Background: Hepatitis C virus (HCV) induces autophagosomes in its host cells.
Results: The HCV RNA replication complex colocalizes with autophagosomes, which are induced by HCV via a Class III PI3K-independent pathway.
Conclusion: HCV induces autophagosomes and uses their membranes for its RNA replication.
Significance: The perturbation of the autophagic pathway by HCV may have important consequences in HCV pathogenesis.
Keywords: Autophagy, Hepatitis C Virus, RNA Viruses, Viral Replication, Virology
Abstract
Previous studies indicated that hepatitis C virus (HCV) perturbs the autophagic pathway to induce the accumulation of autophagosomes in cells. To understand the role of autophagosomes in the HCV life cycle, we established a stable Huh7 hepatoma cell line that contained an HCV subgenomic RNA replicon and also expressed a GFP-LC3 fusion protein. The GFP-LC3 protein is localized to autophagosomes during autophagy and served as a convenient marker for autophagosomes. Our results indicate that the silencing of the expression of LC3 or Atg7, two protein factors critical for the formation of autophagosomes, suppresses the replication of HCV RNA. Confocal microscopy studies revealed the localization of HCV NS5A and NS5B proteins, which are two important components of the HCV RNA replication complex, and nascent HCV RNA to autophagosomes. The association of the HCV RNA replication complex with the autophagosomal membranes was further confirmed by co-immunoprecipitation and immunoelectron microscopy studies. Interestingly, inhibition of Class III PI3K activity had no effect on the autophagosomes induced by HCV. These results indicate that HCV induces autophagosomes via a Class III PI3K-independent pathway and uses autophagosomal membranes as sites for its RNA replication.
Introduction
Hepatitis C virus (HCV)2 is an important human pathogen that can cause severe liver diseases, including liver cirrhosis and hepatocellular carcinoma. This virus belongs to the Flaviviridae family and has a positive-strand RNA genome of ∼9.6 kb. The HCV genome codes for a polyprotein that is ∼3000 amino acids in length. The translation of this polyprotein is mediated by an internal ribosomal entry sequence. This polyprotein is cleaved by cellular and viral proteases to generate 10 mature protein products named core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. Among these proteins, core, E1, and E2 are structural proteins; p7, NS2, and NS5A are required for viral assembly and release; and NS3, NS4A, NS4B, NS5A, and NS5B can form a complex to mediate the replication of the HCV genomic RNA (for a review, see Ref. 1).
HCV is a hepatotropic virus. Recent studies indicated that HCV perturbs the autophagic pathway to induce the accumulation of autophagosomes in hepatoma cells (2–9). Autophagy plays an important role in the removal of damaged organelles and protein aggregates from cells for recycling and is important for maintaining cellular homeostasis. It is initiated by the formation of membrane crescents known as isolation membranes or phagophores in the cytoplasm. These isolation membranes will grow into enclosed membrane vesicles known as autophagosomes. Autophagosomes are characterized by their double- and sometimes multiple-membrane structures. They mature by fusing with lysosomes, and their cargos will be digested by lysosomal enzymes. Many protein factors that control autophagy have been identified. For examples, Class III PI3K (PI3KC3) is important for the initiation of autophagy; Atg5, Atg12, and Atg16 are important for the formation of isolation membranes; and Atg3, Atg4, and Atg7 are important for the conjugation of LC3 to phosphatidylethanolamine for the formation of autophagosomes. LC3 is a cytosolic protein. However, it is localized to the autophagosomal membranes after its lipidation (for a review, see Ref. 10).
Some viruses such as HSV-1, human cytomegalovirus, and Kaposi sarcoma-associated herpesvirus can suppress autophagy (11–13). In the case of HSV-1, this suppression of autophagy can enhance viral replication and pathogenesis (11). Some other viruses such as poliovirus and hepatitis B virus can induce autophagy to enhance their replication (Refs. 14 and 15; for a review, see Ref. 16). HCV has also been shown to perturb the autophagic pathway to cause the accumulation of autophagosomes (2–8). This perturbation is mediated by the unfolded protein response, which is activated by endoplasmic reticulum stress (3, 4). The induction of autophagosomes by HCV is independent of its genotypes (2, 3). It also does not require the HCV coding sequence upstream of the NS3 sequence, as the HCV subgenomic RNA replicon that expresses only the NS3-NS5B sequence is sufficient to induce autophagosomes (3).
