chimera does not bind to F-actin. Right gel, lanes 1 and 2, the Vh1-Vt chimera in complex with α-cateninΔFABD does not aggregate. Lanes 3 and 4, the Vh1-Vt chimera in complex with α-cateninΔFABD binds F-actin via Vt. Lanes 5 and 6), α-cateninΔFABD does not aggregate. Lanes 7 and 8, α-cateninΔFABD does not bind to F-actin. α-cat or α, α-catenin (residues 144–906); F-act or F, F-actin. Molecular weight markers are shown for the left gel.
RESULTS
Structure of Unfurled VBD of α-Catenin in Complex with Vh1
Binding studies have suggested that α-catenin residues 327–402 were necessary for binding of α-catenin to vinculin (38, 39), and more recently, the minimal VBS was determined as residues 325–360 (40) or 326–377 (41). We determined the α-catenin VBD as residues 277–382 based on secondary structure analyses and homology model prediction (42) and co-crystallized the Vh1-VBD complex. The crystals belong to the orthorhombic space group P21212 with unit cell dimensions of a = 57.2 Å, b = 73.9 Å, and c = 107.4 Å (Table 1) with one heterodimer in the asymmetric unit (Z = 4) resulting in a solvent content of 59% and a crystal volume per unit of protein molecular weight, VM (43), of 3.03 Å3/Da. The Vh1-VBD structure was determined to 2.7 Å resolution. The regions corresponding to Vh1 residues 219–222 and α-catenin 277–289 were not modeled due to lack of discernible electron density, and although the electron density for α-catenin residues 354–361 is weak, the connectivity is unambiguous (supplemental Fig. S1). The final model comprises one Vh1-VBD, nine acetates, and 82 water molecules with a crystallographic and free R-factor of 0.19 and 0.23, respectively (Table 1). All residues fall in allowed regions of the Ramachandran plot as calculated by MolProbity (44) with clash scores of 1.64 (100th percentile) and 7.48 (99th percentile), respectively.
The structure of the Vh1 domain of vinculin when bound to α-catenin resembles that of other VBS-bound Vh1 structures (supplemental Fig. S2A) where the two four-helix bundle (4HB) subdomains are connected by one centrally shared α-helix (α4) (Fig. 1A). As first reported for Vh1 bound to talin-VBS3 (22), the minimal α-catenin VBS (residues 328–352) forms an α-helix that binds the N-terminal 4HB of Vh1 by helix bundle conversion, as subsequently also seen for the VBSs of talin (22, 28, 45–48), α-actinin (49), and the bacterial IpaA (32, 50, 51) and sca4 (52) in complex with Vh1. Interestingly, although α-catenin residues 328DRRERIVAECNAVRQALQDLLS349 align with talin VBS3 residues 1949ELIECARRVSEKVSHVLAALQA1970 (41), talin residues 1948ELIESARKVSEKVSHVLALAQA1969 (Protein Data Bank (PDB) entry 1rkc) are structurally equivalent to α-catenin residues 332RIVAECNAVRQALQDLLSEYMG353 i.e. shifted in register by 4 residues.
FIGURE 1.
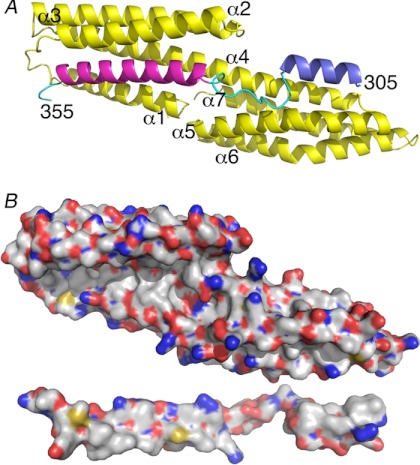
α-Catenin is unfurled when bound to vinculin. A, the crystal structure of Vh1 (residues 1–258; yellow) in complex with α-catenin VBD shows an extended conformation of α-catenin with extensive interactions with both subdomains of Vh1. Vh1 α-helices and α-catenin termini are labeled. Residues 305–316 are shown in blue, 328–351 are in magenta, and the remainder of the α-catenin residues are in cyan. For clarity, residues 290–304 and 362–382 are not shown. B, surface representation showing the respective interfaces (white, carbon; red, oxygen; blue, nitrogen; yellow, sulfur). Vh1 is in the same orientation as shown in A, and α-catenin is rotated 180° down with respect to its orientation shown in A.