The use of RNAi to suppress the expression of genes important for the formation of autophagic vacuoles has been shown to have a negative effect on HCV (3, 6, 17). However, there are controversies regarding how the autophagic pathway may assist HCV replication, as some studies suggested that it enhanced HCV RNA replication, whereas others suggested that it might affect the release of HCV particles or the translation of the HCV RNA (3, 5–7, 17). These controversies may be caused by a variety of reasons, including the use of different experimental systems such as transfection using the HCV genomic RNA or infection using the HCV particles, and the selection of different autophagic cellular factors for the RNAi knockdown experiments. Our previous studies indicated that the knockdown of LC3 or Atg7, which suppresses the formation of autophagosomes, significantly reduces the HCV RNA level in cells productively replicating HCV (3), suggesting a possible role of autophagosomes in HCV RNA replication. These results were supported by a recent report that demonstrated that HCV NS3 and NS5A proteins co-fractionated with the lipidated LC3 protein on a sucrose gradient and that the double-stranded HCV RNA (i.e. the HCV replicative intermediate RNA) was detected on autophagosome-like membrane vesicles (8). However, in this recent report, it was unclear whether the co-fractionation of NS3 and NS5A with lipidated LC3 on the gradient was incidental and whether those autophagosome-like vesicles were indeed autophagosomes.
Thus, in an attempt to further understand the relationship between autophagosomes and HCV RNA replication, we produced an HCV subgenomic RNA replicon cell line that is devoid of HCV structural proteins for our studies. Our results demonstrate that HCV RNA replication takes place primarily on autophagosomal membranes in these replicon cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Nutrient Starvation
Huh7 is a human hepatoma cell line, and Huh7.5 is a subline of Huh7. These cell lines were maintained in DMEM supplemented with 10% FBS. For nutrient starvation, cells were incubated in Hanks' balanced salt solution for 30 min prior to lysis for Western blot analysis. HuhHyg replicon cells are Huh7 cells that contain the subgenomic RNA replicon of the HCV-N strain, which belongs to genotype 1b (18). The production of this cell line has been described previously (19). This cell line was maintained in DMEM containing 10% FBS and 150 μg/ml hygromycin B. To generate the GFP-LC3 replicon (GLR) cells, HuhHyg replicon cells were transfected with plasmid pEGFP-LC3, which expresses the GFP-LC3 fusion protein, followed by selection with 0.7 μg/ml G418 and 150 μg/ml hygromycin B. Sg-PC2 is another Huh7 cell line that contains the HCV Con1 subgenomic RNA replicon. This cell line has also been described previously (20).
siRNA Knockdown
Atg7, LC3(1), LC3(2), Beclin-1, human Vps34 (hVps34), and negative control siRNAs were purchased from Qiagen. LC3(3) siRNA, which was described previously (21), was synthesized at the Genomics Core of the USC Norris Comprehensive Cancer Center. The siRNA knockdown was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Confocal Microscopy
Cells fixed with 4% formaldehyde were incubated with mouse anti-NS5A antibody 9E10 (a gift from Dr. Charles Rice, Rockefeller University), rabbit anti-NS5B antibody (a gift from Dr. Soon Hwang, Hallym University, Gangwon-do, South Korea), mouse anti-Rab7 antibody (Sigma), or mouse anti-bromouridine antibody (Sigma), followed by rhodamine-conjugated goat anti-mouse or Alexa Fluor 405-conjugated goat anti-rabbit antibody for confocal microscopy. Cell nuclei were stained with DAPI.
BrUTP Labeling
Cells pretreated with 5 μg/ml actinomycin D for 1 h at 37 °C were washed with buffer A (50 mm Tris-HCl (pH 8), 4.5 mm magnesium acetate, 20 mm KCl, 5 mm NaCl, and 150 mm sucrose) and incubated with buffer A containing 100 μg/ml lysolecithin for 90 s on ice. Cells were then treated for 40 min with buffer B (50 mm Tris-HCl (pH 8), 6 mm magnesium acetate, 20 mm KCl, 44 mm NaCl, 150 mm sucrose, 1 mm ATP, 200 μm GTP, 200 μm CTP, 500 μm BrUTP, 10 μm dTTP, 12 μm creatine phosphate, 200 μm spermidine, 10 μg/ml actinomycin D, 100 μg/ml creatine phosphokinase, and 1 mm DTT) at 37 °C for the labeling of nascent HCV RNA.
Isolation of Autophagosomes
Cells scraped in 20 mm HEPES (pH 7) and 0.25 mm sucrose were lysed using a 27.5-gauge syringe needle, followed by a brief centrifugation in a microcentrifuge for the removal of nuclear debris. The supernatant was incubated with the mouse anti-GFP antibody or control mouse IgG, followed by incubation with BioMag goat anti-mouse IgG beads. The protein-antibody complex was separated with a magnetic separator and subjected to analysis by in vitro RNA replication as described below and by Western blotting for the presence of HCV proteins, GFP-LC3, and Rab7.
In Vitro RNA Replication Assay
Autophagosomes affinity-purified as described above were resuspended in a reaction mixture containing 50 mm HEPES, 5 mm MgCl2, 0.5 mm MnCl2, 10 mm KCl, 1 mm ATP, 1 mm GTP, 1 mm UTP, 10 μm CTP, 5 μg/ml actinomycin D, and 30 μCi of [α-32P]CTP and incubated for 90 min at 30 °C. The HCV RNA synthesized was then isolated using TRIzol (Invitrogen) following the manufacturer's instructions and analyzed by agarose gel electrophoresis and autoradiography.
Immunoelectron Microscopy
Cells were fixed in 2% glutaraldehyde in neutral phosphate buffer, post-fixed in osmium tetraoxide, and embedded in Epon. Sections were cut at 80 nm and examined under a Philips Tecnai 10 electron microscope. For EM-gold, cells were incubated with mouse anti-bromouridine and rabbit anti-GFP antibodies, followed by incubation with 30-nm gold-conjugated goat anti-mouse and 15-nm gold-conjugated goat anti-rabbit antibodies.
RESULTS
Reduction of HCV RNA Levels in Cells by siRNAs Directed against LC3 and Atg7
We have previously demonstrated that the suppression of LC3 and Atg7 expression reduces the HCV RNA level in Huh7.5 hepatoma cells transfected with the HCV JFH1 genomic RNA (3), indicating a possible role of autophagosomes in HCV RNA replication. Because we had previously also demonstrated that the HCV subgenomic RNA replicon induces autophagosomes, we decided to use the HCV subgenomic RNA replicon to further examine the relationship between autophagosomes and HCV RNA replication. HuhHyg cells, which carry the bicistronic subgenomic RNA replicon of the HCV-N strain (Fig. 1A), were transfected with plasmid pEGFP-LC3 to produce a stable HCV replicon cell line that expresses the GFP-LC3 fusion protein. The GFP-LC3 fusion protein is a cytosolic protein, but it is lipidated and associated with autophagosomal membranes during autophagy (3). This GFP-LC3-expressing stable HCV replicon cell line, which we named the GLR cell line, contained the replicating HCV RNA, as demonstrated by its stable expression of the HCV NS5A protein (Fig. 1B) and by the ability of the cell lysates to direct HCV RNA replication in vitro (see below). In agreement with the previous report, this stable cell line also had elevated levels of autophagosomes, as evidenced by the detection of lipidated GFP-LC3, an increased level of lipidated LC3 (Fig. 1B), and an increased number of GFP-LC3 autophagosomal puncta (see below). This GLR cell line was therefore used in our subsequent studies.
FIGURE 1.
Establishment of stable cells that contain HCV subgenomic RNA replicon. A, illustration of the bicistronic HCV subgenomic RNA replicon. In this replicon, the expression of hygromycin phosphotransferase (Hygror) is under the control of the HCV internal ribosomal entry sequence, whereas the expression of HCV NS3-NS5B nonstructural proteins is under the control of the encephalomyocarditis virus internal ribosomal entry sequence (EMCV IRES). B, Western blot analysis of HCV NS5A, GFP-LC3, and LC3 expressed in HCV replicon cells. The control (Cont) Huh7 cells and the HCV GLR cells were lysed for Western blot analysis. Non-lipidated GFP-LC3 (GFP-LC3-I) and lipidated GFP-LC3 (GFP-LC3-II) were detected using the anti-GFP antibody, and non-lipidated LC3 (LC3-I) and lipidated LC3 (LC3-II) were detected using the anti-LC3 antibody. The β-actin protein was also analyzed as a loading control.
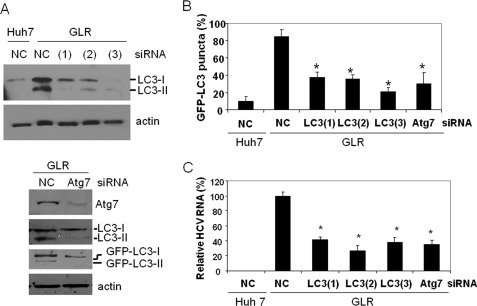
To determine whether autophagosomes are indeed required for HCV RNA replication, we performed the siRNA knockdown experiment to suppress the expression of LC3, a protein critical for the formation of autophagosomes. To rule out the possibility of off-target effects, we used three different LC3 siRNAs. As shown in Fig. 2A, the siRNAs directed against LC3 efficiently reduced its expression level. These siRNAs also reduced the number of autophagosomal puncta in GLR cells (Fig. 2B), confirming the role of LC3 in the formation of autophagosomes. In agreement with our previous studies using the full-length HCV JFH1 genomic RNA, the suppression of LC3 expression led to the reduction of the HCV RNA level in replicon cells (Fig. 2C). In addition to LC3, we also performed an Atg7 knockdown experiment using its specific siRNA. As shown in Fig. 2A, the silencing of Atg7 expression greatly inhibited the lipidation of endogenous LC3 and GFP-LC3. This result is consistent with the biological activity of Atg7, which is to mediate the lipidation of LC3. Similarly, the suppression of Atg7 expression also reduced the number of autophagosomal puncta and the HCV RNA level in GLR replicon cells (Fig. 2, B and C). The reduction of the HCV RNA level after the inhibition of autophagy was not due to the reduction of cell viability (supplemental Fig. 1), nor was it due to the induction of the interferon response, as analysis of the 2′,5′-oligoadenylate synthetase, an interferon-stimulated gene, revealed no induction of expression of this gene after the silencing of Atg7 expression (supplemental Fig. 2).
FIGURE 2.