Unexpectedly, a second α-helix (α-catenin residues 305–316) binds the C-terminal 4HB of Vh1 in a novel fashion, by packing its hydrophobic face against the first (α4) and second (α5) α-helices of the C-terminal Vh1 4HB (Fig. 1A). Collectively, these dual interactions create a large hydrophobic interface that buries nearly 25% of the VBD solvent-accessible surface area (over 2,300 Å2) and about 14% of that present on Vh1 (over 1,900 Å2). These interactions include 12 hydrogen bonds (vinculin residues Thr-8 with α-catenin Arg-330, Pro-15 with α-catenin Lys-358, Gln-19 with α-catenin Gln-345 and Ser-349, His-27 with α-catenin Tyr-351, Thr-61 and Thr-64 with α-catenin Arg-329, Arg-136 with α-catenin Gly-315, and Lys-170 with α-catenin Ala-317 and Met-319) and novel hydrophobic interactions (vinculin residues Val-137, Gly-140, Ile-141, and Gly-167 with α-catenin Leu-318) (Fig. 1B, supplemental Fig. S3A). In comparison, the buried solvent-accessible surface area in other Vh1-VBS structures are only about 1,200 Å2 (22).
Although two IpaA-VBSs have been shown to simultaneously bind to Vh1, where the second binding site is located between two different α-helices (α5 and α6) in the C-terminal 4HB (supplemental Fig. S2B), these were short IpaA-VBS peptides (22 residues) that bind with 2:1 molar ratio (32) and that bury less than 1,300 Å2 of solvent-accessible surface area. Further, no hydrogen bonds were observed for this interaction (32). In contrast, our Vh1-VBD structure is the first report of an entire helix bundle (116 residues) having good electron density that connects two individual α-helices that bind to vinculin. Importantly, in our Vh1-VBD structure, the binding sites for α-catenin on both subdomains of Vh1 are located on the surface of full-length vinculin. Thus, these regions are accessible for α-catenin binding to the full-length vinculin in its inactive, closed conformation (supplemental Fig. S4).
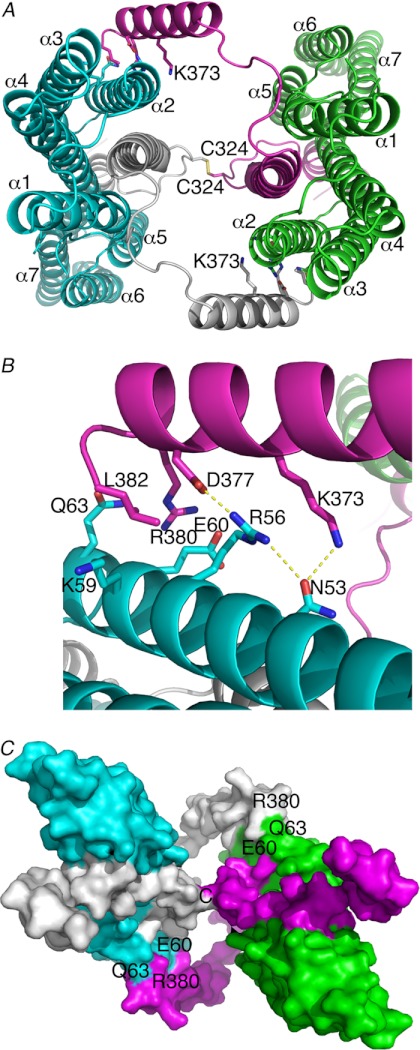
Two additional α-helices of the α-catenin VBD seen in our crystal structure, one on each terminus, engage in further and novel interactions with vinculin (supplemental Fig. S5). Although the N-terminal α-helix is quite short (residues 293–300) (supplemental Fig. S4) and makes crystal contacts with α-helices α5 and α6 of a symmetry-related Vh1 molecule (supplemental Figs. S3B and S5B), the C-terminal α-helix lies almost parallel (at about 15°) to α-helix α2 from a two-fold related Vh1 molecule (Fig. 2B, supplemental Fig. S4). The resulting dimer seems further stabilized by a disulfide bond that is formed between Cys-324 and its symmetry-related mate (Fig. 2, A and C). Although α-catenin is predominantly monomeric but also found as a dimer in solution (53), we found that our Vh1-VBD complex, as well as unbound VBD, is either monomeric or dimeric in solution (Fig. 3). Furthermore, the elution of VBD is consistent with a compact globular domain in solution. Thus, the VBD unfurls upon binding to vinculin in accord with calorimetry data showing that α-catenin (residues 273–510) bound to the VH domain (residues 1–718) with nm affinity and that the binding was endothermic and entropy-driven, consistent with a major conformational change (39). Collectively, our data are consistent with the notion that α-catenin unfurls to bind to vinculin, allowing dimerization.