Suppression of LC3 and Atg7 expression results in reduction of HCV RNA levels in replicon cells. A, Western blot analysis of LC3 and Atg7. HCV GLR replicon cells were transfected with the negative control siRNA (NC), LC3 siRNA, or Atg7 siRNA and then lysed for Western blot analysis. (1), (2), and (3) indicate GLR cells transfected with LC3(1), LC3(2), and LC3(3) siRNAs, respectively. Huh7 cells transfected with the control siRNA were used as the control in the LC3 knockdown experiment (upper panels). β-Actin was also analyzed as a loading control. For Atg7 knockdown (lower panels), the lipidation of both endogenous LC3 and ectopically expressed GFP-LC3 was analyzed. GFP-LC3-I, non-lipidated GFP-LC3; GFP-LC3-II, lipidated GFP-LC3; LC3-I, non-lipidated LC3; LC3-II, lipidated LC3. B, reduction of GFP-LC3 puncta by LC3 and Atg7 siRNA knockdown. Percentages of cells that were positive for more than five GFP-LC3 puncta are shown. Approximately 100–200 cells were counted from different viewing fields under the microscope. C, quantitative analysis of HCV RNA levels in replicon cells. Total cellular RNA was isolated 3 days after the siRNA treatment for quantitative real-time RT-PCR analysis of the HCV RNA. The GAPDH RNA was also analyzed as an internal control. The HCV RNA level in cells treated with the negative control siRNA was arbitrarily defined as 100%. *, statistically significant (p < 0.005).
PI3KC3-independent Induction of LC3 Lipidation by HCV
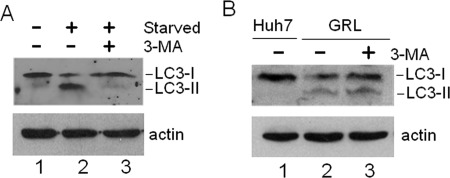
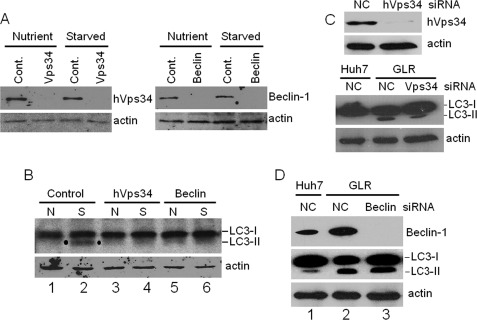
Previous studies have generated discordant results regarding the possible role of autophagy in HCV RNA replication (3, 5–7, 17). As some of these past studies targeted PI3KC3 for its effect on HCV RNA replication, we decided to investigate the possible role of this particular factor in the lipidation of LC3 induced by HCV. PI3KC3 phosphorylates phosphatidylinositol to generate phosphatidylinositol 3-phosphate and is important for the initiation of autophagy induced by nutrient starvation (10). As shown in Fig. 3A, 3-methyladenine (3-MA), which is an inhibitor of PI3KC3, indeed suppressed the lipidation of LC3 induced by nutrient starvation. Interestingly, as shown in Fig. 3B, 3-MA did not abolish the lipidation of LC3 induced by HCV, indicating that HCV likely induced the autophagic response in a PI3KC3-independent manner. To confirm this possibility, we performed the siRNA knockdown experiment to suppress the expression of hVps34, the catalytic subunit of PI3KC3, and Beclin-1, a protein factor that activates the PI3KC3 complex (22). As a positive control, we tested the effects of hVps34 and Beclin-1 siRNAs on autophagy induced by nutrient starvation. As shown in Fig. 4A, these two siRNAs efficiently suppressed the expression of their respective target proteins in Huh7.5 cells with or without nutrient starvation. As expected, these two siRNAs also suppressed the lipidation of LC3 in cells that were nutrient-starved (Fig. 4B). In contrast, the suppression of expression of hVps34 with its specific siRNA had no significant effect on the lipidation of LC3 induced by HCV (Fig. 4C). Similarly, the inhibition of Beclin-1 expression also had no effect on the lipidation of LC3 in GLR replicon cells (Fig. 4D). These results together provide a strong argument that HCV induces autophagosomes in a PI3KC3-independent manner.
FIGURE 3.
Effect of 3-MA on LC3 lipidation induced by HCV. A, suppression of LC3 lipidation by 3-MA in nutrient-starved cells. Huh7.5 cells without or with nutrient starvation for 30 min (see “Experimental Procedures”) were lysed for Western blot analysis. In lane 3, cells were treated with 10 mm 3-MA for 16 h and nutrient-starved for 30 min prior to the end of the 3-MA treatment. B, lack of effect of 3-MA on LC3 lipidation induced by HCV. Lane 1, control Huh7 cells; lane 2, HCV GLR replicon cells; lane 3, HCV GLR cells treated with 10 mm 3-MA for 16 h. LC3-I, non-lipidated LC3; LC3-II, lipidated LC3.
FIGURE 4.