FIGURE 2.
α-Catenin-vinculin complex crystal structure. Vh1 is shown in cyan or green, and its respective bound α-catenin is shown in white or magenta. A, α-catenin Cys-324 forms a disulfide bond with a two-fold related molecule. Likewise, the C-terminal α-catenin α-helix binds to the second Vh1 α-helix of a two-fold related heterodimer burying over 500 Å2 of solvent-accessible surface area with a shape correlation statistic derived using the CCP4 program SC (55) of over 0.8 for this interface, a significant value where a value of 1 indicates a perfect fit versus 0.35, which indicates the mismatch of an artificial association. Vh1 α-helices are labeled, as well as some α-catenin residues 324 and 373. B, close-up view of the dimer interface. Vinculin residues are shown in cyan, and α-catenin are in magenta as in A and in a slightly rotated view from A. C, space-filling representation of the dimer of heterodimers (same color coding as shown in A) and rotated about 180° about the horizontal axis of the A image. Some contact residues are indicated.
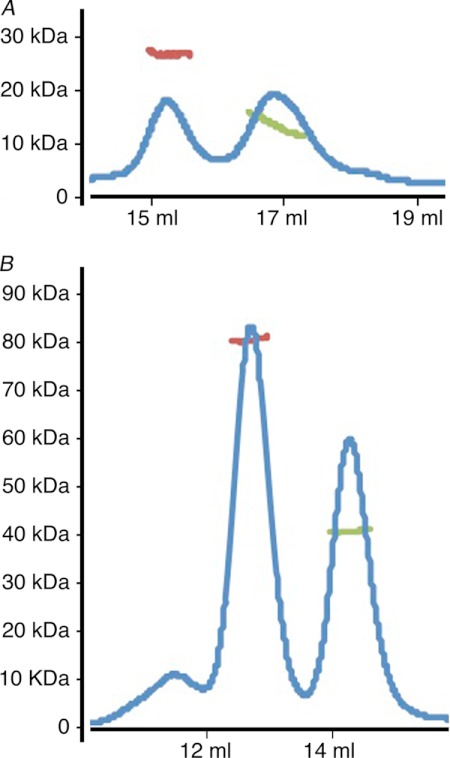
FIGURE 3.
VBD alone or in complex with Vh1 is a monomer and dimer in solution. A and B, the oligomeric states of the α-catenin VBD either alone (A) or in complex with vinculin (B) are estimated by multiangle light scattering combined with size exclusion chromatography, and the respective profiles are provided. The blue line represents the UV absorbance (without units), whereas the green and red lines denote the molecular masses (shown in kDa on the ordinate) as calculated by ASTRA (software provided by Wyatt Technology) as follows: A, 13.6 and 27.2 kDa where the molecular mass of VBD is 12.3 kDa; B, 40.9 and 80.5 kDa where the molecular mass of the 1:1 Vh1-VBD complex is 41.1 kDa.
Vinculin-α-Catenin Binding Is Facilitated by Severing Vinculin Head-Tail Interaction
To avoid potential nonspecific effects on the binding, we assessed the binding properties of untagged α-catenin and vinculin proteins. Because deletion of residues 1–81 increases the stability of α-catenin (20), we thus cloned α-catenin residues 82–906 (hereafter referred to as full-length) for our binding studies. α-Catenin was reported to bind weakly to full-length vinculin (16, 39). Indeed, only a fraction of full-length α-catenin and vinculin formed a complex in solution, as seen in our native gel shift assays (Fig. 4A). Thus, full-length α-catenin in its inactive conformation is not sufficient to activate vinculin. However, the minimal α-catenin-VBS is sufficient for severing the vinculin Vh1-Vt interaction to form a novel Vh1-VBS complex (Fig. 4B), as seen for the VBSs of talin (22, 28, 45–48), α-actinin (49), or the bacterial IpaA (32, 50, 51) and sca4 (52).