Effects of PI3KC3 and Beclin-1 on LC3 lipidation induced by HCV. A, Western blot analysis of Huh7 cells treated with the negative control siRNA (Cont.) or the siRNA directed against hVps34 or Beclin-1. Cells were transfected with the siRNA for 2 days, followed by nutrient starvation for 30 min. Cells were then lysed for Western blot analysis. Nutrient and Starved indicate cells without and with nutrient starvation, respectively. B, effects of hVps34 and Beclin-1 knockdown on LC3 lipidation induced by nutrient starvation. Cells transfected with the control, hVps34, or Beclin-1 siRNA as described for A were lysed for Western blot analysis for LC3. N and S indicate cells without and with nutrient starvation, respectively. Although the control siRNA had no effect on lipidation induced by starvation (lane 2), this lipidation was inhibited by the hVps34 (lane 4) or Beclin-1 (lane 6) siRNA. Dots highlight the lipidated LC3 band (LC3-II). LC3-I, non-lipidated LC3. C, effect of hVps34 silencing on LC3 lipidation in GLR cells. Upper panels, Western blot analysis of hVps34 in GLR cells treated with the negative control siRNA (NC) or the hVps34 siRNA. Lower panels, Western blot analysis of LC3. In all of the panels, β-actin was also analyzed as a loading control. D, effect of Beclin-1 knockdown on LC3 lipidation. Lane 1, Huh7 cells treated with the control siRNA; lane 2, GLR cells treated with the control siRNA; lane 3, GLR cells treated with the Beclin-1 siRNA.
Colocalization of HCV NS5A, HCV NS5B, and Nascent HCV RNA with Autophagosomes
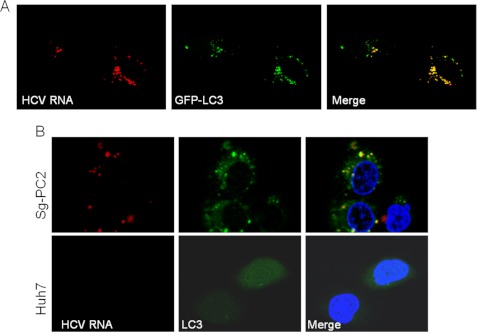
To further understand the role of autophagosomes in HCV RNA replication, we performed confocal microscopy analysis of GLR cells, the HCV replicon cells that stably express the GFP-LC3 fusion protein. As shown in Fig. 5A, a small fraction of the HCV NS5A protein was found to colocalize with autophagosomes, which were revealed by the punctate GFP-LC3 signals. When NS5A and NS5B were analyzed together by confocal microscopy, a small fraction of these two proteins were again found to colocalize on autophagosomes (Fig. 5B), suggesting the possible formation of the HCV RNA replication complex on autophagosomal membranes.
FIGURE 5.
Confocal microscopy analysis of HCV NS5A, HCV NS5B, and autophagosomes. A, colocalization of the HCV NS5A protein with autophagosomes. HCV replicon cells were immunostained with the mouse anti-NS5A antibody, followed by the rhodamine-conjugated goat anti-mouse secondary antibody. Red, NS5A; green, GFP-LC3 puncta, which represent autophagosomes. The boxed area in panel c is enlarged in panel d. Arrows in panel d denote the colocalization of NS5A with autophagic puncta. B, colocalization of HCV NS5A, HCV NS5B, and autophagosomes. The experiments were done as described for A, with the exception that the rabbit anti-NS5B primary antibody and the Alexa Fluor 405-conjugated goat anti-rabbit antibody were included to stain NS5B (blue). The boxed area in panel d is enlarged in panel e. Arrows denote the colocalization of both NS5A and NS5B with autophagic puncta.
To determine whether HCV RNA replication is indeed associated with autophagosomes, we treated HCV replicon cells with actinomycin D to suppress cellular RNA synthesis and labeled nascent HCV RNA with BrUTP. As shown in Fig. 6A, essentially all of the HCV RNA signals labeled by BrUTP were associated with autophagosomes. This labeling was specific to HCV RNA, as treatment of GLR cells with 2′-C-methyladenosine, an inhibitor of HCV NS5B RNA polymerase, resulted in the loss of BrUTP signals and the GFP-LC3 puncta (supplemental Fig. 3). To further confirm this result, we conducted another confocal microscopy experiment using the Sg-PC2 cell line, which contains the HCV Con1 subgenomic RNA replicon without the stable expression of GFP-LC3 (20). In this particular experiment, the anti-LC3 antibody was used to detect endogenous LC3. Similarly, as shown in Fig. 6B, BrUTP-labeled nascent HCV RNA colocalized with punctate LC3 signals. These results provide further support that the replication of HCV RNA takes place on autophagosomes.
FIGURE 6.