Co-localization studies of α-catenin proteins have suggested that an inhibitory domain (α-catenin residues 510–697) prevents vinculin binding (40). However, full-length α-catenin harboring the 510–697 region readily bound to VH (the globular head domain of vinculin, residues 1–843) (Fig. 4C). Surprisingly, the Vt efficiently displaced full-length α-catenin (Fig. 4C) and more truncated versions of α-catenin (residues 82–634, Fig. 4D; or residues 263–634, Fig. 4E) from preformed VH-α-catenin complexes. Thus, intramolecular interactions of vinculin are preferred over those with α-catenin.
F-actin Binds α-Catenin-Vinculin Complex via the C Terminus of Either α-Catenin or Vinculin
The binding of full-length vinculin to F-actin has been reported to be enhanced by α-catenin (residues 273–510) (54). These findings, together with Vt displacement of α-catenin from α-catenin-VH complexes, suggested that once bound to F-actin, Vt cannot displace α-catenin. To test this notion, F-actin co-sedimentation studies were performed. The vinculin VH domain (residues 1–840) formed a ternary complex with α-catenin (residues 144–906; this shorter construct was used to distinguish VH from α-catenin) and F-actin (Fig. 5A), indicating that vinculin-bound α-catenin can bind to F-actin via its C-terminal domain (19). Similarly, α-catenin lacking its F-actin binding domain (α-catenin-ΔFABD, residues 82–634), can form a complex with a vinculin head-tail chimera (Fig. 5B) lacking the vinculin Vt2 domain shown to keep vinculin in its closed conformation (56) and F-actin (Fig. 5C), indicating that F-actin binding blocks the ability of Vt to displace α-catenin. Furthermore, α-catenin activates vinculin lacking the Vt2 domain. Collectively, these findings suggest that F-actin stabilizes the α-catenin-vinculin interaction by binding to the C terminus of α-catenin and/or vinculin.
DISCUSSION
Stabilization of AJs to the actin cytoskeleton is necessary for maintenance of cell-cell contacts (17), yet the mechanism by which this occurs is not resolved. One clear contributor to the response is α-catenin, which is necessary for the formation and stabilization of AJs, for recruitment of vinculin to these sites, and for linking cadherin complexes to the actin cytoskeleton (16–19). α-Catenin also controls branched actin polymerization (53) and directly associates with F-actin during junction reinforcement (40). Our findings suggest that a complex interplay of vinculin, α-catenin, and the F-actin network are together necessary to stabilize AJs.
The structure of the vinculin Vh1-α-catenin VBD complex shows that α-catenin unfurls upon complex formation, allowing dimerization (Fig. 2), and that it has a novel vinculin binding mode, where two α-helices in the VBD bind simultaneously to the four-helix bundle subdomains of the Vh1 domain. Further, the α-catenin binding sites are fully accessible in full-length vinculin (supplemental Fig. S4), and the entire globular head VH domain readily binds to α-catenin (Fig. 4, C–E). However, the two full-length proteins interact only weakly in solution (Fig. 4A), and competitive binding studies support a model whereby vinculin in its open conformation binds to F-actin and then binds to and unfurls α-catenin to form a stable ternary complex.
Vinculin recruitment by α-catenin is force-dependent (40). However, the intramolecular head-tail interaction of vinculin impairs α-catenin-vinculin interaction. Furthermore, the formation and/or stabilization of the α-catenin-vinculin complex is greatly stabilized by the binding of either partner to F-actin through their C-terminal domains. Here proximity rules may come into play, where F-actin-bound α-catenin promotes the association of vinculin with F-actin as vinculin is a highly dynamic protein and exists in equilibrium between its closed and open states. Collectively, these findings suggest that cytoskeletal tension stabilizes the α-catenin-vinculin complex at AJs.
α-Catenin deletion mutants that retain the VBD but cannot bind to the cytoskeleton can recruit vinculin to AJs, which led to the hypothesis that a region C-terminal of the VBD prevents α-catenin binding to vinculin and that this inhibition is relieved when α-catenin is bound to the actin cytoskeleton (40). Furthermore, binding studies with truncated proteins suggested the hypothesis that α-catenin activates vinculin (41). While α-catenin binds vinculin weakly in native gel assays, our competition binding assays show that α-catenin readily binds to VH, yet is displaced by Vt. Thus, the nanomolar affinity of the intramolecular VH-Vt interaction prevents α-catenin binding and vinculin needs to be in its open state to bind to α-catenin. Collectively, the data indicate that α-catenin does not activate vinculin but rather that F-actin bound-vinculin unfurls and binds to α-catenin to stabilize AJs.