Colocalization of HCV RNA replication complex with autophagosomes. A, HCV subgenomic RNA replicon cells (GLR) were permeabilized with lysolecithin and labeled with BrUTP as described under “Experimental Procedures.” Nascent HCV RNA (red) was then analyzed with the mouse anti-bromouridine primary antibody and the rhodamine-conjugated goat anti-mouse secondary antibody. As shown, the nascent HCV RNA signals colocalized with the GFP-LC3 puncta. B, HCV subgenomic RNA replicon cells (Sg-PC2) were labeled with BrUTP. The HCV RNA and endogenous LC3 were analyzed with the mouse anti-bromouridine primary antibody and the rhodamine-conjugated goat anti-mouse secondary antibody and with the rabbit anti-LC3 antibody and the fluorescein-conjugated goat anti-rabbit secondary antibody, respectively. Note that LC3 in Sg-PC2 cells displayed a punctate staining pattern in the cytoplasm, whereas in control Huh7 cells, it displayed a diffuse staining pattern throughout the entire cell.
To investigate whether HCV RNA replication can also take place on autophagosomes in HCV-infected cells, we infected stable GFP-LC3 cells with the HCV JFH1 virus and conducted a similar labeling experiment. As shown in supplemental Fig. 5, nascent HCV RNA was also found to partially colocalized with autophagosomal puncta in HCV-infected cells.
Co-immunoprecipitation of HCV RNA Replication Complex with Autophagosomes
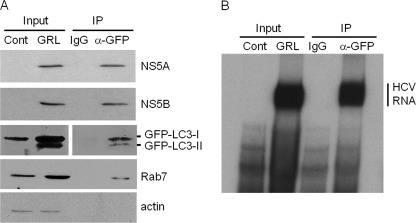
To confirm that the HCV RNA replication complex is indeed associated with autophagosomes, GLR cells were lysed in hypotonic buffer, and autophagosomes were immunoprecipitated by anti-GFP antibody. As shown in Fig. 7A, the anti-GFP antibody, but not the control antibody, was able to precipitate the GFP-LC3 fusion protein and its lipidated form. This anti-GFP antibody could also precipitate Rab7, a small GTPase that is important for the maturation of autophagosomes (23) and for HCV RNA replication (24). However, this antibody could not precipitate the nonspecific β-actin protein (Fig. 7A). In addition, in support of the confocal microscopy results shown in Fig. 5, which indicated that HCV NS5A and NS5B were associated with autophagosomes, both NS5A and NS5B were also co-immunoprecipitated by the anti-GFP antibody. Similarly, in agreement with the results shown in Fig. 6, autophagosomal membranes precipitated by the anti-GFP antibody were able to direct the synthesis of HCV RNA in vitro. To ensure the specificity of these results, we conducted the same experiment with Sg-PC2 cells, which contain the HCV subgenomic RNA replicon without the GFP-LC3 protein. As shown in supplemental Fig. 5, in the absence of GFP-LC3, the anti-GFP antibody was not able to precipitate autophagosomes, as evidenced by the lack of LC3 signals in the Western blot analysis. This antibody was also not able to precipitate the HCV RNA replication complex. Taken together, these results strongly indicate the association of the HCV RNA replication complex with autophagosomes.
FIGURE 7.
Co-immunoprecipitation of HCV RNA replication complex with autophagosomes. HCV replicon cells were lysed with hypotonic buffer, and autophagosomal membranes were immunoprecipitated (IP) with the control antibody (IgG) or the anti-GFP antibody as described under “Experimental Procedures.” A, Western blot analysis. A fraction of total lysates of naïve Huh7 cells or HCV GLR replicon cells was used for Western blot analysis. GLR cell lysates immunoprecipitated with either the control IgG or the anti-GFP antibody were also analyzed. As shown, the HCV subgenomic RNA replicon induced the lipidation of GFP-LC3 (GFP-LC3-II). GFP-LC3-I, non-lipidated GFP-LC3. Also, the anti-GFP antibody, but not the control antibody, immunoprecipitated HCV NS5A and NS5B proteins as well as Rab7. The anti-GFP antibody did not precipitate β-actin, indicating the specificity of this antibody in the immunoprecipitation reaction. B, in vitro HCV RNA replication assay. The immunoprecipitates of GLR cells and a fraction of the total lysates of Huh7 or GLR cells were used for the in vitro HCV RNA replication assay. Details of the experimental procedures are described under “Experimental Procedures.” The location of the replicated HCV RNA is indicated.
Localization of Nascent HCV RNA on Autophagosomal Membranes
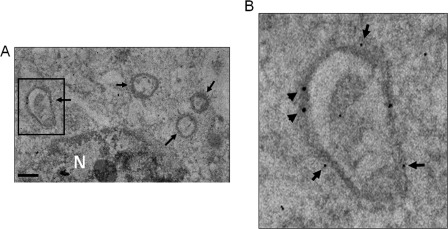
To further understand the relationship between the HCV RNA replication complex and autophagosomes, we performed double-labeling immunoelectron microscopy analysis. In this study, HCV replicon cells were permeabilized with lysolecithin, and nascent HCV RNA was labeled with BrUTP. The localization of nascent HCV RNA in cells was then analyzed using the mouse anti-bromouridine primary antibody and the secondary antibody conjugated with 30-nm gold particles. The localization of GFP-LC3 was analyzed using the rabbit anti-GFP primary antibody and the secondary antibody conjugated with 15-nm gold particles. These gold particle-conjugated secondary antibodies are specific and allow the identification of the locations of nascent HCV RNA and GFP-LC3. As shown in Fig. 8A, multiple autophagosome-like vesicles ranging from 300 to 700 nm in diameter were detected. These membrane vesicles were positive for GFP-LC3, as evidenced by the association of 15-nm gold particles (see the boxed area in Fig. 8A), and for nascent HCV RNA, as evidenced by the association of 30-nm gold particles. These results demonstrate that the synthesis of nascent HCV RNA takes place on autophagosomal membranes.
FIGURE 8.
Immunogold double-staining analysis of nascent HCV RNA and autophagosomes. The arrows in A denote the autophagosome-like vesicles. The boxed area in A is enlarged in B to reveal the gold particles. 30-nm gold particles were used for nascent HCV RNA and 15-nm gold particles were used for GFP-LC3. N, nucleus. Scale bar = 500 nm.
DISCUSSION
Previous studies indicated that HCV perturbs the autophagic pathway to induce the accumulation of autophagosomes. In this study, we have demonstrated that suppression of the expression of LC3 and Atg7, two protein factors critical for the formation of autophagosomes, reduces the HCV RNA replication level (Fig. 2). We further demonstrated the colocalization of HCV NS5A, HCV NS5B and nascent HCV RNA with autophagosomes (Figs. 5 and 6) and showed that the HCV RNA replication complex can be co-immunoprecipitated with autophagosomes (Fig. 7). The association of nascent HCV RNA with autophagosomes was also observed in HCV-infected cells (supplemental Fig. 4). In addition, by conducting immunoelectron microscopy, we demonstrated the localization of the HCV RNA replication complex on autophagosomal membranes (Fig. 8). Taken together, our results strongly indicate that the replication of HCV RNA takes place on autophagosomal membranes. It remains to be determined, however, how the HCV RNA replication complex is assembled on autophagosomal membranes and whether the lipidated LC3 or other cellular proteins associated with autophagosomes directly participate in the replication of HCV RNA.
It has been reported that the HCV RNA replication complex is associated with membranous webs (25, 26), which are clusters of tightly associated membrane vesicles with varying sizes. The relationship between these membranous webs and autophagosomes is unclear at this moment. It is possible that membranous webs may be the result of clustering and coalescing of autophagosomes. The possibility that these membranous webs serve as the germinating center for the production of autophagosomes also cannot be ruled out.
Although independent reports indicated that HCV could can autophagosomes (2–8), the roles of these autophagosomes in the HCV life cycle have been controversial. Our previous results demonstrated that the suppression of LC3 or Atg7 expression with RNAi inhibits replication of the HCV JFH1 genomic RNA in Huh7.5 cells. Mizui et al. (7) also reported that chemical compounds that suppress autophagy or the silencing of Atg5, Atg7, or LC3 expression can inhibit the replication of the HCV subgenomic RNA replicon. In contrast, Dreux et al. (5) reported that the silencing of Beclin-1 suppresses the infection of Huh7 cells with HCV, but this silencing has no effect on HCV in cells containing the stable HCV subgenomic RNA replicon or in cells previously infected with HCV. Dreux et al. provided further results to suggest that the effect of autophagy on HCV is on the translation of incoming HCV RNA. Some of the controversies can be resolved by the fact that different cellular factors were targeted for silencing. As shown in Figs. 3 and 4, we found that HCV induced the autophagic response independently of PI3KC3, an enzyme that is critical for autophagy induced by nutrient starvation (27). Because Beclin-1 is a component of the PI3KC3 complex (22), its silencing would not be expected to affect the autophagosomes induced by HCV. Indeed, as we showed in Fig. 4D, the suppression of Beclin-1 expression did not affect the lipidation of LC3. Because the silencing of expression of autophagic protein factors prior to HCV infection often has a more pronounced effect on HCV RNA replication than when the silencing is conducted after the HCV infection has been established (5, 6, 17), it is also possible that autophagosomes may serve as the site for HCV RNA replication in the early stage of HCV infection, and further alterations of cellular membranes by HCV, such as the induction of membranous webs, in the later stage of infection result in the generation of membranes for HCV RNA replication in an autophagy-independent manner. In this regard, it is interesting to note that some of the nascent HCV RNA in HCV-infected cells was not found to be associated with autophagosomes (supplemental Fig. 4).
How HCV induces autophagosomes in a PI3KC3-independent manner remains unclear. It is likely that it may involve the alteration of the endoplasmic reticulum membranes, as we and another group had reported that HCV induces autophagosomes through the unfolded protein response (3, 9). Further research in this area will likely generate interesting results.
Our finding that the HCV RNA replicates on autophagosomal membranes puts HCV on the list with other positive-strand RNA viruses, including poliovirus, coxsackieviruses, dengue viruses, and possibly also rhinoviruses and nidoviruses, which use the membranes of autophagic vacuoles for their RNA replication (for a review, see Ref. 16). Of particular interest are dengue viruses, which are also members of the Flaviviridae family. It will be interesting to determine whether other members of the Flaviviridae family also use autophagic vacuoles for their RNA replication.