What is still not resolved is what trigger activates vinculin at AJs. One possibility is that this response simply relies on an equilibrium of vinculin in its open versus activated state, where the latter can bind to F-actin that then facilitates interactions with α-catenin. Alternatively, vinculin may bind to other partners at AJs to augment this response, for example, to the Arp2/3 complex, as Arp2/3-directed actin polymerization is suppressed by α-catenin (11).
Supplementary Material
Acknowledgments
We are indebted to our colleagues at Scripps Florida: John Cleveland for discussions and critical review of the manuscript, Zhen Wu and Philippe Bois for sequencing, and Philippe Bois for fruitful discussions. V. Nayak generated the α-catenin 82–906, 144–906, and 277–382 constructs, and HaJeung Park and Ewa Folta-Stogniew (Yale) provided SLS data. We are grateful to the staff at the SER-CAT (beamline BM22) for synchrotron support.
This is Publication Number 21520 from the Scripps Research Institute.

This article contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 4ehp) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AJ
- adherens junction
- 4HB
- four-helix bundle
- FABD
- F-actin binding domain
- LS
- light scattering
- VBS
- vinculin binding site
- VBD
- vinculin binding domain
- VH
- vinculin head
- Vh1
- first vinculin head domain
- Vt
- vinculin tail domain.
REFERENCES
- 1. Braga V. M., Balda M. S. (2004) Regulation of cell-cell adhesion. Semin. Cell Dev. Biol. 15, 631–632 [DOI] [PubMed] [Google Scholar]
- 2. Pokutta S., Weis W. I. (2002) The cytoplasmic face of cell contact sites. Curr. Opin. Struct. Biol. 12, 255–262 [DOI] [PubMed] [Google Scholar]
- 3. Clevers H. (2006) Wnt/β-Catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 4. Wahl J. K., 3rd, Kim Y. J., Cullen J. M., Johnson K. R., Wheelock M. J. (2003) N-cadherin-catenin complexes form prior to cleavage of the proregion and transport to the plasma membrane. J. Biol. Chem. 278, 17269–17276 [DOI] [PubMed] [Google Scholar]
- 5. Haegel H., Larue L., Ohsugi M., Fedorov L., Herrenknecht K., Kemler R. (1995) Lack of β-catenin affects mouse development at gastrulation. Development 121, 3529–3537 [DOI] [PubMed] [Google Scholar]
- 6. Radice G. L., Rayburn H., Matsunami H., Knudsen K. A., Takeichi M., Hynes R. O. (1997) Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181, 64–78 [DOI] [PubMed] [Google Scholar]
- 7. Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O., Kemler R. (2001) Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264 [DOI] [PubMed] [Google Scholar]
- 8. Huelsken J., Vogel R., Erdmann B., Cotsarelis G., Birchmeier W. (2001) β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 105, 533–545 [DOI] [PubMed] [Google Scholar]
- 9. Pokutta S., Drees F., Takai Y., Nelson W. J., Weis W. I. (2002) Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pappas D. J., Rimm D. L. (2006) Direct interaction of the C-terminal domain of α-catenin and F-actin is necessary for stabilized cell-cell adhesion. Cell Commun. Adhes. 13, 151–170 [DOI] [PubMed] [Google Scholar]
- 11. Drees F., Pokutta S., Yamada S., Nelson W. J., Weis W. I. (2005) α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 123, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J. (2005) Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weis W. I., Nelson W. J. (2006) Re-solving the cadherin-catenin-actin conundrum. J. Biol. Chem. 281, 35593–35597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pokutta S., Drees F., Yamada S., Nelson W. J., Weis W. I. (2008) Biochemical and structural analysis of α-catenin in cell-cell contacts. Biochem. Soc. Trans. 36, 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abe K., Takeichi M. (2008) EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. U.S.A. 105, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiss E. E., Kroemker M., Rüdiger A. H., Jockusch B. M., Rüdiger M. (1998) Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 141, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasioukhin V., Bauer C., Yin M., Fuchs E. (2000) Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100, 209–219 [DOI] [PubMed] [Google Scholar]
- 18. Maddugoda M. P., Crampton M. S., Shewan A. M., Yap A. S. (2007) Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell-cell contacts in mammalian epithelial cells. J. Cell Biol. 178, 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rimm D. L., Koslov E. R., Kebriaei P., Cianci C. D., Morrow J. S. (1995) α1(E)-Catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. U.S.A. 92, 8813–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pokutta S., Weis W. I. (2000) Structure of the dimerization and β-catenin-binding region of α-catenin. Mol. Cell 5, 533–543 [DOI] [PubMed] [Google Scholar]