Supplementary Material
Acknowledgments
We thank Dr. Charles Rice for the anti-NS5A antibody, Dr. Soon Hwang for the anti-NS5B antibody, the Confocal Microscopy Core of the USC Research Center for Liver Diseases for the confocal microscopy, and the EM Core of the USC Norris Comprehensive Cancer Center and Ivy Hsieh (San Francisco Veterans Affairs Medical Center) for the EM-gold experiments. We are also indebted to Dr. Michael Lai and his laboratory members for assisting us in establishing the GLR cell line.
This work was supported, in whole or in part, by National Institutes of Health Grants AI083025, DK094652, and CA123328.

This article contains supplemental Figs. 1–5.
- HCV
- hepatitis C virus
- PI3KC3
- Class III PI3K
- GLR
- GFP-LC3 replicon
- hVps34
- human Vps34
- 3-MA
- 3-methyladenine.
REFERENCES
- 1. Lemon S. M., Walker C. M., Alter M. J., Yi M. (2007) in Fields Virology (Knipe D. M., Howley P. M., eds) 5th Ed., pp. 1253–1304, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Ait-Goughoulte M., Kanda T., Meyer K., Ryerse J. S., Ray R. B., Ray R. (2008) Hepatitis C virus genotype 1a growth and induction of autophagy. J. Virol. 82, 2241–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sir D., Chen W. L., Choi J., Wakita T., Yen T. S., Ou J. H. (2008) Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48, 1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sir D., Liang C., Chen W. L., Jung J. U., Ou J. H. (2008) Perturbation of autophagic pathway by hepatitis C virus. Autophagy 4, 830–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreux M., Gastaminza P., Wieland S. F., Chisari F. V. (2009) The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 106, 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanida I., Fukasawa M., Ueno T., Kominami E., Wakita T., Hanada K. (2009) Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5, 937–945 [DOI] [PubMed] [Google Scholar]
- 7. Mizui T., Yamashina S., Tanida I., Takei Y., Ueno T., Sakamoto N., Ikejima K., Kitamura T., Enomoto N., Sakai T., Kominami E., Watanabe S. (2010) Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J. Gastroenterol. 45, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferraris P., Blanchard E., Roingeard P. (2010) Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91, 2230–2237 [DOI] [PubMed] [Google Scholar]
- 9. Ke P. Y., Chen S. S. (2011) Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 121, 37–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D. A., Levine B. (2007) HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin-1 autophagy protein. Cell Host Microbe 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 12. Chaumorcel M., Souquère S., Pierron G., Codogno P., Esclatine A. (2008) Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy 4, 46–53 [DOI] [PubMed] [Google Scholar]
- 13. Lee J. S., Li Q., Lee J. Y., Lee S. H., Jeong J. H., Lee H. R., Chang H., Zhou F. C., Gao S. J., Liang C., Jung J. U. (2009) FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 11, 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor M. P., Kirkegaard K. (2007) Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81, 12543–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sir D., Tian Y., Chen W. L., Ann D. K., Yen T. S., Ou J. H. (2010) The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U.S.A. 107, 4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sir D., Ou J. H. (2010) Autophagy in viral replication and pathogenesis. Mol. Cells 29, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guévin C., Manna D., Bélanger C., Konan K. V., Mak P., Labonté P. (2010) Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 405, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J. T., Bichko V. V., Seeger C. (2001) Effect of α-interferon on the hepatitis C virus replicon. J. Virol. 75, 8516–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu H. M., Aizaki H., Choi K. S., Machida K., Ou J. J., Lai M. M. (2009) SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein) is a host factor involved in hepatitis C virus RNA replication. Virology 386, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi J., Lee K. J., Zheng Y., Yamaga A. K., Lai M. M., Ou J. H. (2004) Reactive oxygen species suppress hepatitis C virus RNA replication in human hepatoma cells. Hepatology 39, 81–89 [DOI] [PubMed] [Google Scholar]
- 21. Zhu J. H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C. T. (2007) Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 170, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Funderburk S. F., Wang Q. J., Yue Z. (2010) The Beclin-1-VPS34 complex: at the crossroads of autophagy and beyond. Trends Cell Biol. 20, 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E. L. (2004) Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117, 4837–4848 [DOI] [PubMed] [Google Scholar]
- 24. Manna D., Aligo J., Xu C., Park W. S., Koc H., Heo W. D., Konan K. V. (2010) Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398, 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H. E., Bienz K., Moradpour D. (2003) Identification of the hepatitis C virus RNA replication complex in Huh7 cells harboring subgenomic replicons. J. Virol. 77, 5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Egger D., Wölk B., Gosert R., Bianchi L., Blum H. E., Moradpour D., Bienz K. (2002) Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tassa A., Roux M. P., Attaix D., Bechet D. M. (2003) Class III phosphoinositide 3-kinase-Beclin-1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem. J. 376, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.