- 21. Yang J., Dokurno P., Tonks N. K., Barford D. (2001) Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 20, 3645–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Izard T., Evans G., Borgon R. A., Rush C. L., Bricogne G., Bois P. R. (2004) Vinculin activation by talin through helical bundle conversion. Nature 427, 171–175 [DOI] [PubMed] [Google Scholar]
- 23. Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., Izard T. (2004) Crystal structure of human vinculin. Structure 12, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 24. Ylänne J., Scheffzek K., Young P., Saraste M. (2001) Crystal structure of the α-actinin rod reveals an extensive torsional twist. Structure 9, 597–604 [DOI] [PubMed] [Google Scholar]
- 25. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lecuit T. (2010) α-Catenin mechanosensing for adherens junctions. Nat. Cell Biol. 12, 522–524 [DOI] [PubMed] [Google Scholar]
- 27. Park H., Rangarajan E. S., Sygusch J., Izard T. (2010) Dramatic improvement of crystal quality for low-temperature-grown rabbit muscle aldolase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yogesha S. D., Sharff A., Bricogne G., Izard T. (2011) Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin. Protein Sci. 20, 1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D. Biol. Crystallogr. 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 31. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 32. Nhieu G. T., Izard T. (2007) Vinculin binding in its closed conformation by a helix addition mechanism. EMBO J. 26, 4588–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vagin A., Teplyakov A. (1997) MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 34. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek P., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Folta-Stogniew E., Williams K. R. (1999) Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10, 51–63 [PMC free article] [PubMed] [Google Scholar]
- 37. Bois P. R., O'Hara B. P., Nietlispach D., Kirkpatrick J., Izard T. (2006) The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 281, 7228–7236 [DOI] [PubMed] [Google Scholar]
- 38. Imamura Y., Itoh M., Maeno Y., Tsukita S., Nagafuchi A. (1999) Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144, 1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bakolitsa C., Cohen D. M., Bankston L. A., Bobkov A. A., Cadwell G. W., Jennings L., Critchley D. R., Craig S. W., Liddington R. C. (2004) Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 [DOI] [PubMed] [Google Scholar]
- 40. Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M. (2010) α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 [DOI] [PubMed] [Google Scholar]
- 41. Peng X., Maiers J. L., Choudhury D., Craig S. W., DeMali K. A. (2012) α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 287, 7728–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 43. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 44. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Izard T., Vonrhein C. (2004) Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 279, 27667–27678 [DOI] [PubMed] [Google Scholar]
- 46. Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 47. Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 48. Yogesha S. D., Rangarajan E. S., Vonrhein C., Bricogne G., Izard T. (2012) Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin. Protein Sci. 21, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bois P. R., Borgon R. A., Vonrhein C., Izard T. (2005) Structural dynamics of α-actinin-vinculin interactions. Mol. Cell. Biol. 25, 6112–6122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 50. Izard T., Tran Van Nhieu G., Bois P. R. (2006) Shigella applies molecular mimicry to subvert vinculin and invade host cells. J. Cell Biol. 175, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park H., Valencia-Gallardo C., Sharff A., Tran Van Nhieu G., Izard T. (2011) Novel vinculin binding site of the IpaA invasin of Shigella. J. Biol. Chem. 286, 23214–23221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park H., Lee J. H., Gouin E., Cossart P., Izard T. (2011) The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J. Biol. Chem. 286, 35096–35103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benjamin J. M., Kwiatkowski A. V., Yang C., Korobova F., Pokutta S., Svitkina T., Weis W. I., Nelson W. J. (2010) αE-Catenin regulates actin dynamics independently of cadherin-mediated cell-cell adhesion. J. Cell Biol. 189, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Janssen M. E., Kim E., Liu H., Fujimoto L. M., Bobkov A., Volkmann N., Hanein D. (2006) Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell 21, 271–281 [DOI] [PubMed] [Google Scholar]
- 55. Lawrence M. C., Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 56. Cohen D. M., Chen H., Johnson R. P., Choudhury B., Craig S. W. (2005) Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280, 17109–17117 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.